Copper interactions with DNA of chromatin and its role in neurodegenerative disorders
M. Govinrju, H.S. Shkr, S.B. Stsh, P. Vsuv Rju,K.R. Smsiv Ro, K.S.J. Ro, A.J. Rjmm
aMolecular Biophysics Unit, Indian Institute of Science, Bangalore, India
bDepartment of Pharmacy Practice, KIMS Hospital and Research Center, VIPS, Bangalore, India
cDepartment of Pharmaceutics, Acharya & B.M. Reddy College of Pharmacy, Bangalore 560107, India
dDepartment of Neuroscience, Medical University of South Carolina, Charlton, USA
eDepartment of Biotechnology, Acharya Nagarjuna University, Nagarjuna Nagar, Guntur, India
fCentre for Neuroscience, Institute for Scientific Research and Technological services, INDICASAT-AIP, City of Knowledge,Republic Panama, Germany
gDepartment of Pharmacognosy, KLEU's College of Pharmacy, Bangalore 560010, India
1. Introduction
Chromatin is the combination of DNA and proteins that make up the contents of the nucleus of eukaryotic cells. The primary functions of chromatin are:to package DNA into a smaller volume to fit in the cell, to strengthen DNA to allow mitosis and meiosis and prevent DNA damage.It also helps to control gene expression and DNA replication [1]. An alteration in the chromatin organization may lead to the neuronal cell death and loss of regulation of DNA methylation, leading to altered gene expression as observed in many neurodegenerative disorders [2].
The chromatin structure is susceptible to change under conditions of ionic strength,pH,temperature and interaction of divalent metal ions with DNA [3]. The interaction of copper ions with DNA has been of particular interest because of the involvement of copper ions in regular activities such as cellular respiration and neurotransmitter biosynthesis etc.
Copper works as cofactor for numerous enzymes and plays an important role in the development of central nervous system.However, excessive levels or perturbation of copper metabolism can lead to accumulation of copper preferentially in heterochromatin regions and cause intracellular toxicity [4,5]. The redox properties of copper can cause oxidative damage to DNA. Copper induced DNA damage may probably lead to neuronal dysfunction, critical failure of biological functions and ultimately cell death contribute to neurological disease [6]. Copper functions as a “double whammy”in the brain by generating large number of DNA attacking reactive oxygen species (ROS) via a Fenton reaction, which causes catastrophic damage to lipids, proteins and DNA [7].
Copper can also directly bind to protein and DNA leading to structural and functional modifications and is involved in chromatin condensation. For this reason, copper is one of the strongly suspected etiological factors in neurodegenerative disorders such as, Alzheimer's disease, Parkinson's disease, Huntington disease and familial amyotrophic lateral sclerosis [8].
Here we have studied the properties of chromatin with respect to conformational changes and damage as a consequence of interaction with copper.Structural and conformational aspects of the interaction of copper with chromatin are studied by a variety of spectroscopic techniques including UV-visible spectrophotometry, optical melting studies, circular dichroism (CD), circular dichroism melting studies and fluorescence spectroscopy. Temperature studies provide the basis to understand the factors that dictate the stability and structure of chromatin in the presence of copper.
2. Materials and methods
2.1. Reagents
Copper chloride dihydrate, CuCl2·2H20 (Merck Schuchard), ethidium bromide (EB), C21H20BrN3, (Amersham Life Sciences) and Tris buffer (Sigma), were purchased and used without further purification.
The stock solutions of copper chloride (50 mM) and EB (5 mg/mL)were prepared using Milli-Q water and stored at 4°C in the dark until use.The stock solution of Tris buffer was prepared by dissolving 2.4 g of Tris in 200 mL of Milli-Q water. The pH of the buffer was set by the addition of dilute HCl and measured using digital pH meter with a combined glass electrode (EUTECH Instruments).
2.2. Isolation of the nuclei from the brain samples
Nuclei were isolated from the cortex region of the human brain(Brain bank, JSS Medical College, Mysore) according to the method described by Usha Rani et al., 1986 [9]. In brief, brain tissue was weighed (5 g) and perfused with normal saline to remove any blood in the tissue. Brain tissue was cut into small pieces and minced thoroughly. The minced brain tissue was homogenized in 0.34 M sucrose in buffer-A (50 mM Tris—HCl pH 7.5, 25 mM KCl, 5 mM MgCl2, and 0.5 mM PMSF) using homogenizer. The homogenate was filtered through 2 layers of cheese cloth and the filtrate was centrifuged at 1000 g (3500 rpm)for 10 min at 4°C.The supernatant was decanted carefully and the pellet was resuspended in 1 M sucrose in buffer-A.The suspended pellet was homogenized using hand held homogenizer and centrifuged at 100,000 g (42,000 rpm) in ultracentrifuge for 1 h.The pellet obtained was washed with 1 M sucrose in buffer-A,0.34 M sucrose and 0.34 M sucrose with 0.1% Triton X-100,respectively.The pellet obtained was dissolved in Tris—HCl buffer and the concentration of the nuclear suspension was determined by taking absorbance at A260in 1 mL of 2 M NaCl or 5 M urea.
2.3. Preparation of nuclei and soluble chromatin
Soluble chromatin was prepared from the isolated nuclei by limited digestion of nuclei with micrococcal nuclease [10].Nuclear suspension (100 μL) was mixed with 0.1 M CaCl2and incubated at 37°C for 2 min. After incubation period, the nuclear suspension was digested with micrococcal nuclease (50 units) by incubating at 37°C for 1 min.The reaction was stopped by 0.25 M EDTA and centrifuged at 5000 rpm for 5 min.The pellet obtained was suspended in 10 mM NaHSO3and 1 mM EDTA (pH 7.5).The soluble chromatin was used for the copper interaction studies.
2.4. Copper-chromatin binding studies
2.4.1. Spectrophotometric method
Spectrophotometric binding studies of copper with chromatin were performed to understand the nature of the conformational changes of chromatin on binding with copper.The electronic absorption studies were investigated at pH 7.4 using a Jasco V-530 Spectrophotometer equipped with a Peltier temperature controller. Chromatin sample was prepared in Tris—HCl buffer (5 mM, pH 7.4, 400 μL) in the presence and absence of CuCl2(50 μM and 100 μM). Absorbance spectrum was measured at wavelength between 210 nm and 320 nm with a matched set of 1 cm path length quartz cuvettes. Buffer baseline was subtracted with the Jasco software and the resultant spectrum was recorded.
2.4.2. Circular dichroism studies
Circular dichroism (CD) spectroscopy is one of the most sensitive techniques available for monitoring conformational properties of DNA in solution. Copper induced conformational change of chromatin was measured on a Jasco J-715 Spectro polarimeter at 25°C. Spectra were recorded using a path length of 1 mm quartz cuvette at 1 nm intervals in the wavelength between 200 and 320 nm. An average of four repetitive scans using a scan speed of 20 nm/min was taken into consideration. Chromatin sample was prepared in Tris—HCl buffer(5 mM,300 μL,pH 7.4)in the absence and presence of different concentrations of CuCl2(50, 100 and 500 μM). Buffer background was subtracted by using the built-in feature of Jasco software and the resultant spectrum was recorded.
2.5. Thermal denaturation studies
2.5.1. Spectrophotometric method
UV-thermal denaturation of chromatin in the presence and absence of CuCl2was measured with Jasco V-530 spectrophotometer equipped with Jasco ETC-505T temperature controller and cell holder that permits temperature control using the temperature control program.
2.5.2. Circular dichroism melting studies
Melting studies of chromatin in the presence and absence of CuCl2were performed on a Jasco J-715 spectropolarimeter connected to model PTC-348WI, peltier type temperature control system.Samples were recorded at wavelengths between 200 nm and 320 nm by varying the temperature from 20oC to 100oC.
2.6. Fluorescence studies of DNA structure and dynamics
EB replacement experiment was carried out to verify the local structural information due to the interaction of copper with chromatin. Fluorescence emission studies were carried out using equimolar concentrations (1:1) of chromatin and EB. The EB binding pattern of chromatin and the effect of different concentrations of CuCl2(100—500 μM) on the EB fluorescence were analyzed. Samples of DNA/EB solutions were excited at 530 nm and emission spectra were recorded from 550 nm to 650 nm using Jasco J-600 spectrofluorimeter.
3. Results
3.1. Copper-chromatin binding studies
Spectrophotometric binding studies help in understanding ability of chromatin to bind to copper and the nature of conformational changes that occur to chromatin upon binding with copper. The absorbance spectra of chromatin show the absorption maximum at 265 nm as shown in Fig.1A.Upon addition of CuCl2(50 μM and 100 μM),there was an increase in the absorbance with blue shift to 260 nm.On further addition of copper, no change in the intensity was noticed indicating saturation.
Circular dichroism (CD) is a useful technique for studying conformational changes and the degree of asymmetry of bases of DNA in chromatin [11]. Structural transition of chromatin in the absence of copper, exhibits a characteristic positive peak at 275 nm(Fig.2A). CD spectra of chromatin between 250 and 300 nm are dominated by DNA and proteins contribute very little to CD spectra of this region[12—14].Upon addition of copper to chromatin,a decrease in the magnitude of both the positive and the negative bands was noticed.This is due to complex formation between Cu2+and the bases of DNA. This was accompanied by a conformational change with a cross over point at 245 nm as illustrated in Fig.2A.
3.2. Thermal denaturation studies of chromatin
The thermal behavior of chromatin was monitored using UV/vis absorbance spectroscopy. The melting temperature curves of DNA are enhanced as the temperature increases and stabilized the DNA in chromatin as shown in Fig.1B and C. Identical melting curves are produced free chromatin and chromatin with 50 μM copper.Melting profile of chromatin shown in Fig.1D reveals two transitions, the first transition at 58.8°C and the second at 70.27°C.Upon addition of 100 μM copper, Tm shifts to 60°C and 72.5°C.
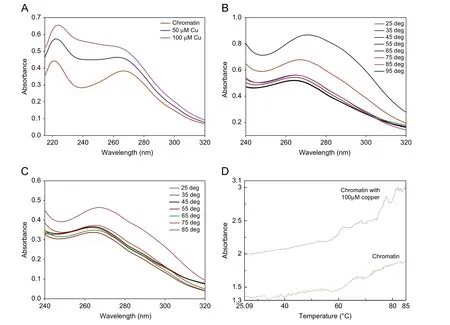
Fig.1 (A)Absorbance spectra of chromatin with varied concentrations of copper.(B)Absorbance spectra of free chromatin at varied temperature.(C)Absorbance spectra of chromatin with 50 μM copper at varied temperature. (D) Melting profile of free chromatin and chromatin with 100 μM copper.

Fig.2 (A) Circular dichroism studies of chromatin with varied concentrations of copper. (B) Circular dichroism studies of chromatin at varied temperature. (C) Circular dichroism studies of chromatin with 50 μM copper at varied temperature. (D) Circular dichroism studies of chromatin with 100 μM copper at varied temperature. (E) Melting profile of free chromatin and chromatin with 500 μM copper at 275 nm wavelength.
Thermal denaturation studies of chromatin in the absence of copper at variable temperature from 20°C to 105°C are shown in Fig.2B. Effect of 50 μM copper is shown in Fig.2C, and 100 μM copper is shown in Fig.2D, the melting profile of chromatin is also shown in Fig.2E. There is an increase in intensity of both positive and negative bands at the isodichroic point at 227 nm for chromatin and at 229 nm in the presence of copper.These results indicated that the CD transition happens in two states and the structural change was cooperative. It also confirms identical melting profiles of native chromatin and chromatin with 50 μM and 100 μM copper [15].The CD melting profile in the presence and absence of copper at 275 nm shown in Fig.2E is evidence for the above results.
3.3. Fluorescence studies of DNA structure and dynamics
The fluorescence spectra of chromatin—EB complex excited at 530 nm and emission spectra were scanned from 550 to 650 nm as shown in Fig.3A. Upon addition of copper to chromatin, the fluorescence intensity of chromatin—EB complex is decreased with increasing concentration of copper.
4. Discussion
4.1. Copper-chromatin binding studies
The absorbance spectra of chromatin in this wavelength region(Far UV)of the absorption spectra are sensitive to π—π*transitions of the electrons of the purine and pyrimidine rings. This is due to the increased positive base pair tilting of conformational change in DNA [16]. The hyperchroism and blue shift (hypsochromic shift)is due to binding of copper to the bases of DNA in chromatin.Hydrogen bonds are disrupted by the process of partial unwinding which induces structural changes such as loosening of base—base interaction, base tilting and destabilization of the DNA double helix leading to DNA denaturation [17]. The destabilization was due to most probable binding sites in DNA such as Cu2+to N7of guanine and N3of cytosine in line with our present results and with the predictions of Eichhorn and Clark [18,19].
CD spectra of chromatin showed a characteristic positive peak at 275 nm (Fig.2A) due to base stacking and a negative peak at 210 nm due to protein content. CD spectra of chromatin between 250 and 300 nm are dominated by DNA, chromatin as proteins contribute very little to CD spectra of this region. Upon gradual addition of copper to chromatin, a decrease in the magnitude of both the positive and negative bands was noticed. These data indicate that cationic copper binds to the anionic phosphate ions of the DNA back bone,consequently,the lengthening of DNA which induced the loss of conformation. Copper first binds to anionic phosphate [20] groups of the backbone and subsequently to the bases, specifically to guanine [21] and cytosine [22] of DNA by competing with hydrogen bands. This disrupts the interactions between the bases of DNA and thereby weakening base stacking
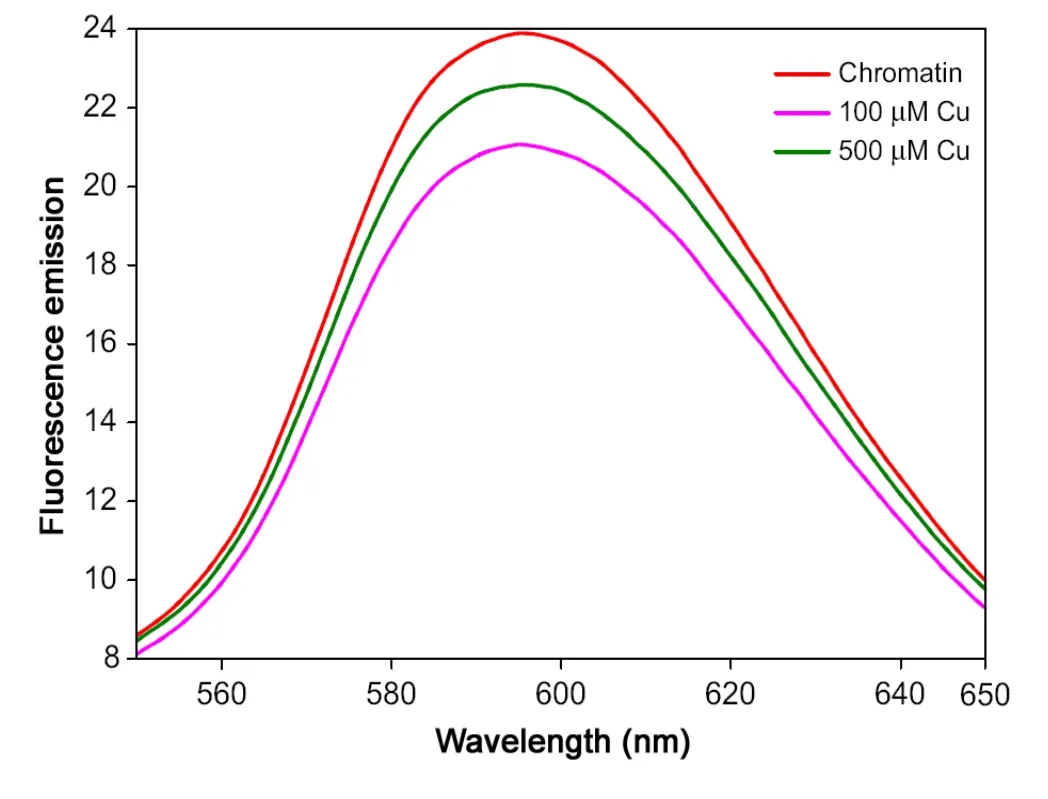
Fig.3 Fluorescence emission spectra of free chromatin and chromatin with copper.
[23] by tilting the bases leads to change in the winding angle.As the winding angle increases, the magnitude of the band decreases with a greater degree of twisting of the chain.Hydrogen bonding forces play an essential role in the binding [24,25] which may affect sugar puckering and change in the conformation of guanine from anti to syn [19]. The negative super helical tension and rearrangement in coordination caused by copper may drive local transitions to alternate conformational change in the DNA structures in chromatin.Binding of Cu2+with adenine is similar in interaction with the N7 position and the phosphate site would represent a third copper-base binding site.Copper does not bind to thymine but both the bases of GC pair are known to involve in the copper complexes. DNA conformation is an important aspect for the gene expression. This study provides evidence of copper induced DNA damage in the chromatin organization and neuronal cell death, which is implicated in many neurological disorders.
4.2. Thermal denaturation studies
This study provides information about binding affinity of copper with DNA in chromatin and subsequent conformational changes to DNA.It is known that double stranded DNA gradually dissociates to single strands with increasing temperature [26]. Tm is strictly related to the stability of the double helix and the interaction of copper with DNA.
The thermal denaturation experiments by both UV and CD are represented by two transitions. The small variation in Tm is evidence that copper binds to anionic phosphate backbone and to the bases depending upon the accessibility of DNA in chromatin.The results of CD melting studies show that protein bound to DNA in chromatin stabilizes DNA. Higher stabilization is due to the compact binding of histones. Both CD and UV melting results indicate that there is not much effect of temperature on DNA in chromatin. This may be because the DNA bases in chromatin are protected by associating with the chromatin proteins, so that the bases are accessible for copper [27]. It is concluded that the basic conformation of DNA in native chromatin is determined largely by histones and nonhistone proteins. It is also seen that DNA is greatly stabilized against thermal melting in the DNA—histone complex. The small increase in Tm indicates that copper interacts with DNA in chromatin and changes the conformation of chromatin structure. It is also evidence that DNA is greatly stabilized against thermal melting in the DNA—histone complex.
4.3. Fluorescence studies of DNA structure and dynamics
Fluorescence spectroscopy is an important technique for probing the structure and dynamics of nucleic acids. The utility of fluorescence techniques stems the ability of fluorophores to reflect changes in their molecular environment through measurable alterations in emission properties. The decrease in chromatin—EB emission with the addition of copper indicates that binding of Cu2+ions with chromatin —EB complex forms a new nonfluorescent complex of Cu—chromatin—EB, which causes the fluorescence quenching of chromatin—EB complex [28]. These data show that Cu2+ions bind to DNA in chromatin, resulting in chemical DNA denaturation and the binding of copper is mainly concentration dependent.
5. Conclusion
Our study showed that copper binds to bases of DNA in chromatin by changing the winding angle of the helix. This induces the DNA damage and alters the B-conformation. Altered B-DNA conformation will alter the integrity of DNA which may affect the normal process of DNA replication and transcription.Copper induced DNA damage in the brain may cause neurotoxicity and the neuronal cell death and is implicated in Alzheimer's disease and other neurological disorders.Hence copper is expected to become one of the key factors for causing neurodegeneration. It is also concluded that histone and nonhistone proteins present in chromatin protect DNA from oxidative DNA damage and slowdown the age related diseases.
We thank the Chairman, Prof. S.P. Siddhartha, Molecular Biophysics Unit,Indian Institute of Science,Bangalore,for helpful discussions and valuable comments of this research and suggestions.The authors of this publication wish to thank Government of India for financial support to Molecular Biophysics Unit, Indian Institute of Science, Bangalore.
[1] R.D. Kornberg, Structure of chromatin, Annu. Rev. Biochem. 46(1977) 931—954.
[2] R.D. Kornberg, Chromatin structure: a repeating unit of histones and DNA, Science 184 (1974) 868—871.
[3] J.L.Sagripanti,P.L.Goering,A.Lamanna,Interaction of copper with DNA and antagonism by other metals,Toxicol.Appl.Pharmacol.110(1991) 477—485.
[4] S. Scarpa, A.D. Fuso, F. Anselmi, et al., Presenilin 1 gene silencing by S-adenosylmethionine: a treatment for Alzheimer disease, FEBS Lett. 541 (2003) 145—148.
[5] B. Halliwell, J.M.C. Gutteridge, Oxygen toxicity, oxygen radicals,transition metals and disease, Biochem. J. 219 (1984) 1—14.
[6] D. Strausak, J.F. Mercer, H.H. Dieter, et al., Multhaup, Copper in disorders with neurological symptoms: Alzheimer's, Menkes, and Wilson diseases, Brain Res. Bull. 55 (2001) 175—185.
[7] D.E.Hartter,A.Barnea,Brain tissue accumulates copper by two liganddependent saturable processes, J. Biol. Chem. 263 (1988) 799—805.
[8] T.A. Rouault, Systemic iron metabolism: a review and implications for brain iron metabolism, Pediatr. Neurol. 2 (2001) 130—137.
[9] B. Usha Rani, K.S. Rao, DNA and DNase in isolated neuronal,astrocyte and oligodendrocyte cell enriched fraction from young and old chick brain, Indian J. Biochem. Biophys. 23 (1986) 279—282.
[10] J.R.Korenberg,S.M.Pulst,R.L.Neve,et al.,The Alzheimer amyloid precursor protein maps to human chromosome 21 bands q21.105-q21.05, Genomics 5 (1) (1989) 124—127.
[11] P. Vasudevaraju, T. Bharathi, Jyothsna et al. New evidence on iron,copper accumulation and zinc depletion and its correlation with DNA integrity in aging human brain regions, Indian J. Psychiatry 52 (2)(2010) 140—144.
[12] A.J. Adler, G.D. Fasman, L.J. Wangh, et al., Altered conformational effects of naturally acetylated Histone f2a1(IV) in f2al-DNA complexes, J. Biol. Chem. 249 (1974) 2911—2914.
[13] I. Sissoeff, J. Grisvard, E. Guille, Studies on metal ions-DNA interactions: specific behaviour of reiterative DNA sequences, Prog.Biophys. Mol. Biol. 31 (1976) 165—199.
[14] F.E.Rosetto,E.Nieboer,The Interaction of metal ions with synthetic DNA:Induction of conformational and structural transitions,J.Inorg.Biochem. 54 (1994) 167—186.
[15] J. Dugoid, V.A Bloomfield, J. Benevides, Raman spectroscopy of DNA—metal complexes.I.Interactions and conformational effects of the divalent cations: Mg,Ca,Sr,Mn,Co,Ni,Cu,Pd and Cd, Biophys. J.65 (1993) 1916—1928.
[16] J.L Mergny, Li Jing, Lacroix Laurent, et al., Thermal difference spectra:a specific signature for nucleic acid structures,Nucleic. acids Res. 33 (16) (2005) e138.
[17] C.Zimmer,G.Luck,H.Fritzsche,DNA—copper(II)complex and the DNA conformation, Biopolymers 10 (1971) 441—463.
[18] G.L. Eichhorn, P. Clark, Interactions of metal ions with polynucleotides and related compounds. v. the unwinding and rewinding of DNA strands under the influence of copper(II)ions,Proc.Natl.Acad.Sci. USA 53 (1965) 586—593.
[19] P. Clark, G.L. Eichhorn, A Simple probe for DNA accessibility in chromatin, J. Inorg. Biochem. 19 (1995) 765—772.
[20] H. Fritzsche, C. Zimmer, IR studies of DNAs, their constituents and analogues.4.The binding sites of Cu (II) in DNA, Eur. J. Biochem. 5(1968) 42—44.
[21] M.N.Dehkord,A.K.Bordbar,P.Lincoln,Spectroscopic study on the interaction of ct-DNA with manganese salen complex containing triphenyl phosphonium groups, Spectrochim. Acta A Mol. Biomol.Spectrosc. 90 (2012) 50—54.
[22] W. Forster, E. Bauer, H. Schut, Thermodynamics and kinetics of the interaction of copper (II)ions with Native DNA, Biopolymers 18(1979) 625—661.
[23] Y. Courtois, P. Fromageot, W. Guschlbaue, Protonated Polynucleotide Structures. 3. An Optical Rotatory Dispersion Study of the Protonation of DNA, Eur. J. Biochem. 56 (4) (1968) 493—501.
[24] G. Goutam, B. Manju, V. Sashishekaran, Conformational flexibility of DNA: Polymorphism and handedness,Proc. Natl.Acad.Sci.USA 77 (11) (1980) 6486—6490.
[25] G.L. Eichhorn, P. Clark, Interactions of metal ions with polynucleotides and related compounds v. The unwinding and rewinding of DNA strands under the influence of copper(II)ions,Proc.Nat.Acad.Sci. USA 53 (1965) 586—593.
[26] R. Mandel, G.D. Fasman, Thermal denaturation of DNA and polypeptide complexes. Simultaneous absorption and circular dichroism measurements,Biochem.Biophys.Res.Commun.59(20)(1974)672—679.
[27] C. Zimmer, G. Luck, H. Triebel, Conformation and reactivity of DNA, 4. Base binding ability of transition metal ions to native DNA and the effect of helix conformation with special reference to the DNA- Zn (II) complex, Biopolymers 13 (3) (1974) 425—454.
[28] John Olmsted 111, R.K. David, Mechanism of ethidium bromide fluorescence enhancement on binding to nucleic acids, Biochemistry 16 (16) (1977) 3647—3654.
 Journal of Pharmaceutical Analysis2013年5期
Journal of Pharmaceutical Analysis2013年5期
- Journal of Pharmaceutical Analysis的其它文章
- A novel and rapid microbiological assay for ciprofloxacin hydrochloride
- Determination of sodium hyaluronate in pharmaceutical formulations by HPLC-UV
- A critical quality parameter in quantitative fused-core chromatography: The injection volume
- Application of LC-MS/MS for quantitative analysis of glucocorticoids and stimulants in biological fluids
- In situ modified screen printed and carbon paste ion selective electrodes for potentiometric determination of naphazoline hydrochloride in its formulation
- Kinetic performance comparison of fully and superficially porous particles with sizes ranging between 2.7 μm and 5 μm: Intrinsic evaluation and application to a pharmaceutical test compound
