Establishment of the integrated applied core collection and its comparison with mini core collection in soybean(Glycine max)
Yong Guo,Yinghui Li,Huilong Hong,Li-Juan Qiu*
National Key Facility for Crop Gene Resources and Genetic Improvement/Institute of Crop Science,Chinese Academy of Agricultural Sciences,Beijing 100081,China
1.Introduction
Plant germplasm denotes the genetic resources for plant breeding.A large number of germplasm accessions have been collected in gene banks all over the world,but methods for managing and utilizing such a large collection efficiently remain a challenging task for breeders.Frankel and Brown first proposed sampling the collections to yield a manageable sample or so-called “core collection” [1,2].A core collection(CC) consists of a limited set of accessions derived from the collection(about 10%of the full collection),and represents the genetic diversity of a species and its relatives with a minimum of repetitiveness.Owing to the reduced size,CC can be studied extensively and the derived information can be used to guide more efficient utilization of the much larger reserve collection.To date,CCs have been developed in many crops including rice[3],wheat[4],soybean[5],cotton[6]and peanut[7].
Usually the number of accessions in a CC is still too large for meaningful replicated evaluations at different locations,given the enormous sizes of the full collections (FCs) of many crops.To address this problem,Upadhyaya and Ortiz postulated the concept of the “mini core collection” [8].Usually a mini core collection (MCC) consists of 10% of the accessions from the CC,so that the number of accessions is only about 1% of that of the FC.MCC still represents the diversity of the FC,but the number of accessions is sharply reduced.MCC is thus suitable for evaluation and utilization in breeding and genetic studies.Following this strategy,MCCs of rice,maize,soybean,peanut,chickpea (Cicer arietinum L.) and pigeonpea (Cajanus cajan) have been developed [9–14].Some accessions with desirable agronomic and nutritional traits have been identified using these MCCs,including chickpea with drought-avoidance root traits [13],pigeonpea with drought tolerance [14],pigeonpea with multiple disease resistance [15],peanut with high-quality [16] and wheat with high-molecular-weight glutenin subunits [17].A high proportion of high-yielding hybrids have been produced by crossing between alfalfa (Medicago sativa subsp.sativa L.) populations derived from previously selected high-yielding accessions from a CC[18].The identification of accessions with desirable agronomic and nutritional traits from CCs and MCCs has confirmed the representative character of these collections.
China has the most abundant genetic resources for soybean,and more than 23,000 cultivated soybean accessions are maintained in the Chinese National Soybean GeneBank(CNSGB).The primary CC of Chinese cultivated soybean,which consists of 2794 accessions (about 11.8% of accessions in FC) and represents 73.6% of the genetic diversity,has been developed based on the characterization of selected phenotypes and on molecular markers [5,19].A MCC of cultivated soybean has also been developed,based on the established primary CC,and represents 94.5% of the phenotypic diversity and 63.5% of the genetic diversity of the FC [20].The soybean accessions in the MCC provide trait-specific resources for soybean improvement programs and may be used for crossing or backcrossing with elite varieties in specific eco-regions.Previous results showed that some disadvantageous traits such as lodging,disease sensitivity,and low-quality could be improved after backcrossing only twice with elite varieties[21,22].The soybean accessions in the MCC may also be used for basic studies including gene discovery,allele mining,marker-trait associated analysis,and gene functional study.Upon validating the association between polymorphic molecular markers and segregating phenotypic traits,plants with desirable characters such as optimal height,growth duration,100-seed weight,protein content,and fat content may be selected based on the associated markers.QTLs underlying tolerance to cold and drought stresses have also been identified by the use of backcross introgression lines developed from soybean accessions in the MCC [23,24].Moreover,soybean accessions in the MCC have been used for genetic diversity and allelic variation analysis of the Dt1/GmTfl1 locus,the primary controller of determinate growth habit in soybean,suggesting that human selection for determinacy took place in early stages of landrace radiation [25].Soybean accessions in MCC have also been used for allele mining at other two loci (GmF3′H and GmF3′5′H) that encode key enzymes involved in coloration of tissues and organs,and three lowfrequency new alleles have been identified,further confirming the abundant genetic variation of the accessions in MCC[26].
However,accessions with desirable agronomic or nutritional traits in a MCC are rare,owing to the need to include as many accessions as possible with various traits while preserving appropriate sample sizes.For this reason,accessions with specific traits identified in MCCs could be used only as indicators for directional identification of more elite accessions from CCs or FCs.For example,for studying disease resistance,stress tolerance and other traits,soybean accessions in MCC can first be used to characterize the distribution of different traits,and then a large number of accessions may be evaluated based on these distributions.This strategy has been used to identify elite accessions in previous studies of salt tolerance and soybean cyst nematode(SCN)resistance[27,28].
In the present study,we developed an integrated applied core collection (IACC) for soybean that consists of accessions with cold tolerance,drought tolerance,salt tolerance,SCN resistance,soybean mosaic virus (SMV) resistance,high protein content and high fat content.Here,IACC was defined as a core collection of accessions with different desirable agronomic and nutritional traits fulfilled with interest to genetic research and breeding programs.The diversity of this newly formed IACC was compared with that of the established MCC of soybean.IACC of soybean developed in this study lays a foundation for selection of crossing parents fulfilling various breeding goals in different eco-regions.
2.Materials and methods
2.1.Plant materials
Soybean accessions were obtained from the Chinese National Soybean GeneBank (CNSGB) at the Institute of Crop Science,Chinese Academy of Agricultural Sciences.A large-scale evaluation of agronomic traits of the conserved accessions of cultivated soybeans was performed.A total of 159 soybean accessions,including 18 from the MCC and 141 from the FC of soybean,were selected.These 159 accessions accounted for about 10% of accessions in the FC carrying at least one of seven desirable agronomic or nutritional traits of particular interest to genetic research and breeding programs.This selection strategy was aimed at obtaining similar proportions of accessions with each desirable trait from the FC based on the phenotypic data.The seven traits included cold tolerance(rank 0 or 1),drought tolerance (rank 0 or 1),salt tolerance(high tolerance,relative salt injury index less than 20 in bud or/and injured leaf area less than 10% in seedling stage),SCN resistance (rank 0 or 1 to race 1,2,3,4 and/or 5),SMV resistance (rank 1),high seed protein content(at least 50.0%),and high seed oil content(at least 23.0%).
2.2.SSR genotyping
To genotype the 159 soybean accessions,55 SSR markers were selected based on genome location and degree of polymorphism,as in a previous study [29].DNA extraction,PCR amplification,and SSR genotyping were performed as previously described [5,30].PCR amplification was performed on a PTC-200 Thermocycler (MJ Research/Bio-Rad,USA)with 5′ fluorescent end-labeled primers and PCR products were visualized by silver staining after separation by 6%SDS-polyacrylamide gel electrophoresis.The products were used for genotypic analysis on a Mega BACETM 1000(Amersham Biosciences,USA) and allele fragment sizes were obtained with software BioCalculator 2.0(QIAGEN,Germany).

Table 1-Category analysis of soybean accessions with desirable traits in different collections.
2.3.Statistical analysis
A total of 14 phenotypic traits (nine qualitative and five quantitative traits)were used for phenotypic diversity analysis.The proportions of different classes of nine qualitative phenotypic traits(seed coat color,cotyledon color,seed shape,growth habit,stem termination,pubescence color,flower color,leaf shape and hilum color) in the 159 accessions and a PIC(polymorphic information content) value for each trait were calculated.Chi-square tests were used for detecting similarity of distribution with the accessions in the established MCC.Seed coat has five colors including yellow,green,black,brown and di-color,designated as 1–5.Cotyledon has yellow and green colors,designated as 1 and 2.The codes for seed shape are 1–6 and refer to spherical,spherical flattened,ellipse,flat ellipse,long ellipse and reniform.Codes 1–4 of growth habit refer to erect,semi-erect,semi-rampant,and rampant,and codes 1–3 of stem termination refer to determinate,semi-determinate,and indeterminate.Codes 1–2 of pubescence color and flower color refer to gray and tawny pubescence and to white and purple flower,respectively.The four leaf shapes (lanceolate,ovoid,ellipse and round) are designated as 1–4 and six hilum colors(yellow,buff,brown,dark brown,blue,imperfect black and black) as 1–6.Mean value,standard deviation (SD) and coefficient of variation (CV) of five quantitative phenotypic traits(growth duration,100-seed weight,plant height,protein content and fat content)were calculated using Microsoft Excel software.A large-sample Z-test was used for detecting the similarity of distributions to those of accessions in the MCC.Numbers of observations,allele number,gene diversity,observed heterozygosity,and PIC-value of molecular markers were calculated with PowerMarker V3.25 [31].The PIC-value was calculated as:where Piis the frequency of the ith allele.The chi-square value was calculated as
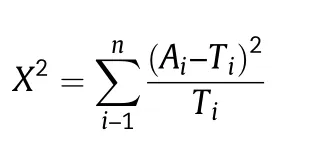
where Aiis the frequency of the ith allele among soybean accessions in IACC and Tiis the frequency of the ith allele among soybean accessions in MCC.
The Z-value was calculated as:
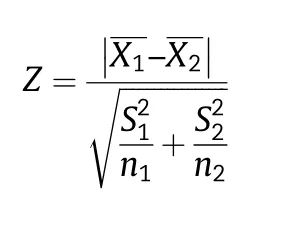
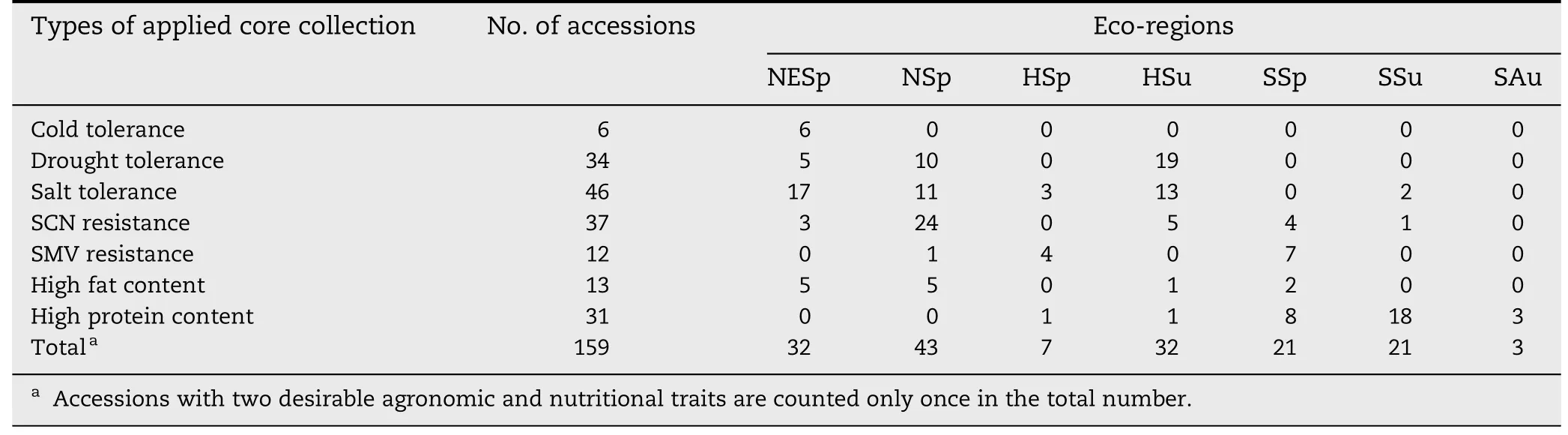
Table 2-Eco-regional distribution of soybean accessions in IACC.
3.Results
3.1.Development of IACC for soybean germplasm resources
For effective utilization of soybean germplasm resources,an IACC was developed by selection of soybean accessions with desirable agronomic and nutritional traits based on the evaluation data.This collection encompasses 6,34,46,37,12,13,31 accessions with cold tolerance,drought tolerance,salt tolerance,SCN resistance,SMV resistance,high protein content and high fat content,respectively.The sampled number of accessions accounted for about 10% of accessions carrying at least one of these seven traits especially useful to soybean breeders in the FC.Category analysis of accessions with desirable traits in this newly formed core collection showed that the proportion of accessions in each category was much higher than that of the accessions in the FC and the established MCC of soybean(Table 1).
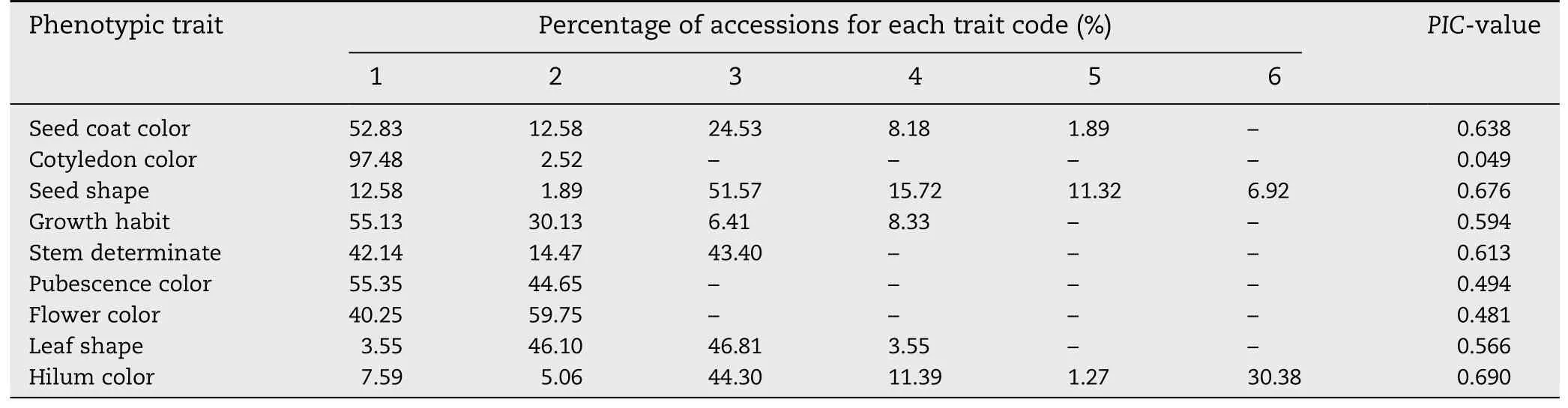
Table 3-Diversity and frequency of nine qualitative phenotypic traits for soybean accessions in IACC.
3.2.Distribution of soybean accessions in IACC
Eco-region analysis of soybean accessions in IACC showed that these accessions originated in all seven eco-regions of China (northeast spring sowing,NESp; north spring sowing,NSp; Huanghuaihai spring sowing,HSp; Huanghuaihai summer sowing,HSu; south spring sowing,SSp; south summer sowing,SSu; and south autumn sowing,SAu).Among these,accessions from the NSp region were the most common accessions in IACC,followed by accessions from the NESp and HSu regions.Accessions from SAu region were the rarest in this core collection (Table 2).With respect to the specific traits,accessions with different desirable agronomic and nutritional traits were distributed unequally.For example,all accessions with cold tolerance were from the NESp eco-region.Most accessions with drought tolerance,salt tolerance,SCN resistance and high protein content were from the HSu,NESp,NSp and SSu eco-regions,respectively.This unequal distribution of accessions in different ecoregions satisfies the need for desirable traits in different regions of China.
The number of desirable agronomic and nutritional traits for each soybean accession was also different in IACC.Most(139 of 159) accessions had only one desirable trait and 20 accessions had two desirable traits.However,no accession had three or more desirable agronomic or nutritional traits,indicating that the integration of desirable traits is very important for soybean breeding.
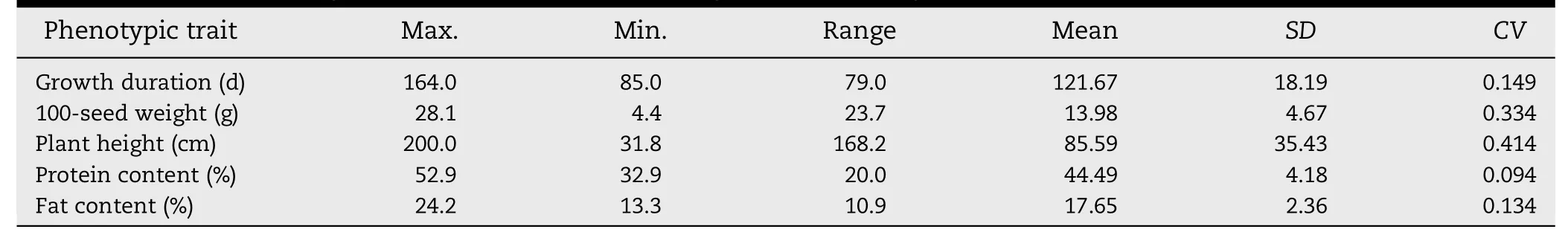
Table 4-Statistical analysis of five quantitative phenotypic traits for soybean accessions in IACC.
3.3.Phenotypic diversity of soybean accessions in IACC
With the aim of characterizing the phenotypic diversity in IACC of soybean,the diversity of nine qualitative and five quantitative traits exhibiting phenotypic diversity was calculated.For the nine qualitative phenotypic traits,the frequencies of accessions with each rank of each trait were determined and PIC-values were calculated as the index of diversity.The results showed that 52.83%,24.53% and 12.58%of the accessions in the new collection had yellow,black,and green seed coats,respectively.The other two seed coat colors were associated with less than 10% of the collection.Most(97.48%) cotyledon color of the collection was yellow,with only a few (2.52%) green cotyledons noted.As to seed shape,51.57% and 15.72% of the accessions had ellipse and flat ellipse seed shape,and the remaining other accessions had the other four seed shapes.The growth habits of the accessions were mainly erect and semi-erect,with frequencies of these two higher than those of the other two growth habits.Similarly,stem terminations were mainly determinate and indeterminate,with only 14.47% of accessions having semi-determinate stem termination.Both pubescence color and flower color were evenly distributed among these accessions.Leaf shape and hilum color were of two main types (Table 3).The diversity index of each qualitative trait was relatively high except for cotyledon color,owing to the high proportion of accessions with yellow cotyledons.This result was consistent with the high proportion of yellow cotyledon color in the full soybean collection.
For the five quantitative phenotypic traits including growth duration,100-seed weight,plant height,protein content,and fat content,the maximum value,minimum value,range,mean value,standard deviation(SD) and coefficient of variation (CV)for each trait of soybean accessions in IACC were all high.The CV and range of plant height were 41.4% and 168.2 cm,respectively.The range of 100-seed weight (33.4%) was also wide in comparison to growth duration(14.9%),protein content(9.4%),and fat content(13.4%)(Table 4).These results indicated that soybean accessions in the new core collection had diverse phenotypic traits and high diversity.
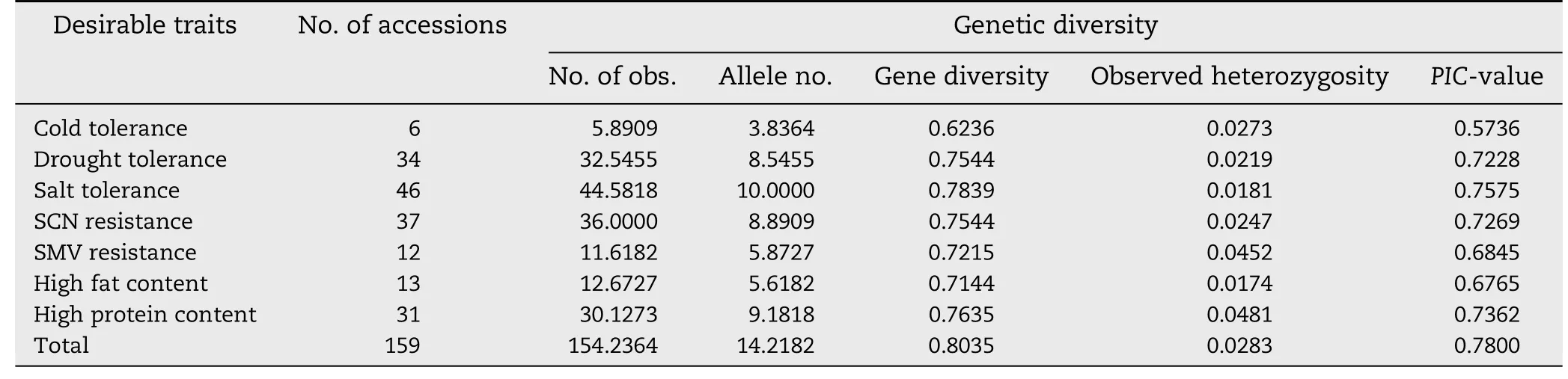
Table 5-Genetic diversity analysis of soybean accessions in IACC by SSR markers.
3.4.Genetic diversity analysis of soybean accessions in IACC based on molecular markers
To evaluate at the molecular level the diversity of the soybean accessions in IACC,55 SSR markers were used to genotype the 159 accessions.A total of 782 alleles were detected,with fragment lengths ranging from 101 to 393 bp.The effective number of alleles at each locus ranged from 2 (Satt387 and Sct_188) to 30 (Satt462),with a mean of 14.2 alleles per locus(Table 5).The proportion of the most common allele at each locus ranged from 10.9% (Satt462) to 63.6% (Satt230),with a mean of 31.9%.The mean diversity among 55 SSR markers was 0.80 and the diversity at individual loci ranged from 0.50(Sct_188)to 0.94(Satt462).The mean heterozygosity among all loci was 0.028 and the heterozygosity of individual loci ranged from 0 (Satt373,Satt390 and Satt556) to 0.129 (Satt453).The PIC-values of loci ranged from 0.374 (Sct_188) to 0.938(Satt462),with a mean of 0.780.
The genetic diversity of accessions with each specific trait was also compared with that of IACC.The results suggested that although the mean allele number was lower,the mean gene diversity and PIC-value in each trait class in the accessions were similar to those of IACC,with cold tolerance the only exception.The mean observed heterozygosity rates of all trait classes were low;indicating that the IACC developed in this study was broadly representative of each set of accessions with desirable agronomic and nutrient traits.The difference in the accessions with cold tolerance may be due to the small number of selected accessions.
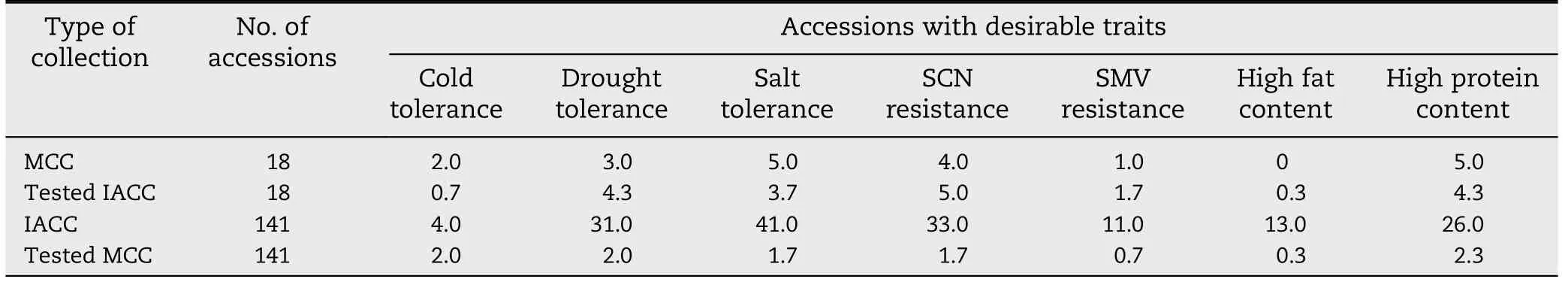
Table 6-Comparison of soybean accessions with desirable traits between IACC and MCC by random sampling.
3.5.Comparison between the newly formed IACC and the established MCC of soybean
Among the 159 soybean accessions in the IACC,18 accessions were from MCC and 141 were from FC of soybean.The 18 from MCC included all types of accessions except those with high oil content.The 18 accessions were randomly selected from IACC three times to assess its representativeness.The average number of accessions with each desirable agronomic and nutritional trait was calculated from three independently selected sets (Table 6).The results showed that the distribution of the accessions with each desirable trait was similar to that of the 18 accessions from MCC,indicating that the representativeness of the IACC was similar to that of MCC.The 141 accessions were also randomly selected three times from the full MCC.The average numbers of accessions with each desirable trait were all strikingly lower than those of accessions in IACC,except for accessions with cold tolerance(Table 6),indicating that few accessions with extremely desirable traits were present in MCC of soybean.Thus the development of IACC is favorable to the utilization of accessions with desirable traits.
The phenotypic diversities of accessions in the newly formed IACC were also compared with those in MCC of soybean.The distribution of accessions with each of the nine qualitative phenotypic traits in IACC was similar to that in MCC,with no significant difference by chi-square test(Table 7).The means,standard deviations,and coefficient of variations of five quantitative phenotypic traits were also similar to those of MCC.Z-tests showed that 100-seed weight,protein content and fat content had no significant difference between these two collections,whereas differences in growth duration and plant height were significant(P<0.05).
The genetic diversity of soybean accessions in the newly formed IACC was also compared with that of MCC by random sampling,the same strategy as used for comparison of phenotype (Table 8).The test also used 18 accessions randomly selected from the whole sample and 141 accessions randomly selected from MCC collection.All the random selections were performed three times and the means of the genetic diversity indices were calculated.The results showed that the mean allele number,gene diversity,observed heterozygosity,and PIC-value of 18 randomly selected accessions were similar to those of 18 accessions from MCC,indicating that the IACC was similarly representative to the MCC at the molecular level.As with the analysis of the desirable traits,the mean allele number,gene diversity,hererozygosity,and PIC-value of 141 randomly selected accessions were different from those of 141 accessions not included in the MCC but included in the IACC of soybean.These results were consistent with the different numbers of soybean accessions with desirable traits in IACC and MCC.
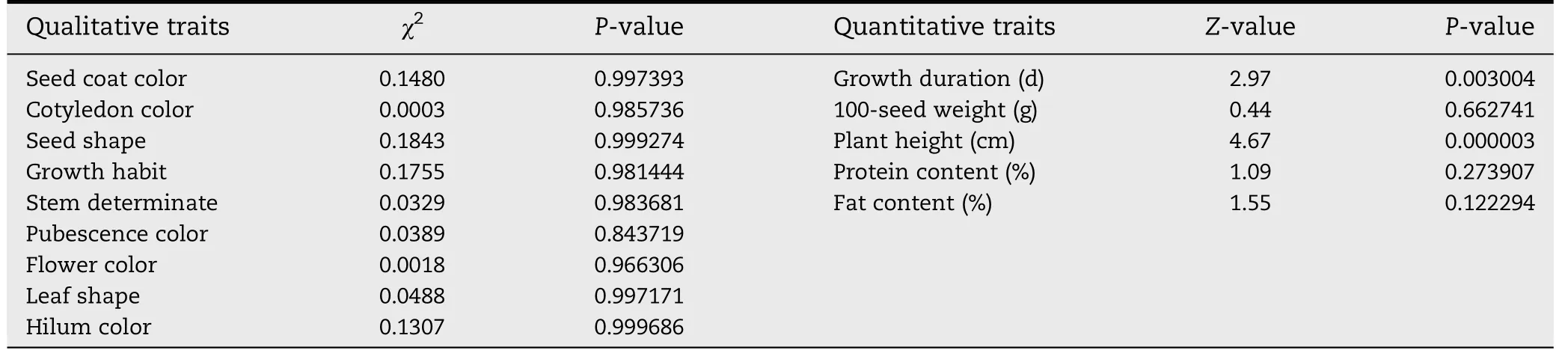
Table 7-Phenotypic diversity comparison between IACC and MCC of soybean accessions.
4.Discussion
4.1.Further expansion of the CC and the MCC in the IACC of soybean
The main tasks for soybean breeders worldwide are expanding the genetic background of crossing parents,discovering desirable alleles,and improving soybean varieties.The greatest obstacle to accomplishing these tasks is the lack of detailed evaluation and genetic characterization of large amounts of soybean germplasm resources[32,33].The development of CCs and MCCs provides an effective way to solve these problems.However,there is always a tradeoff between maintaining genetic diversity and integrating desirable traits,owing to the relatively narrow adaptability of soybean varieties that has resulted from their sensitive light and temperature responses.Accordingly,the direct utilization of the CCs and MCCs encounters limitations in soybean breeding practice.The screening of soybean accessions to develop an IACC was based on the strategy of MCCs,which selects a set of accessions with defined numbers and high genetic diversity.The IACC of soybean is composed of accessions with desirable agronomic and nutritional traits and will meet the demand for accessions with traits useful to soybean breeders.Thus the development of the IACC further expands the concepts of CC and MCC.
4.2.Broad representativeness in the IACC of soybean
The CC and MCC of soybean have broad representativeness.The analysis of nine qualitative and five quantitative phenotypic traits of soybean accessions from the Huanghuaihai eco-region in the primary CC showed that the coefficients of variation of these traits were similar to those in the FC [34].The diversities of these 14 phenotypic traits in the CC and FC were not significantly different.These results suggested that the CC of soybean represents the diversity of the FC.Analysis of the population structure and genetic diversity of soybean accessions in the MCC showed that the MCC of soybean has several features including small sample size,broad representation,low redundancy,and rich diversity [20].In addition,both common and specific alleles were observed among soybean accessions from different eco-regions.The genetic background could accordingly be broadened by incorporation of soybean accessions of different types.In this study,the concept of the IACC was based on the evaluation of soybean germplasm resources.A collection of soybean accessions with specific desirable agronomic and nutritional traits (including cold tolerance,drought tolerance,salt tolerance,SCN resistance,SMV resistance,high protein content,and high fat content) and high diversity of other traits was selected and formed an IACC.This collection showed a high level of diversity and a wide range of representativeness,based on analysis of eco-regions,agronomic traits,and molecular background.Soybean accessions in this IACC can serve as a supplement to the MCC and promote the effective use of crossing parents in soybean breeding.
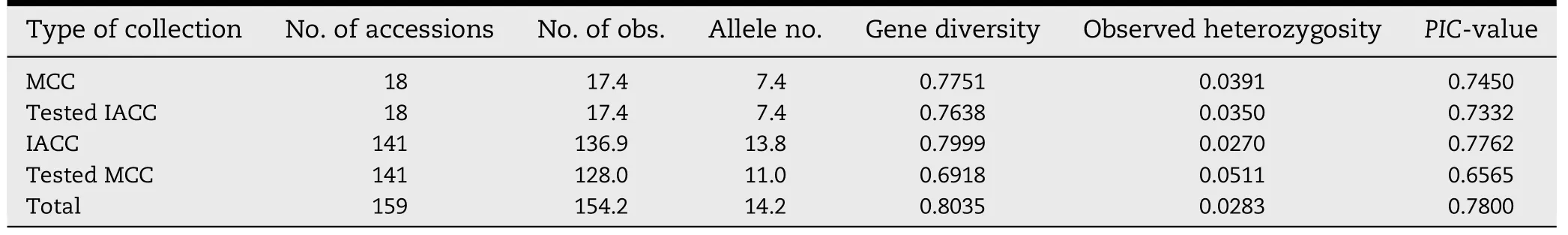
Table 8-Genetic diversity comparison of soybean accessions between IACC and MCC by random sampling.
4.3.Balance of sample size with desirable traits and genetic diversity in the IACC of soybean
Soybean accessions with specific traits in CCs have been developed in a previous study for utilization of soybean germplasm resources with desirable traits.A core SCN resistance collection containing 28 representative accessions was developed by evaluation and diversity analysis of 432 soybean accessions with immunity or high resistance to SCN [35].This core collection encompassed 70.8% of the allelic variation present in the overall resistance collection.However,the sample size would increase dramatically if each of the individual specific traits were assigned to a specific core collection.But such a step would be inconvenient for researchers and would contravene the principle of core collection.For this reason,the soybean IACC developed in this study was assembled from accessions with different desirable agronomic and nutritional traits.These accessions showed a high level of diversity with respect to target traits,non-target traits,and molecular markers.Comparative analysis revealed that the diversity of phenotype and genetic background did not differ significantly between this newly formed IACC and the established MCC.However,the number of accessions with specific desirable traits is substantially greater in the IACC.Thus the concept of the IACC resolves the conflict between reducing sample size and concentrating genetic diversity.Furthermore,the strategy of integrating various desirable traits in the IACC of soybean is consistent with the goal of soybean breeding.Some accessions with more than one specific trait can be used directly for breeding elite varieties.However,our study also showed that the diversity of small numbers of accessions with specific desirable traits (such as cold tolerance) differed from that of MCC.The number of such accessions should be increased in future studies.
This work was supported by the State Key Basic Research and Development Plan of China (973) (2010CB125900,2009CB118400),the Fundamental Research Funds for Excellent Young Scientists of ICS-CAAS (Grant to Y.G.),the State High-tech Research and Development Program (863 Program) (No.2012AA101106),and the Crop Germplasm Conservation Program (NB2010-2130135-25-05).The authors thank Dr.Chengguo Yao at the University of California,Irvine,USA for critical reading of the manuscript and the reviewers for constructive comments on earlier versions of this manuscript.
[1] A.H.D.Brown,Core collections: a practical approach to genetic resources management,Genome 31(1989) 818–824.
[2] O.H.Frankel,A.H.D.Brown,Current plant genetic resources–a critical appraisal,Genetics: New Frontiers,vol.IV,Oxford and IBH Publishing,1984.
[3] Z.C.Li,H.L.Zhang,Y.S.Cao,Z.E.Qiu,X.H.Wei,S.X.Tang,P.Yu,X.K.Wang,Studies on the sampling strategy for primary core collection of Chinese ingenious rice,Acta Agron.Sin.29(2003) 20–24(in Chinese with English abstract).
[4] Y.C.Dong,Y.S.Cao,X.Y.Zhang,S.C.Liu,L.F.Wang,G.X.You,B.S.Pang,L.H.Li,J.Z.Jia,Establishment of candidate core collections in Chinese common wheat germplasm,J.Plant Genet.Resour.4 (2003) 1–8(in Chinese with English abstract).
[5] L.X.Wang,Y.Guan,R.X.Guan,Y.H.Li,Y.S.Ma,Z.M.Dong,X.Liu,H.Y.Zhang,Y.Q.Zhang,Z.X.Liu,R.Z.Chang,H.M.Xu,L.H.Li,F.Y.Lin,W.J.Luan,N.Z.Ya,X.C.Ning,L.Zhu,Y.H.Cui,R.H.Piao,Y.Liu,P.Y.Chen,L.J.Qiu,Establishment of Chinese soybean (Glycine max) core collections with agronomic traits and SSR markers,Euphytica 151 (2006) 215–223.
[6] H.Xu,Y.Mei,J.Hu,J.Zhu,P.Gong,Sampling a core collection of island cotton (Gossypiumbar badense L.) based on the genotypic values of fiber traits,Genet.Resour.Crop.Evol.53(2006) 515–521.
[7] C.C.Holbrook,W.F.Anderson,Evaluation of a core collection to identify resistance to late leafspot in peanut,Crop Sci.35(1995) 1700–1702.
[8] H.D.Upadhyaya,R.Oritz,A mini core subset for capturing diversity and promoting utilization of chickpea genetic resources in crop improvement,Theor.Appl.Genet.102(2001) 1292–1298.
[9] R.Wang,Y.Yu,J.Zhao,Y.Shi,Y.Song,T.Wang,Y.Li,Population structure and linkage disequilibrium of a mini core set of maize inbred lines in China,Theor.Appl.Genet.117 (2008) 1141–1153.
[10] K.Ebana,Y.Kojima,S.Fukuoka,T.Nagamine,M.Kawase,Development of mini core collection of Japanese rice landrace,Breed.Sci.58(2008) 281–291.
[11] K.R.Kottapalli,M.D.Burow,G.Burow,J.Burke,N.Puppala,Molecular characterization of the US peanut mini core collection using microsatellite markers,Crop Sci.47(2007)1718–1727.
[12] L.J.Qiu,Y.H.Li,R.X.Guan,Z.X.Liu,L.X.Wang,R.Z.Chang,Establishment representative testing and research progress of soybean core collection and mini core collection,Acta Agron.Sin.35 (2009) 571–579(in Chinese with English abstract).
[13] J.Kashiwagi,L.Krishnamurthy,H.D.Upadhyaya,H.Krishna,S.Chandra,V.Vadez,R.Serraj,Genetic variability of drought-avoidance root traits in the mini core germplasm collection of chickpea(Cicer arietinum L.),Euphytica 146(2005)213–222.
[14] H.D.Upadhyaya,K.N.Reddy,C.L.L.Gowda,S.Singh,Phenotypic diversity in the pigeonpea (Cajanus cajan) core collection,Genet.Resour.Crop.Evol.54(2007) 1167–1184.
[15] M.Sharma,A.Rathore,U.N.Mangala,R.Ghosh,S.Sharma,H.D.Upadhyay,S.Pande,New sources of resistance to Fusarium wilt and sterility mosaic disease in a mini core collection of pigeonpea germplasm,Eur.J.Plant Pathol.133(2012) 707–714.
[16] Y.Chu,L.Ramos,C.C.Holbrook,P.Ozias-Akins,Frequency of a loss-of-function mutation in oleoyl-PC desaturase(ahFAD2A)in the mini core of the US peanut germplasm collection,Crop Sci.47 (2007) 2372–2378.
[17] X.Y.Zhang,B.S.Pang,G.X.You,L.F.Wang,J.Z.Jia,Y.C.Dong,Allelic variation and genetic diversity at Glu-1 loci in Chinese wheat (Triticum aestivum L.) germplasms,Sci.Agric.Sin.35(2002) 1302–1310(in Chinese with English abstract).
[18] H.S.Bhandari,C.A.Pierce,L.W.Murray,I.M.Ray,Combining abilities and heterosis for forage yield among high-yielding accessions of the alfalfa core collection,Crop Sci.47(2007)665–673.
[19] L.J.Qiu,Y.S.Cao,R.Z.Chang,X.A.Zhou,G.X.Wang,J.Y.Sun,H.Xie,B.Zhang,X.H.Li,Z.Y.Xu,L.H.Liu,Establishment of Chinese soybean (Glycine max) core collection: I.Sampling strategy,Sci.Agric.Sin.36(2003) 1442–1449(in Chinese with English abstract).
[20] X.E.Song,Y.H.Li,R.Z.Chang,P.Y.Guo,L.J.Qiu,Population structure and genetic diversity of mini core collection of cultivated soybean(Glycine max(L.)Merr.)in China,Sci.Agric.Sin.43(2010) 2209–2219(in Chinese with English abstract).
[21] J.W.Zhang,F.X.Han,J.M.Sun,F.K.Yu,L.Ma,M.Y.Chen,J.Y.Zhang,S.R.Yan,H.Yang,Genetic variation of the protein content and correlation between protein content and main agronomic characters of soybean,J.Plant Genet.Resour.12(2011) 501–506(in Chinese with English abstract).
[22] L.Yan,Y.Feng,C.Y.Yang,X.L.Shi,M.C.Zhang,The character of protein and oil content distribution among introgression lines using Jidou 12 as recipient parent,Acta Agric.Boreali Sin.27(2012) 87–92(in Chinese with English abstract).
[23] H.W.Jiang,C.D.Li,C.Y.Liu,W.B.Zhang,P.C.Qiu,W.F.Li,Y.L.Gao,G.H.Hu,Q.S.Chen,Genotype analysis and QTL mapping for tolerance to low temperature in germination by introgression lines in soybean,Acta Agron.Sin.35(2009)1268–1273(in Chinese with English abstract).
[24] P.C.Qiu,W.B.Zhang,C.D.Li,H.W.Jiang,C.Y.Liu,D.M.Fan,Q.L.Zeng,G.H.Hu,Q.S.Chen,Genetic overlap of drought-tolerance loci between germination stage and seedling stage analyzed using introgression lines in soybean,Acta Agron.Sin.37(2011) 477–483(in Chinese with English abstract).
[25] Z.Tian,X.Wang,R.Lee,Y.Li,J.E.Specht,R.L.Nelson,P.E.McClean,L.Qiu,J.Ma,Artificial selection for determinate growth habit in soybean,Proc.Natl.Acad.Sci.U.S.A.107(2010) 8563–8568.
[26] Y.Guo,L.J.Qiu,Allele-specific marker development and selection efficiencies for both flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase genes in soybean subgenus soja,Theor.Appl.Genet.126 (2013) 1445–1455.
[27] C.P.Yuan,Y.H.Li,Z.X.Liu,R.X.Guan,R.Z.Chang,L.J.Qiu,DNA sequence polymorphism of the Rhg4 candidate gene conferring resistance to soybean cyst nematode in Chinese domesticated and wild soybeans,Mol.Breed.30(2012)1155–1162.
[28] G.Y.Liu,R.X.Guan,R.Z.Chang,L.J.Qiu,Correlation between Na+contents in different organs of soybean and salt tolerance at the seedling stage,Acta Agron.Sin.37(2011)1266–1273(in Chinese with English abstract).
[29] Y.H.Li,M.J.M.Smulders,R.Z.Chang,L.J.Qiu,Genetic diversity and association mapping in a collection of selected Chinese soybean accessions based on SSR marker analysis,Conserv.Genet.12(2011) 1145–1157.
[30] H.Xie,R.Z.Chang,Y.S.Cao,M.H.Zhang,Z.F.Feng,L.J.Qiu,Selection of core SSR loci by using Chinese autumn soybean,Sci.Agric.Sin.36(2003)360–366(in Chinese with English abstract).
[31] K.Liu,S.V.Muse,PowerMarker: an integrated analysis environment for genetic marker analysis,Bioinformatics 21(2005) 2128–2129.
[32] J.Y.Gai,T.J.Zhao,Z.L.Cui,J.X.Qiu,Genetic base of 651 soybean cultivars in China,J.Agric.Sci.Technol.1(1999)26–30(in Chinese with English abstract).
[33] J.Y.Gai,T.J.Zhao,The core ancestors of soybean cultivars in China,J.Nanjing Agric.Univ.24(2) (2001) 20–23(in Chinese with English abstract).
[34] Y.H.Cui,L.J.Qiu,R.Z.Chang,W.H.Lv,A study of genetic diversity of Huanghuai summer sowing soybean in China,Sci.Agric.Sin.37(2004)15–22(in Chinese with English abstract).
[35] Y.S.Ma,W.H.Wang,L.X.Wang,F.M.Ma,P.W.Wang,R.Z.Chang,L.J.Qiu,Genetic diversity of soybean and the establishment of a core collection focused on resistance to soybean cyst nematode,J.Intergr.Plant Biol.48(2006)722–731(in Chinese with English abstract).
- The Crop Journal的其它文章
- Effect of nitrogen fertilizer on distribution of starch granules in different regions of wheat endosperm
- Induced defense responses in rice plants against small brown planthopper infestation
- Overexpression of GmDREB1 improves salt tolerance in transgenic wheat and leaf protein response to high salinity
- Impacts of nighttime post-anthesis warming on rice productivity and grain quality in East China
- Differential microRNA expression between shoots and rhizomes in Oryza longistaminata using high-throughput RNA sequencing
- Genome-wide analysis of the WRKY transcription factor gene family in Gossypium raimondii and the expression of orthologs in cultivated tetraploid cotton

