Overexpression of GmDREB1 improves salt tolerance in transgenic wheat and leaf protein response to high salinity
Qiyan Jiang,Zheng Hu,Hui Zhang*,Youzhi Ma**
Institute of Crop Science,National Key Facility for Crop Gene Resources and Genetic Improvement,Chinese Academy of Agricultural Sciences,Beijing 100081,China
1.Introduction
High levels of salt,drought and freezing induce the dehydration of plant cells and thereby impair plant growth,biomass production,and crop productivity [1].To protect cells from stress,plants generally respond to these abiotic stresses in a complex,integrated manner that involves many genes,several cellular signal transduction pathways,and many stress-related proteins and enzymes [2].Given the polygenic nature of abiotic stress responses,the development of abiotic stress tolerance in crop plants by conventional approaches has been a challenge for plant breeders [3].The genetic engineering of plants with individual genes gives promise of achieving abiotic stress tolerance with less effort and time.These genes include primarily those governing the accumulation of compatible solutes; those encoding detoxification enzymes,late embryogenesis abundant (LEA) protein,and protein kinases related to the signal transduction of this protein;and those encoding transcription factors[4].
Of these gene products,the DREB transcription factors have gained much attention,owing to their involvement in the regulation of many stress-related genes that play an important role in producing a cascade of responses to environmental stimuli [5,6].Since the DRE cis elements were identified in Arabidopsis [7],approximately 40 homologs of the DREB gene from nearly 20 types of plants have been reported,and one DREB gene can be induced by multiple stress factors [4,8,9].Owing to the important role of DREBs in abiotic stress tolerance,plants have been transformed with more than 20 different DREB transcription factors induced by the constitutive promoter CaMV35S or by the stress-inducible promoter rd29A,which confers multiple abiotic stress tolerance to plants[4,8].
The genetic engineering of plants for abiotic stress tolerance can be achieved by the expression of DREB transcription factors that,in turn,regulate the expression of abiotic stress-related downstream genes by binding to DRE/CRT cis-acting elements in the promoter regions of these genes [7,10].Most of these downstream genes have been found to encode proteins including osmoprotectants,LEA proteins,protease inhibitors,lysophospholipase C,cold acclimation proteins,glucose transporter proteins,and transcription factors.These genes were identified using cDNA microarrays and play important roles in plant stress tolerance [4,8,11–13].
A proteomic approach was used to investigate the protein expression profiles of wild-type and transgenic plants overexpressing DREB2C under mild heat stress (37 °C) for 24 h.Eleven protein spots were identified as being differentially regulated in 35S:DREB2C plants.Of these 11 proteins,four were up-regulated at both translation and transcriptional levels.Moreover,one or more DRE/CRT sequences (5′-A/GCCGAC) (the recognition sequence of DREB2C) were found in the 1000-bp promoter regions of these four proteins.Thus four genes encoding peptidyl-prolyl isomerase ROC4,glutathione transferase 8,elongation factor Tu (EF-Tu),and pyridoxal biosynthesis protein PDX1 are potential targets of DREB2C[14].The expression of seven other proteins that do not contain the DRE/CRT motif in their promoter region was also affected by the overexpression of DREB2C [14].Savitch et al.[15]reported the overexpression of two Brassica CBF/DREB1-like transcription factors (BNCBF5 and BNCBF17),the presence of accumulated COR gene mRNA and the accumulation of GLK1-and GLK2-like transcription factors,cyclophilin ROC4,β-amylase,and triose-P/Pi translocator in transgenic Brassica plants.In addition to producing changes in the transcript levels of these proteins,transgenic plants showed improved photosynthetic capacity,enhanced activity of enzymes involved in the Calvin cycle,and increased sucrose and starch biosynthesis.In transgenic Arabidopsis,DREB1/CBF also regulated the expression of genes encoding transcription factors [11] that may regulate the expression of other downstream genes,thus forming a regulatory network of DREB responses to stress.
However,the members of this regulatory network vary with the DREB gene and/or with the type of stress [4,8].Transgenic crops overexpressing the DREB gene show significantly increased tolerance to stress under laboratory or greenhouse conditions.However,it remains undetermined whether these transgenic plants show enhanced stress tolerance under complex field conditions.In certain transgenic plants,the overexpression of the DREB gene under a constitutive CaMV35S promoter enhanced stress tolerance.However,simultaneously,negative effects on the plant phenotype were observed in these transgenic plants[16–18].For example,the constitutive expression of SbDREB2 led to pleiotropic effects in rice,and these transgenic plants did not set seed [19].Certain transgenic plants constitutively overexpressing the DREB gene showed better growth parameters than the wild type without growth retardation[20,21].Thus the stress tolerance of transgenic plants grown in the field,the physiological and biochemical mechanisms of improving salt tolerance in transgenic plants,and the regulatory network of DREB genes require further study.
The GmDREB1 gene(GenBank accession number AF514908),which encodes a stress-inducible transcription factor,was cloned by screening a cDNA library of Glycine max cv.Jinong 27 using the yeast one-hybrid method [22].The stress-inducible expression of GmDREB1 conferred salt tolerance on transgenic alfalfa plants [23].T1transgenic lines of wheat with Ubi::GmDREB1 and with rd29A::GmDREB1 showed better drought and salt tolerance than wild-type plants[22].
In the present study,the advanced-generation transgenic wheat lines T349 and T378 with Ubi::GmDREB1 and the wild-type Jimai 19 were used to evaluate the salt tolerance of these plants at the germination and seedling stages and throughout the growing season.Using a comparative proteomic approach,we investigated the mechanisms that underlie high-salinity tolerance in Ubi::GmDREB1 transgenic wheat based on phenotypic characteristics,physiological parameters and protein responses to salt stress.
2.Materials and methods
2.1.Plant growth and salt tolerance evaluation
T349 and T378 are transgenic lines of wheat constitutively expressing the GmDREB1 gene under the control of the maize ubiquitin promoter in wheat variety Jimai 19.Wild-type Jimai 19 was used as the control.
2.1.1.Germination stage
In total,100 seeds of each genotype were germinated on wet filter paper in culture dishes with distilled water(CK)and with a 2.0%NaCl solution under white light(150 μmol Photons m-2s-1;14-h light/10-h dark photoperiod)at 20 °C in a growth chamber.When the coleoptiles were 1/3 or the radicle was 1/2 of the length of the seed,the seed was considered germinated.The percent germination under CK and the treatment was scored at 5 and 10 days,respectively,after seeding.The relative salt injury rate(RSIR)was calculated as
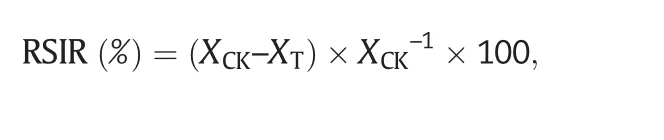
where XCKis the mean germination percentage of the cultivar under normal,non-stressed (control) conditions,and XTis the mean germination percentage of the cultivar under salt stress.The coleoptile length,radicle number,and radicle length of seeds were recorded.The experiment was performed with three replications.
2.1.2.Seedling stage
In total,50 germinated seeds were grown in plastic containers containing complete Kimura B nutrient solution[24]under white light(150 μmol Photons m-2s-1;14-h light/10-h dark photoperiod) at 25 °C in a growth chamber.Ten-day-old seedlings were treated with 300 mmol L-1NaCl in Kimura B nutrient solution for 7 days.The salt injury symptoms of seedlings were investigated and assigned a score of 0–5 following the method used in other studies[25–28],with some modification.The classification criteria of salt injury were as follows: level 0 (no injury),level 1(damage on leaf tips),level 2 (half of the leaf showing injury),level 3 (full leaf showing injury),level 4 (only the youngest leaf surviving),and level 5 (death).The experiment was performed with three replications.The salt injury index(SI)was calculated using the following formula[25–28]:
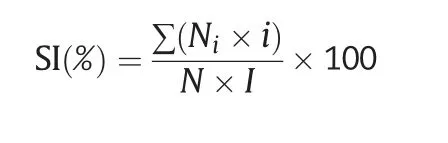
where Niis the number of plants assigned with score i?(from 0 to 5);N is the total number of tested seedlings and I is the highest score.The fresh weight and root length of seedlings were recorded.The salt tolerance score at the germination and seedling stages was assigned according to the RSIR and SI(Table 1).
To determine the numbers of tillers per plant at the seedling stage under salt stress,T349,T378,and Jimai 19 were planted in pots (7 cm × 7 cm × 7 cm) with soil and watered with a 0.3%NaCl solution.Each pot had only one plant,with 12 pots in one plate and three plates for each replication.After growth for 3 months in a 4 °C phytotron,the number of tillers and the fresh weight per plant were investigated.The experiment was performed with three replications.
2.1.3.The entire growing season
T349,T378,and Jimai 19 were grown in saline–alkaline soil in natural fields using a randomized complete block design with six replicates.Each plot consisted of 10 rows 2 m long,with 30 seeds per row.The space between rows was 30 cm and the separation between plots was 50 cm.The average soil salt content was 0.66%.Seedling emergence rate was recorded 65 days after sowing.Other agronomic traits,namely biomass per plant,tillers per plant,effective tillers per plant,plant height,spike length,grain number per spike,grain weight per plant,grain number per plant,and 1000-grain weight,were measured at harvest.

Table 1-Salt tolerance score at germination and seedling stages.
2.2.Determination of physiological indices
The germinated seeds were grown in plastic containers containing complete Kimura B nutrient solution under white light(150 μmol Photons m-2s-1;14-h light/10-h dark photoperiod)at 25 °C in a growth chamber.Ten-day-old seedlings were treated with 300 mmol L-1NaCl in Kimura B nutrient solution.Before treatment and after 1,3,5,and 7 days,the first leaves of seedling samples were harvested for physiological analysis.The entire experiment was independently repeated three times.
2.2.1.Proline content
Proline contents were determined according to the method of Li [29].Wheat leaf samples (0.5 g) from each group were homogenized in 3% (w/v) sulfosalicylic acid,and the residue was removed by centrifugation.The extract(2 mL)was mixed with 2 mL of glacial acetic acid and with 3 mL of acid ninhydrin (1.25 g of ninhydrin was warmed in a mixture of 30 mL of glacial acetic acid and 20 mL of 6 mol L-1phosphoric acid until dissolved) for 1 h at 100 °C; the reaction was terminated in an ice bath.The reaction mixture was extracted with 5 mL of toluene.The chromophore-containing toluene was warmed to room temperature and its optical density was measured at 520 nm.Proline concentrations were determined using calibration curves.
2.2.2.Glycine betaine content
Fresh tissues were ground in liquid nitrogen and 25 mL of 95%ethanol was added.After being heated for 3 h,the concentrate was diluted with 1.5 mL of 0.1 mol L-1HCl and 0.3 mL of petroleum ether was added for extraction.Active carbon was added to decolorize the solution.After centrifugation,the supernatant was heated for 10 min in boiling water.One milliliter of Reinecke's salt was added,and the solution was cooled for 3 h.After centrifugation,the supernatant was precipitated with 1 mL of ethyl ether.The precipitate was redissolved in 1 mL of 70% acetone and the absorbance was read at 525 nm.The glycine betaine content was calculated as follows:
Glycine betaine content= (A525–0.0121)/0.035×1.5×25/0.5.
2.2.3.Malondialdehyde content
Lipid peroxidation was determined by measuring malondialdehyde (MDA) formation using the thiobarbituric acid method described by Madhava Raoand and Sresty [30].One half gram of a leaf sample was homogenized with 2.5 mL of 0.1%trichloroacetic acid(TCA)to extract MDA.The homogenate was centrifuged for 10 min at 10,000 ×g.For every 1 mL of the aliquot,4 mL of 20% TCA containing 0.5% thiobarbituric acid(TBA) was added.The mixture was heated at 95 °C for 30 min and cooled rapidly in an ice bath.The mixture was then centrifuged for 15 min at 10,000 ×g,and the absorbance of the supernatant was read at 450,532,and 600 nm.The MDA content was calculated as follows:
MDA concentration=6.45× (A532–A600)–0.56×A450
MDA content = (MDA concentration × extraction volume) /(sample weight × 1000).
2.2.4.Relative electrolyte leakage
Electrolyte leakage was determined according to the method of Li [29].For each measurement,0.5 g of the first leaves of wheat seedlings was cut into 1 cm long segments,floated in 15 mL of double-distilled water,and vacuum filtered until all of the segments sank.The conductivity of the bathing solution was measured (value A) with an electrolyte leakage apparatus.The solution and segments were then transferred into sealed tubes and boiled for 15 min.After cooling to room temperature,the conductivity of the bathing solution was measured again as value B.For each measurement,ion leakage was expressed as the percentage of leakage,i.e.,(value A/value B) × 100.
2.3.Proteomic analysis
The germinated seeds were grown in plastic containers containing complete Kimura B nutrient solution under white light (150 μmol Photons m-2s-1; 14-h light/10-h dark photoperiod) at 25 °C in a growth chamber.Ten-day-old seedlings were treated with 300 mmol L-1NaCl in Kimura B nutrient solution.After 7 days,the first expanded leaves of seedlings were harvested,frozen in liquid nitrogen,and stored at-80 °C for proteomic analysis.The entire experiment was independently repeated 3 times.
2.3.1.Protein extraction,electrophoresis and image analysis
Proteins were extracted using the protocol of Jiang et al.[31].Approximately 350 mg of protein was loaded onto isoelectrofocusing (IEF) polyacrylamide gels (pH 3.5–10.0).The IEF gels were polymerized in glass tubes to obtain gels 13.5 cm long and 2 mm in diameter according to the method of Komatsu et al.[32].The gel mixture,the equilibration of the IEF gels and the second-dimension SDS-PAGE were performed as described by Jiang et al.[31].The gel was stained with 0.1% (w/v)Coomassie brilliant blue R-250,24% (v/v) ethanol and 8% (v/v) acetic acid.The stained gels were scanned and analyzed using ImageMaster 2D Platinum software 5.0(GE Healthcare Bio-Science)to identify the differentially expressed protein spots,as described by Jiang et al.[31].
2.3.2.In-gel tryptic digestion
The target protein spots were excised from the preparative gels and de-stained with 100 mmol L-1NH4HCO3in 30%ACN.After removal of the de-staining buffer,the gel pieces were lyophilized and rehydrated in 30 μL of 50 mmol L-1NH4HCO3containing 50 ng trypsin (sequencing grade,Promega,USA).After overnight digestion at 37 °C,the peptides were extracted three times with 0.1% TFA in 60% ACN.Extracts were pooled and lyophilized.The resulting lyophilized tryptic peptides were stored at-80 °C for mass spectrometric analysis.A protein-free gel piece was treated as described above and used as a control to identify autoproteolysis products derived from trypsin.
2.3.3.MALDI-TOF/TOF MS analysis and database searching
Mass spectrometry(MS)and MS/MS spectra were obtained with an ABI 4800 Proteomics Analyzer MALDI-TOF/TOF (Applied Biosystems,Foster City)operating in result-dependent acquisition mode.Peptide mass maps were acquired in positive ion reflector mode(20 kV accelerating voltage)with 1000 laser shots per spectrum.Monoisotopic peak masses were automatically determined within the mass range 800–4000 Da,with a signal-to-noise ratio minimum set to 10 and with a local noise window width of m/z 250.Up to five of the most intense ions with a minimum signal-to-noise ratio of 50 were selected as precursors for MS/MS acquisition,excluding common trypsin autolysis peaks and matrix ion signals.In MS/MS positive ion mode,spectra were averaged,collision energy was 2 kV,and default calibration was specified.Monoisotopic peak masses were automatically determined with a minimum signal-tonoise ratio of 5 and with a local noise window width of m/z 250.The MS and MS/MS spectra were searched against the UniProtKB/SwissProt database (v.2009.03.03,release number 14.9/56.9)using the software GPS Explorer,version 3.6(Applied Biosystems)and MASCOT version 2.1(Matrix Science)with the following parameter settings: trypsin cleavage,one missed cleavage allowed,carbamidomethylation set as a fixed modification,oxidation of methionines allowed as a variable modification,peptide mass tolerance set at 0.1 Da,fragment tolerance set at ± 0.3 Da,and minimum ion score confidence interval for MS/MS data set at 95%.
2.4.Statistical analysis
Data for morphology,physiology,and agronomic traits were statistically analyzed using a one-way analysis of variance(ANOVA).The volume changes of protein spots were analyzed using Student's t-test.
3.Results
3.1.Evaluation of salt tolerance
3.1.1.Germination stage
When seeds were grown in 2% NaCl solution,there were no significant differences in RSIR between T349 and Jimai 19 or between T378 and Jimai 19.The transgenic lines and the control all had a salt tolerance score of 2,classifying these plants as salt-tolerant at the germination stage according to the standard in Table 1.When the transgenic wheat lines were compared with the wild type,the coleoptile lengths and the radicle lengths of T349 and T378 were all significantly longer than those of Jimai 19.The radicle number of the transgenic varieties was also significantly greater than that of Jimai 19(Fig.1-A).The radicles of the transgenic wheat seeds were well developed under salt treatment (Fig.1-B).These results indicate that the salt tolerance of the transgenic lines T349 and T378 was higher than that of the wild type Jimai 19 at the germination stage.
3.1.2.Seedling stage
Under salt stress,the leaves of the wild type Jimai 19 turned yellow earlier than the leaves of the transgenic wheat lines T349 and T378,and the roots of wild-type plants were shorter than those of the transgenic lines (Fig.2-A).According to the salt injury symptoms observed in the seedlings,the salt injury index of Jimai 19 was 72%,and the salt tolerance was scored as 4,whereas the salt injury index values of T349 and T378 were 54% and 58%,respectively,and the salt tolerance levels were both scored as 3.The root length and fresh weight of the transgenic lines were significantly greater than those of the wild type(Fig.2-B).
After growing for 40 days in a 4 °C phytotron under salt stress (watering soil with 0.3% NaCl solution),the vernalization and the tiller formation of the wheat seedlings were complete (Fig.2-C).After growing for 3 months under salt stress conditions,the number of tillers and the fresh weight per plant for seedlings were significantly different between the transgenic lines and the wild type.The transgenic lines T349 and T378 had more tillers per plant than the wild type Jimai 19,so that the fresh weight of the transgenic plant was much higher than that of Jimai 19(Fig.2-C,D).The evaluation of salt tolerance at the seedling stage suggested that the salt tolerance of the transgenic lines T349 and T378 was higher than that of the wild-type Jimai 19 at the seedling stage.
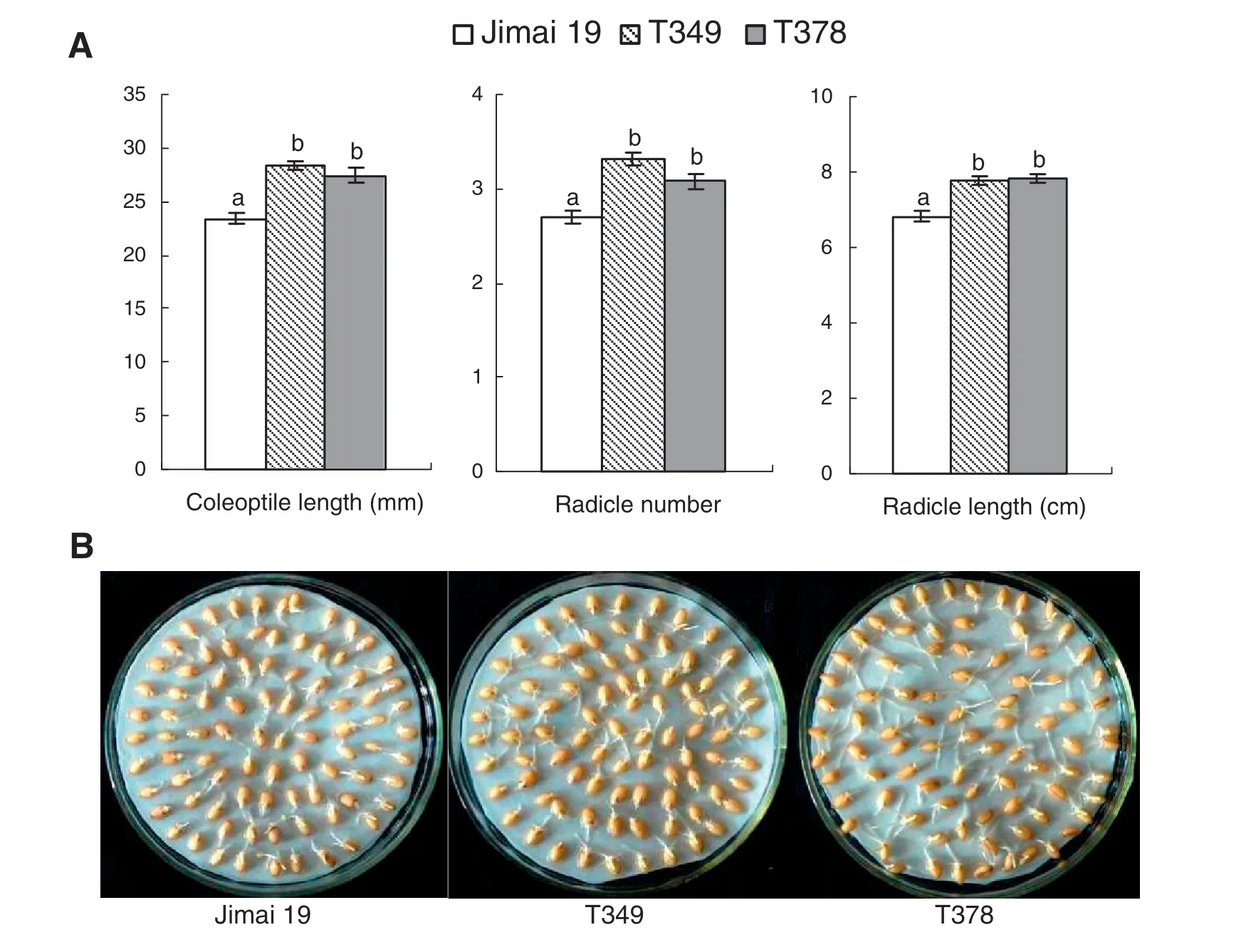
Fig.1-Differences in seed germination between the transgenic wheat lines and the wild type under salt stress.One hundred seeds of the wild type(Jimai 19)and transgenic wheat lines(T349 and T378)were germinated with a 2.0%NaCl solution.Ten days later,the average coleoptile length,radicle number,and radicle length of the transgenic wheat lines were significantly greater than those of the wild type(A),and this increase can be clearly seen(B).The experiment was performed with three replications.The values are the means ± SE(n = 3).Different letters indicate significance at P <0.05(A).
3.1.3.Entire growing season
To analyze the salt tolerance of transgenic wheat throughout the growing season,T349,T378,and Jimai 19 were planted in saline–alkaline soil in natural fields.The average soil salt content was 0.66%.The results showed that the biomass per plant and the number of tillers per plant of transgenic lines were significantly higher than those of the wild type Jimai 19.The seedling emergence rate,effective number of tillers per plant,grain number per plant,and grain weight per plant of the transgenic lines were significantly greater than those of Jimai 19.The spike length of transgenic lines was significantly less than that of Jimai 19.There were no significant differences in plant height,grain number per spike,or 1000-grain weight between the transgenic lines and the wild type (Table 2).
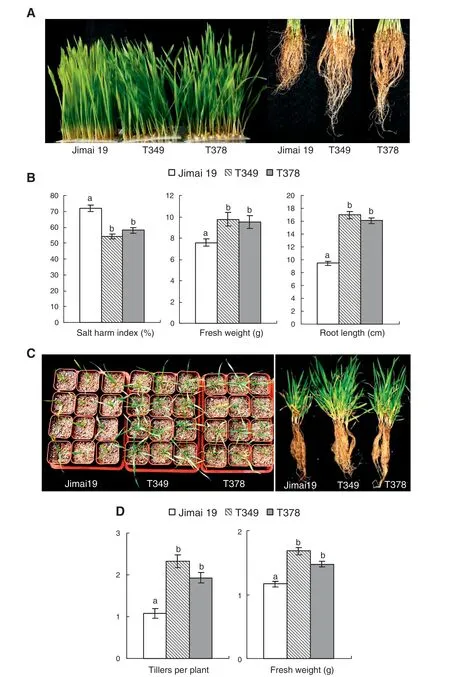
Fig.2-Differences in seedlings between transgenic wheat lines and the wild type under salt stress.When grown in plastic containers containing 300 mmol L-1 NaCl in Kimura B nutrient solution,the leaves of the transgenic lines(T349 and T378)were greener and the roots were longer than those of the wild type(Jimai 19)(A).The differences in salt injury index,root length,and fresh weight(50 plants)between the wild type and the transgenic lines were significant at P <0.05(B).When planted in pots with soil and watered with a 0.3%NaCl solution,the transgenic lines had more tillers compared with the wild type(C),and average tiller numbers and fresh weight per plant were significantly higher in the transgenic lines than in the wild type(D).The experiment was performed with three replications.The values are the means ± SE(n = 3 replications).Different letters indicate significance at P <0.05.
Although the spike length of the transgenic lines was significantly lower,the grain number per spike was not significantly different between transgenic lines and the wild type.Because of the significantly higher number of effective tillers per plant in the transgenic lines,the grain number per plant of the transgenic lines was more than 20% greater than that of Jimai 19,and the grain weight per plant and the biomass per plant were also significantly greater in the transgenic lines.As a result,the salt tolerance of the transgenic lines was greater than that of the wild type Jimai 19 throughout the growing season when the plants were grown in natural fields.This difference is reflected primarily in the increased values per plant of number of effective tillers,biomass,grain number,and grain weight of the transgenic lines.
As indicated in Table 2,the overexpression of the GmDREB1 gene improves the salt tolerance of wheat at the germination stage,the seedling stage and throughout the growing season.Because the salt tolerance of the transgenic line T349 was slightly higher than that of T378,we selected the transgenic line T349 for further investigation of physiological and protein responses to the salt stress.
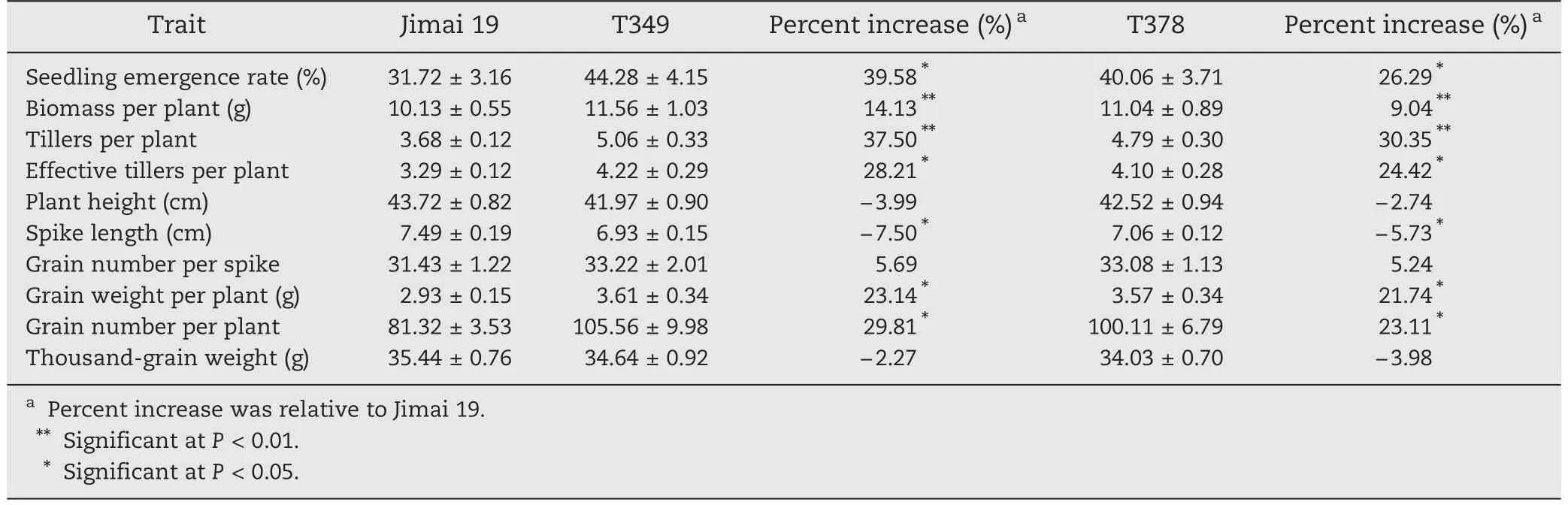
Table 2-Comparison of means for 10 agronomic traits between transgenic wheat lines (T349 and T378)and the wild type(Jimai 19)throughout the growing season under salt stress(means ± SE).
3.2.Physiological analysis
After 0,1,3,5,and 7 days of NaCl treatment,the first leaves of T349 and Jimai 19 seedling samples were harvested for measurement of the betaine,proline,and malondialdehyde(MDA)contents and relative electrolyte leakage.
Although proline and glycine betaine are critical for osmoprotection,there were no significant differences in glycine betaine and proline contents between T349 and Jimai 19 after 0 and 1 day of NaCl treatment.After 3,5,and 7 days of NaCl treatment,glycine betaine,and proline contents were significantly higher in T349 than in Jimai 19(Fig.3).
The MDA content and relative electrolyte leakage are associated with the oxidization of the cell membrane.There were no significant differences in MDA content or relative electrolyte leakage between T349 and Jimai 19 after 0,1,and 3 days of NaCl treatment.After 5 and 7 days of NaCl treatment,MDA content and relative electrolyte leakage were significantly lower in T349 than in Jimai 19(Fig.3).
3.3.Proteomic analysis
Total proteins were extracted from the first expanded leaves of salt-treated seedlings of T349 and Jimai 19.The profiles of wheat leaf proteins were established at a pI range of 3.5 to 10.0 and with a molecular mass range of 13 to 110 kDa (Fig.4).Compared with Jimai 19,17 protein spots(S1-1 to S1-17)were up-regulated in T349 (Fig.5),and all of these proteins were identified by mass spectrometry (Table 3).The significant differences between Jimai 19 and T349 leaves corresponded to their different protein responses to salt stress.
The functional classification analysis according to gene ontology (GO) annotations and PubMed references revealed that the proteins were clustered into several categories.Those 17 differential proteins were involved in osmotic stress,oxidative stress,photosynthesis,and lipid metabolism.Osmotic stress-related proteins include methionine synthase (S1-11)and glyceraldehyde-3-phosphate dehydrogenase (GPD) (S1-6).Oxidative stress-related proteins include NADP-dependent malic enzyme (S1-12),glutathione transferase (S1-3) and 2-cys peroxiredoxin (S1-10).Photosynthesis-related proteins include Rubisco large subunit (RLS),Rubisco activase (S1-16)and chlorophyll a–b binding proteins(S1-9).Spots S1-7,S1-8,S1-13,S1-14,and S1-15 were all identified as Rubisco large subunits with different molecular masses and isoelectric points corresponding to their spot positions on the gel.Lipases (S1-17) directly catalyze the hydrolysis or synthesis of lipids.
Spots S1-1,S1-2,S1-4,and S1-5 were identified as predicted proteins of barley.According to NCBI BLAST results,spot S1-1(gi|326503994)contains the region PLN00128,which is annotated as a succinate dehydrogenase (ubiquinone) flavoprotein subunit,and has 94% identity with the Triticum urartu protein succinate dehydrogenase(ubiquinone)flavoprotein subunit(sequence ID:gb|EMS46614.1|).Spot S1-2 (gi|326511988) contains the region MopB_Res-Cmplx1_Nad11,which is annotated as the second domain of the Nad11/75-kDa subunit of the NADH-quinone oxidoreductase,and has 98%identity with the T.urartu protein NADH-ubiquinone oxidoreductase 75 kDa subunit(sequence ID:gb|EMS48685.1|).Spot S1-4 (gi|326493416) contains the region PLN02300,which is annotated as lactoylglutathione lyase,and has 98% identity with the Aegilops tauschii protein lactoylglutathione lyase(sequence ID:gb|EMT08036.1|).Spot S1-5(gi|326491885) contains the region WD40,a domain found in many eukaryotic proteins that cover a wide variety of functions,including adaptor/regulatory modules in signal transduction,pre-mRNA processing and cytoskeleton assembly.
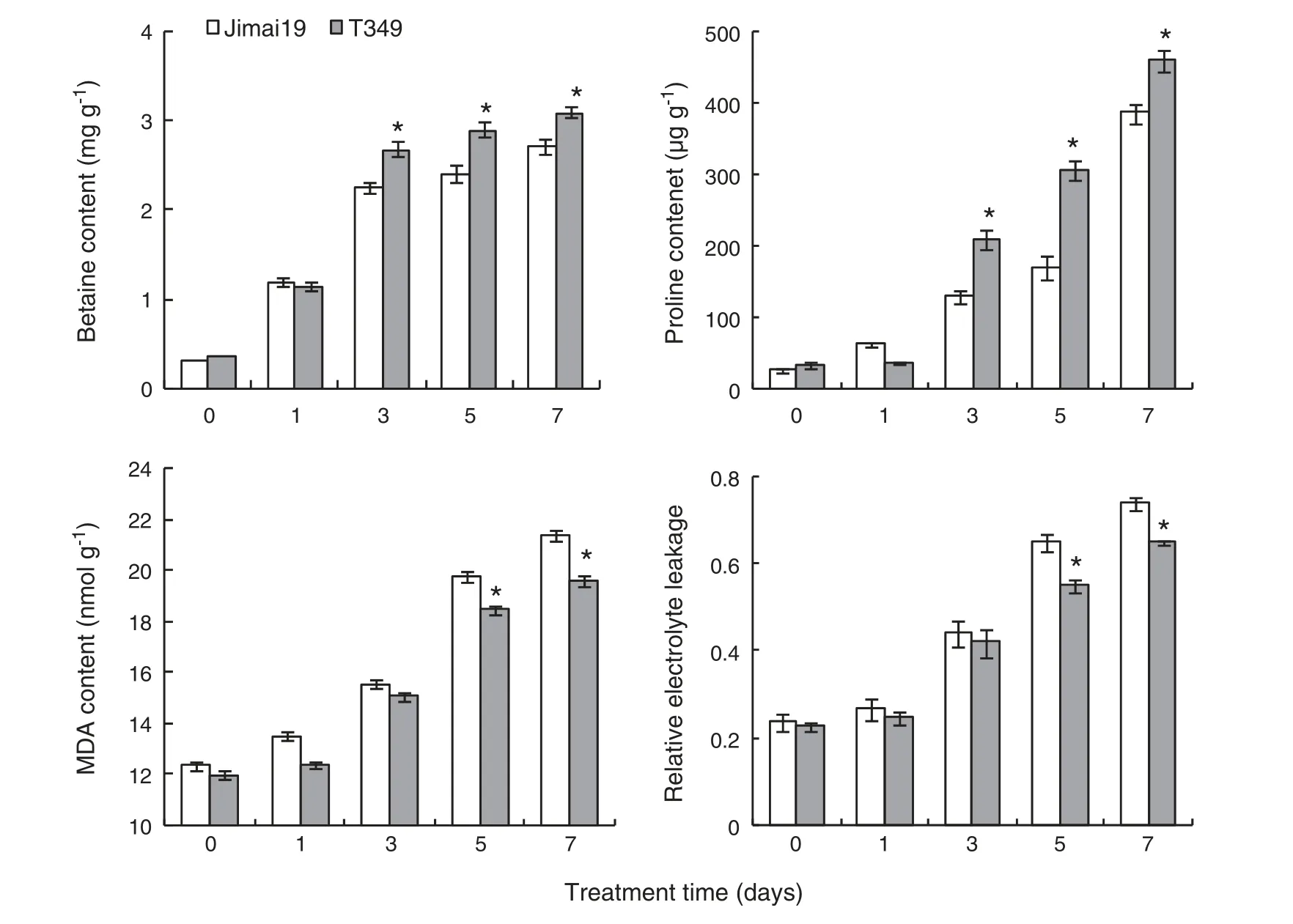
Fig.3-Physiological index of the transgenic line T349 and wild type Jimai 19 treated with salt at 0,1,3,5,and 7 days.Fifty germinated seeds of the wild-type Jimai 19 and transgenic wheat line T349 were grown in plastic containers containing complete Kimura B nutrient solution,and then ten-day-old seedlings were treated with 300 mmol L-1 NaCl in Kimura B nutrient solution.After treatment for 0,1,3,5,and 7 days,the first leaves of seedling samples were harvested for physiological analysis.The experiment was independently repeated three times.The values are the means of three replicates ± SE.*Transgenic line T349 was significantly different at P <0.05 when compared with Jimai 19.

Fig.4-2-DE of total protein extracts from T349 and Jimai 19 leaves with salt stress.Ten-day-old seedlings of Jimai 19 and T349 were treated for 7 days with 300 mmol L-1 NaCl in Kimura B nutrient solution.Total proteins were extracted from the first expanded leaves.Comparing the total leaf protein expression of Jimai 19 and T349,17 protein spots,S1-1 to S1-17,were up-regulated in the transgenic line T349 compared with the wild type Jimai 19.
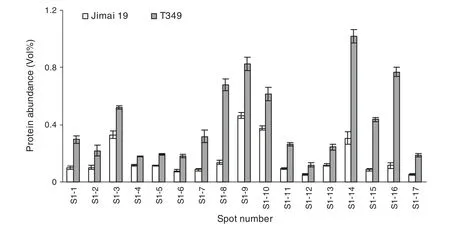
Fig.5-Quantification of salt stress-responsive proteins.Image Master reported the%Vol for every spot on the gel.The average%Vol of a spot was obtained from three replicates of the same segment,and the%Vol was compared between gels of Jimai 19 and T349.Only spots that showed more than 1.5-fold changes in average%Vol between segments,with statistically significant differences,as determined by Student's t-test(P <0.05),were defined as differentially expressed protein spots.The data are means of three independent replicates ± SD.
4.Discussion
4.1.The overexpression of the GmDREB1 gene improves salt tolerance in transgenic wheat
The coleoptile length,radicle length,and radicle number of the GmDREB1 transgenic wheat lines were significantly higher than those of the wild type,suggesting that the overexpression of the GmDREB1 gene improves the growth of wheat seedlings under saline conditions.Early and rapid elongation of roots is an important indicator of the ability of the plant to resist abiotic stresses,such as cold and salt[33,34].
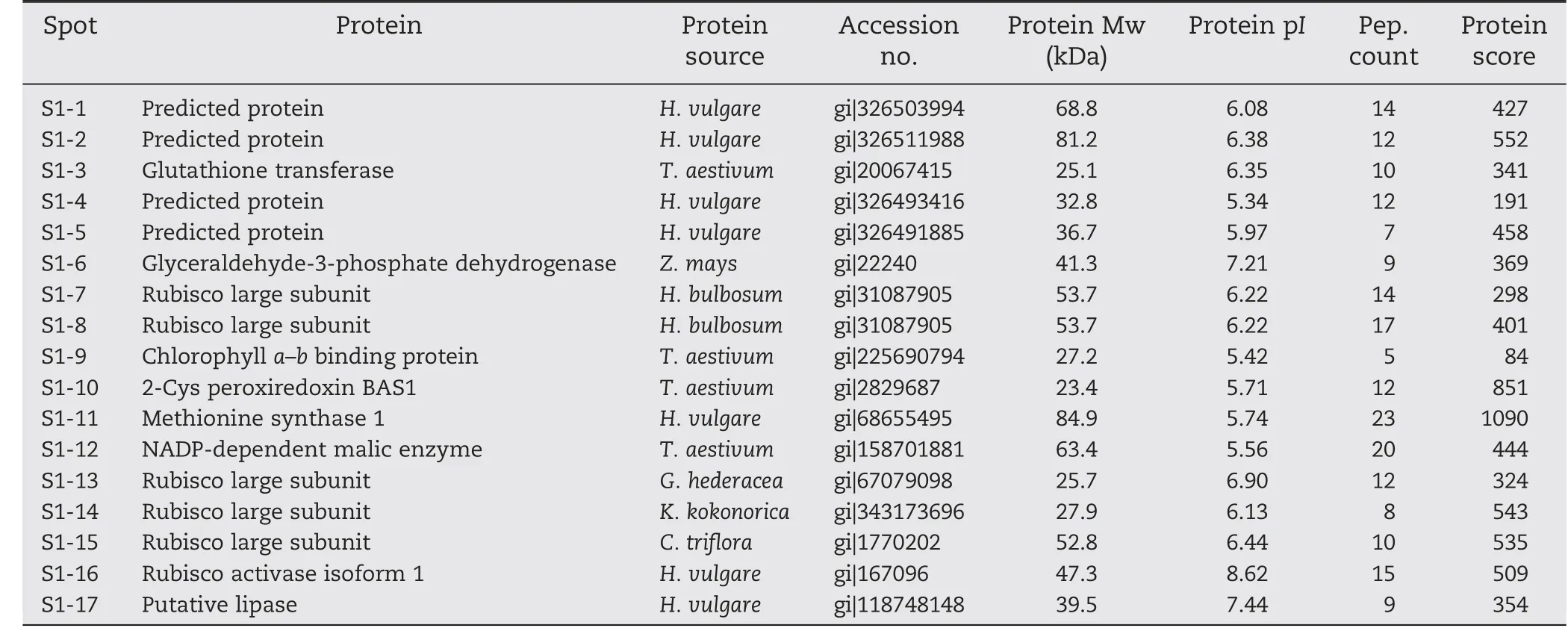
Table 3-Identification of salt-stress-responsive proteins in wheat leaves.
In this study,when seedlings were grown in a nutrient solution with salt in the laboratory,the salt tolerance of Jimai 19 was found to be 4,whereas that of GmDREB1 transgenic wheat lines was found to be 3.The wild type was sensitive to salt,whereas the transgenic varieties had medium tolerance,and the roots of the transgenic varieties were longer than those roots of the wild type.When the seedlings were grown in soil watered with salt in a phytotron,the transgenic lines had more tillers than the wild type.When plants were grown in natural saline–alkaline soil,the seedling emergence rate and the effective tillers per plant of the transgenic lines were also significantly higher than those of the wild type.At the physiological level,the significant amounts of proline and glycine betaine that accumulated in the transgenic line T349and the significant reduction in the relative electrolyte leakage and in the MDA content in T349 suggested that the tolerance of transgenic wheat seedlings to salt stress was enhanced by the GmDREB1 gene transfer.
When the GmDREB1 gene was transferred into alfalfa [23],the transgenic plants also showed enhanced tolerance to salt at the seedling stage.The transgenic wheat overexpressed the GmDREB1 gene and grew normally in culture medium with 0.6%NaCl,whereas the leaves of the wild type were curly,and the roots were slender at the three-leaf stage [22].This observation suggests that the salt tolerance of the transgenic wheat was enhanced by the transfer of the GmDREB1 gene.
The ultimate goal of plant transformation is the introduction of a novel trait without producing detrimental effects on agronomic performance.Evaluation of transgenic plants under field conditions is thus necessary for determining the effects of genetic transformation on crop agronomic traits[35,36].In this study,for the first time,the salt tolerance of DREB transgenic wheat grown in natural fields was investigated.Grown in saline–alkaline soil of natural fields and thus likely facing additional stresses,the transgenic lines showed improvements in some agronomic traits but no growth retardation,sterility,or negative effects on phenotype.
4.2.Leaf protein response of GmDREB1 transgenic wheat to high salinity
In this study,the transgenic lines overexpressing GmDREB1 showed higher salt tolerance than the wild type.DREB expression confers abiotic stress tolerance on transgenic plants because DREB transcription factors bind to DRE/CRT cis-acting elements in the promoter regions of many stress-related genes that play important roles in plant stress tolerance [5,6].However,the increased stress tolerance and plant growth achieved in DREB-transgenic plants maybe due not only to the up-regulation of stress-related gene expression; other genes working in different plant physiological and developmental processes that,in turn,contribute to improved plant growth in DREB transgenic plants may be involved[11,14,15].High salinity can cause osmotic stress and further salt intake,and osmotic stress can produce superabundant reactive oxygen species(ROS) that increase oxidative stress in plants [37,38].In the present study,under salt stress,some osmotic and oxidative stress-related proteins that may be involved in improving the salt tolerance of transgenic wheat were up-regulated in the transgenic line T349.
Methionine synthase catalyzes the formation of methionine by the transfer of a methyl group from 5-methyltetrahydrofolate to homocysteine.This reaction occurs in the activated methyl cycle,which is known as the metabolic source of single carbons[39].In this cycle,methionine is further converted into S-adenosylmethionine(SAM)by S-adenosylmethionine synthetase.SAM provides a methyl group for many metabolites,including important compounds,such as glycine betaine,methylated polyols,and polyamines,under high salinity conditions.Glycine betaine and methylated polyols are compatible solutes that accumulate in the cytoplasm and that regulate osmotic balance under salt stress [40,41].Thus the up-regulation of methionine synthase(S1-11)in T349 may play an important role in improving the ability of transgenic wheat to tolerate salt by regulating the osmotic balance.In barley leaves,the methionine synthase protein and transcript levels all increased under salt stress (200 mmol L-1NaCl for three days) [42].Glyceraldehyde-3-phosphate dehydrogenase (GPD)(S1-6)was also up-regulated in T349 under salt stress.GPD is an important enzyme in the glycolysis and gluconeogenesis pathways.Increased GPD activity mobilizes carbon away from glycerol and into the pathway leading to glycolysis and ATP formation,providing the compatible osmolytes and the energy required for osmotic stress tolerance [43].In other studies,the salt tolerance of transgenic potato plants was improved by the gene transfer of glyceraldehyde-3 phosphate dehydrogenase[44].GPD was transcriptionally up-regulated in Mesembryanthemum crystallinum during salt stress[45].Thus the up-regulation of methionine synthase and GPD in T349 may also play an important role in improving the plant's salt tolerance by regulating the osmotic balance.At the physiological level,after 3,5,and 7 days of NaCl treatment,glycine betaine,and proline contents were significantly higher in T349 than in Jimai 19.Although there is a positive correlation reported between proline accumulation and osmotolerance,the cardinal role of proline as an osmoprotectant under varying conditions of stress has been shown in certain plants [46,47].It is well known that glycine betaine,as an osmolyte and enzyme-protectant,can protect the integrity of the membrane under conditions of salt stress,thereby improving the salt tolerance of the plant[48,49].
ROS function by generating peroxidants that injure membrane lipids,proteins,and nucleic acids under salt stress conditions [50].In the present study,NADP-dependent malic enzyme (S1-12),glutathione transferase (S1-3) and 2-cys peroxiredoxin BAS1(S1-10)were up-regulated in the transgenic line T349 under salt stress.The NADP-dependent malic enzyme catalyzes the oxidative decarboxylation of L-malate,producing pyruvate,CO2,and NADPH.NADPH provides the reducing power required for ROS metabolism [51].Glutathione transferase catalyzes the conjugation of the tripeptide glutathione with compounds containing an electrophilic center to form more soluble,nontoxic peptide derivatives to reduce the lipid peroxidation caused by ROS [52,53].The molecule 2-cys peroxiredoxin BAS1 is a homodimeric thiol-based peroxidase that catalyzes the reduction of H2O2(producing H2O)or reduces the peroxide substrate to the corresponding alcohol,reducing the cell injury caused by oxidative stress [54].The presence of spots S1-1,S1-2,and S1-4,which contain the region of the succinate dehydrogenase (ubiquinone) flavoprotein subunit,NADH-quinone oxidoreductase,and lactoylglutathione lyase,respectively,indicates that these proteins are involved in the oxidative stress response.These proteins were induced by stress,salt/abscission,aluminum or by low temperature [55].Thus,all of these proteins maybe involved in removing superabundant ROS to reduce the lipid peroxidation caused by ROS and thereby improve the salt tolerance of the plant.Rice NADP-dependent malic enzyme genes have been shown to be up-regulated by NaCl stress at the transcriptional level [56,57].The overexpression of glutathione transferase in transgenic tobacco seedlings produced reduced levels of lipid peroxidation[58].These findings indicate that the overexpression of the NADP-dependent malic enzyme and glutathione transferase provides protection from oxidative damage caused by salt stress.After 5 and 7 days of NaCl treatment,MDA contents and relative electrolyte leakage were significantly lower in the transgenic line T349 than in the wild-type Jimai 19.The relative electrolyte leakage reflects the permeability of the cell membrane,so that increased electrolyte leakage is considered a reliable indicator of membrane damage.Malondialdehyde,which is a product of lipid peroxidation,has also been considered to indicate oxidative damage.Both of these proteins have been widely used as indicators of a plant's ability to tolerate salt[59–61].These results at the protein and physiological level suggest that the transgenic wheat line T349 effectively reduces the cell damage caused by oxidative damage,thereby improving its salt tolerance.
This study was supported by the National Transgenic Key Project from the Ministry of Agriculture of China(2014ZX08011-003)and the Agricultural Science and Technology Innovation Program(ASTIP).
[1] E.A.Bray,Plant responses to water deficit,Trends Plant Sci.2(1997) 48–54.
[2] B.Vinocur,A.Altman,Recent advances in engineering plant tolerance to abiotic stress:achievements and limitations,Curr.Opin.Biotechnol.16(2005) 123–132.
[3] P.Langridge,N.Paltridge,G.Fincher,Functional genomics of abiotic stress tolerance in cereals,Brief Funct.Genomic Proteomic 4 (2006) 343–354.
[4] C.Lata,M.Prasad,Role of DREBs in regulation of abiotic stress responses in plants,J.Exp.Bot.62(2011) 4731–4748.
[5] P.K.Agarwal,P.Agarwal,M.K.Reddy,S.K.Sopory,Role of DREB transcription factors in abiotic and biotic stress tolerance in plants,Plant Cell Rep.25(2006) 1263–1274.
[6] S.S.Hussain,M.A.Kayani,M.Amjad,Transcription factors as tools to engineer enhanced drought tolerance in plants,Biotechnol.Prog.27(2011) 297–306.
[7] K.Yamaguchi-Shinozaki,K.Shinozaki,A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought,low-temperature or high-salt stress,Plant Cell 6(1994) 251–264.
[8] M.S.Khan,The role of DREB transcription factors in abiotic stress tolerance of plants,Biotechnol.Biotechnol.Equip.25(2011) 2433–2442.
[9] C.Lata,S.Bhutty,R.P.Bahadur,M.Majee,M.Prasad,Association of a SNP in a novel DREB2-like gene SiDREB2 with stress tolerance in foxtail millet[Setaria talica(L.)],J.Exp.Bot.62 (2011) 3387–3401.
[10] M.Kasuga,S.Miura,K.Shinozaki,K.Yamaguchi-Shinozaki,A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought and low-temperature stress tolerance in tobacco by gene transfer,Plant Cell Physiol.45 (2004) 346–350.
[11] K.Nakashima,Y.Ito,K.Yamaguchi-Shinozaki,Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses,Plant Physiol.149(2009)88–95.
[12] S.Fowler,M.F.Thomashow,Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway,Plant Cell 14(2002)1675–1690.
[13] K.Maruyama,Y.Sakuma,M.Kasuga,Y.Ito,M.Seki,H.Goda,Y.Shimada,S.Yoshida,K.Shinozaki,K.Yamaguchi-Shinozaki,Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems,Plant J.38(2004)982–993.
[14] K.Lee,K.S.Han,Y.S.Kwon,J.H.Lee,S.H.Kim,W.S.Chung,Y.Kim,S.S.Chun,H.K.Kim,D.W.Bae,Identification of potential DREB2C targets in Arabidopsis thaliana plants overexpression DREB2C using proteomic analysis,Mol.Cell 28(2009)383–388.
[15] L.V.Savitch,G.Allard,M.Seki,L.S.Robert,N.A.Tinker,N.P.A.Huner,K.Shinozaki,J.Singh,The effect of overexpression of two Brassica CBF/DREB1-like transcription factors on photosynthetic capacity and freezing tolerance in Brassica napus,Plant Cell Physiol.46(2005)1525–1539.
[16] L.Cong,H.C.Zheng,Y.X.Zhang,T.Yao,Arabidopsis DREB1A confers high salinity tolerance and regulates the expression of GA dioxygenases in Tobacco,Plant Sci.174(2008)156–164.
[17] Y.Ito,K.Katsura,K.Maruyama,T.Taji,M.Kobayashi,M.Seki,K.Shinozaki,K.Yamaguchi-Shinozaki,Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice,Plant Cell Physiol.47(2006) 141–153.
[18] M.T.Pino,J.S.Skinner,E.Park,Z.Jeknic,P.M.Hayes,M.F.Thomashow,T.H.Chen,Use of a stress inducible promoter to drive ectopic AtCBF expression improves potato freezing tolerance while minimizing negative effects on tuber yield,Plant Biotechnol.J.5(2007) 591–604.
[19] P.Bihani,B.Char,S.Bhargava,Transgenic expression of sorghum DREB2 in rice improves tolerance and yield under water limitation,J.Agric.Sci.149 (2011) 95–101.
[20] P.Agarwal,P.K.Agarwal,A.J.Joshi,S.K.Sopory,M.K.Reddy,Overexpression of PgDREB2A transcription factor enhances abiotic stress tolerance and activates downstream stress-responsive genes,Mol.Biol.Rep.37(2010) 1125–1135.
[21] M.Chen,Q.Y.Wang,X.G.Cheng,Z.S.Xu,L.C.Li,X.G.Ye,L.Q.Xia,Y.Z.Ma,GmDREB2,a soybean DRE-binding transcription factor,conferred drought and high-salt tolerance in transgenic plants,Biochem.Biophys.Res.Commun.353(2007) 299–305.
[22] S.Q.Gao,H.J.Xu,X.G.Cheng,M.Chen,Z.S.Xu,L.C.Li,X.G.Ye,L.P.Du,X.Y.Hao,Y.Z.Ma,Improvement of wheat drought and salt tolerance by expression of a stress inducible transcription factor GmDREB of soybean (Glycine max),Chin.Sci.Bull.50(2005) 2714–2723.
[23] T.Jin,Q.Chang,W.Li,D.X.Yin,Z.J.Li,D.L.Wang,B.Liu,L.X.Liu,Stress-inducible expression of GmDREB1 conferred salt tolerance in transgenic alfalfa,Plant Cell Tissue Organ Cult.100 (2010) 219–227.
[24] N.Shimada,in:Y.Noguchi,S.Kawada (Eds.),Dictionary of Agriculture,Yokendo,Tokyo,1987,p.817.
[25] J.Zhu,Z.Bie,Y.Li,Physiological and growth responses of two different salt-sensitive cucumber cultivars to NaCl stress,Soil Sci.Plant Nutr.54 (2008) 400–407.
[26] U.Erturk,N.Sivritepe,C.Yerlikaya,M.Bor,F.Ozdemir,I.Turkan,Responses of the cherry rootstock to salinity in vitro,Biol.Plant.51(2007) 597–600.
[27] J.Shaterian,D.Waterer,H.De Jong,K.K.Tanino,Differential stress responses to NaCl salt application in early-and late-maturing diploid potato (Solanum sp.) clones,Environ.Exp.Bot.54(2005) 202–212.
[28] A.Zhen,Z.Bie,Y.Huang,Z.Liu,Q.Li,Effects of scion and rootstock genotypes on the anti-oxidant defense systems of grafted cucumber seedlings under NaCl stress,Soil Sci.Plant Nutr.56(2010) 263–271.
[29] X.J.Li,M.F.Yang,H.Chen,L.Q.Qu,F.Chen,S.H.Shen,Abscisic acid pretreatment enhances salt tolerance of rice seedlings: proteomic evidence,Biochim.Biophys.Acta 1804(2010) 929–940.
[30] K.V.Madhava Rao,T.V.S.Sresty,Antioxidative parameters in the seedlings of pigeonpea(Cajanus cajan L.Millspaugh)in response to Zn and Ni stresses,Plant Sci.157 (2000) 113–128.
[31] Q.Y.Jiang,H.Chen,X.L.Pan,Y.H.Shi,X.R.Li,G.Y.Zhang,Y.J.Wang,S.G.Xie,S.H.Shen,Proteomic analysis of wheat(Triticum aestivum L.)hybrid necrosis,Plant Sci.175(2008)394–401.
[32] S.Komatsu,A.Muhammad,R.Rakwal,Separation and characterization of proteins from green and etiolated shoots of rice Oryza sativa L.:towards a rice proteome,Electrophoresis 20(1999)630–636.
[33] P.Sharifi,Evaluation on sixty-eight rice germplasms in cold tolerance at germination stage,Rice Sci.17 (2010) 77–81.
[34] K.Çavuşoğlu,S.Kılıç,K.Kabar,Some morphological and anatomical observations during alleviation of salinity (NaCl)stress on seed germination and seedling growth of barley by polyamines,Acta Physiol.Plant.29 (2007) 551–557.
[35] F.Barro,P.Barceló,P.A.Lazzerí,P.R.Shewry,A.Martin,J.Ballesteros,Field evaluation and agronomic performance of transgenic wheat,Theor.Appl.Genet.105 (2002) 980–984.
[36] A.Bahieldin,H.T.Mahfouz,H.F.Eissa,O.M.Saleh,A.M.Ramadan,I.A.Ahmed,W.E.Dyer,H.A.El-Itriby,M.A.Madkour,Field evaluation of transgenic wheat plants stably expressing the HVA1 gene for drought tolerance,Physiol.Plant.123 (2005) 421–427.
[37] Y.C.Boo,J.Jung,Water deficit-induced oxidative stress and antioxidative defenses in rice plants,J.Plant Physiol.155(1999) 255–261.
[38] P.M.Hasegawa,R.A.Bressan,J.K.Zhu,H.J.Bohnert,Plant cellular and molecular responses to high salinity,Annu.Rev.Plant Physiol.Plant Mol.Biol.51 (2000) 463–499.
[39] A.D.Hanson,D.A.Gage,Y.Shachar-Hill,Plant one-carbon metabolism and its engineering,Trends Plant Sci.5(2000)206–213.
[40] H.J.Bohnert,R.G.Jensen,Strategies for engineering water-stress tolerance in plants,Trends Biotechnol.14(1996)89–97.
[41] T.Takabe,T.Nakamura,M.Nomura,Y.Hayashi,M.Ishitani,Y.Muramoto,A.Tanaka,T.Takabe,Glycinebetaine and the genetic engineering of salinity tolerance in plants,in: K.Satoh,N.Murata (Eds.),Stress Responses of Photosynthetic Organisms,Elsevier Science,Tokyo,1998,pp.115–131.
[42] Y.Narita,H.Taguchi,T.Nakamura,A.Ueda,W.Shi,T.Takabe,Characterization of the salt-inducible methionine synthase from barley leaves,Plant Sci.167 (2004) 1009–1016.
[43] H.Sobhanian,N.Motamed,F.R.Jazii,T.Nakamura,S.Komatsu,Salt stress induced differential proteome and metabolome response in the shoots of Aeluropus lagopoides(Poaceae),a halophyte C4plant,J.Proteome Res.9(2010)2882–2897.
[44] M.J.Jeong,S.C.Park,M.O.Byun,Improvement of salt tolerance in transgenic potato plants by glyceraldehyde-3 phosphate dehydrogenase gene transfer,Mol.Cell 12(2001)185–189.
[45] D.M.Vernon,J.A.Ostrem,H.J.Bohnert,Stress perception and responses in a facultative halophyte: the regulation of salinity-induced genes in Mesembryanthemum crystallinum,Plant Cell Environ.16(1993) 437–444.
[46] K.F.McCue,A.D.Hanson,Drought and salt tolerance:towards understanding and application,Trends Biotechnol.8 (1990)358–362.
[47] J.D.Ashton,D.P.S.Verma,Proline biosynthesis and osmoregulation in plants,Plant J.4 (1993) 215–223.
[48] M.Jain,G.Mathur,S.Koul,N.B.Sarin,Ameliorative effects of proline on salt stress-induced lipid peroxidation in cell lines of groundnut (Arachis hypogaea L.),Plant Cell Rep.20(2001)463–468.
[49] R.Munns,Comparative physiology of salt and water stress,Plant Cell Environ.25(2002) 239–250.
[50] S.Meneguzzo,C.L.M.Sgherri,F.Navari-Izzo,R.Izzo,Stromal and thylakoid-bound ascorbate peroxidases in NaCl-treated wheat,Physiol.Plant.104 (1998) 735–740.
[51] R.Mittler,Oxidative stress,antioxidants and stress tolerance,Trends Plant Sci.7 (2002) 405–410.
[52] B.Ezaki,R.C.Gardner,Y.Ezaki,H.Matsumoto,Expression of aluminum-induced genes in transgenic Arabidopsis plants can ameliorate aluminum stress and/or oxidative stress,Plant Physiol.122 (2000) 657–665.
[53] M.Katsuhara,T.Otsuka,B.Ezaki,Salt stress-induced lipid peroxidation is reduced by glutathione S-transferase,but this reduction of lipid peroxides is not enough for a recovery of root growth in Arabidopsis,Plant Sci.169 (2005) 369–373.
[54] P.Costa,N.Bahrman,J.M.Frigerio,A.Kermer,C.Plomion,Water-deficit-responsive proteins in maritime pine,Plant Mol.Biol.38 (1998) 587–596.
[55] T.Matsumoto,T.Tanaka,H.Sakai,N.Amano,H.Kanamori,K.Kurita,A.Kikuta,K.Kamiya,M.Yamamoto,H.Ikawa,N.Fujii,K.Hori,T.Itoh,K.Sato,Comprehensive sequence analysis of 24,783 barley full-length cDNAs derived from 12 clone libraries,Plant Physiol.156 (2011) 20–28.
[56] S.Liu,Y.Cheng,X.Zhang,Q.Guan,S.Nishiuchi,K.Hase,T.Takano,Expression of an NADP-malic enzyme gene in rice(Oryza sativa.L)is induced by environmental stresses;over-expression of the gene in Arabidopsis confers salt and osmotic stress tolerance,Plant Mol.Biol.64(2007) 49–58.
[57] J.C.Cushman,Characterization and expression of a NADP-malic enzyme cDNA induced by salt stress from the facultative crassulacean acid metabolism plant,Mesembryanthemum crystallium,Eur.J.Biochem.208(1992)259–266.
[58] V.P.Roxas,S.A.Lodhi,D.K.Garrett,J.R.Mahan,R.D.Allen,Stress tolerance in transgenic tobacco seedlings that overexpress glutathione S-transferase/glutathione peroxidase,Plant Cell Physiol.41(2000)1229–1234.
[59] B.Stevanovic,J.Sinzar,O.Glisic,Electrolyte leakage differences between poikilohydrous and homoiohydrous species of Gesneriaceae,Biol.Plant.40(1997)299–303.
[60] J.A.Hernández,M.S.Almansa,Short-term effects of salt stress on antioxidant systems and leaf water relations of pea leaves,Physiol.Plant.115 (2002) 251–257.
[61] D.A.Meloni,M.A.Oliva,C.A.Martinez,J.Cambraia,Photosynthesis and activity of superoxide dismutase,peroxidase and glutathione reductase in cotton under salt stress,Environ.Exp.Bot.49(2003)69–76.
- The Crop Journal的其它文章
- Genome-wide analysis of the WRKY transcription factor gene family in Gossypium raimondii and the expression of orthologs in cultivated tetraploid cotton
- Development of highly glyphosate-tolerant tobacco by coexpression of glyphosate acetyltransferase gat and EPSPS G2-aroA genes
- SSR genetic linkage map construction of pea (Pisum sativum L.) based on Chinese native varieties
- Rank correlation among different statistical models in ranking of winter wheat genotypes,
- Effects of exogenous ABA application on post-anthesis dry matter redistribution and grain starch accumulation of winter wheat with different staygreen characteristics
- Genetic characterization and linkage disequilibrium mapping of resistance to gray leaf spot in maize(Zea mays L.)

