Genome-wide Analysis of Ovate Family Proteins in Arabidopsis
Huang Jian-ping,Li Hong-ling,and Chang Ying
College of Life Sciences,Northeast Agricultural University,Harbin 150030,China
Introduction
The ovate gene,a quantitative genetic factor,is first found in tomato (Lycopersicurn esculenturn).Based on classical genetic studies,it is proposed that a single Mendelian recessive gene called pr is responsible for pear-shaped tomato fruit (Hendrick and Booth,1907;Price and Drinkard,1908;Lindstrom,1925;Macarthur 1926;Ku et al.,1999).pr is renamed o as it is partially recessive to one or more allelomorphs in round and oblate tomato sorts (Macarthur,1926;1928;Lindstrom,1926;1927).Recent molecular-marker analysis of a cross between yellow pears,a tomato variety bearing small,pear-shaped fruit,and the roundfruitd,wild species,Lycopersicon pimpinellifolium LA1589,revealed that pear-shaped fruit is determined largely by the major QTL ovate on chromosome 2,and molecular marker TG645 is found to near the ovate locus (Ku et al.,1999).Quantitative trait mapping of an F2population between Long John,a tomato variety displaying an extremely elongated fruit phenotype,and LA1589 revealed four fruit shape QTLs,located on chromosomes 2,3,7 and 11 (ljfs2,ljfs3,ljfs7 and ljfs11),respectively.Furthermore,QTL map position and the effect of the loci on fruit shape suggested that ljfs2 was allelic to ovate (Knaap et al.,2002).By using the molecular marker TG645 on tomato chromosome 2,Liu et al.(2002) cloned the ovate locus from tomato (TA493,L.esculentum cv.Heinz 1706),and further genetic analyses indicated that ovate was expressed in early stages of flower and fruit development and encoded a hydrophilic protein with a putative bipartite nuclear localization signal,Von Willebrand factor type C domains,and an≈70-aa C-terminal domain conserved in tomato,Arabidopsis,and rice.So this domain is also called as OVATE domain.A GTA496-to-TTA493nucleotide polymorphism in the second exon of ovate,leading to a premature stop codon,causes the transition of tomato fruit from round-to pearshaped,and ectopic,transgenic expression of OVATE unevenly reduces the size of floral organs and leaflets (Liu et al.,2002),which is similar to the phenotype of the Arabidopsis OVATE over-expression lines (Wang et al.,2007).Besides,sun (another major loci controlling fruit shape index) and ovate contributed to almost all aspects of tomato fruit shape such as the distal and proximal end features (Gonzalo et al.,2008).However,the ovate allele is not present in all kinds of elongated fruit,for example,Howard German and Banana Legs,two tomato varieties that exhibit extreme fruit shape characteristics,do not contain the ovate gene in the lower half of chromosome 2 of the genomes (Brewer et al.,2007).
Recently,by using a large-scale yeast two-hybrid technology to systematically analyze the TALE protein interactions,Hackbusch et al.(2005) found that nine members of a previously unrecognized plant protein family,which shared a conserved C-terminal domain (domain unknown function,DUF623) with tomato OVATE protein and were accordingly identified as AtOFPs (Arabidopsis thaliana ovate family proteins),were included in this module.Moreover,Hackbusch et al.(2005) found nine members (AtOFP1,AtOFP2,AtOFP3,AtOFP4,AtOFP5,AtOFP6,AtOFP12,AtOFP14,and AtOFP15) of this family involved in the intracellular localization of TALE proteins.Later on,an activation-tagged mutant with reduced length in all aerial organs including hypocotyl,cotyledon,rosette leaf,cauline leaf,inflorescence stem,floral organs and silique was isolated by Wang (2007) and molecular cloning revealed that these phenotypes were caused by elevated expression of the Arabidopsis thaliana Ovate Family Protein 1 (AtOFP1).AtOFP1 is then identified as an active transcriptional repressor that regulates cell elongation in parts by directly controlling the expression of AtGA20ox1,a gene encoding the key enzyme in GA biosynthesis (Wang et al.,2007).Plants over-expressing other closely related AtOFP genes-AtOFP2 and AtOFP7-phenocopied plants over-expressing AtOFP1,indicating there is a possible overlapping function among members of the AtOFP gene family.On the other hand,Gabriela et al.(2007) showed that AtOFP5,which might mediate the suppression of ectopic activity of BELL-KNOX TALE complexes,is required during female gametophyte development and TALE-OVATE protein complexes might be functional during embryo sac early developmental stages.AtOFP2 is also shown to be a good candidate for regulators of ovule development through robust microarray averaging (RMA) analysis of array data from ovule mutants: inner no outer and aintegumenta (Skinner et al.,2009).
In order to understand these potential roles of ovate genes in Arabidopsis systematically,an indepth analysis of 17 AtOFPs genes was reported in this study.By analysis on the sequence information,protein characterization,chromosome localization,phylogenetic relationships,subcellular localization,protein interaction and expression profile of AtOFP family members,we laid a foundation for functional validation exercises aimed at understanding the role of this class of proteins in plant growth and development.
Materials and Methods
Retrieval of AtOFP genes and analysis of AtOFPs amino acid sequence
AtOFP genes were obtained by searching against the Arabidopsis thaliana genome using ATOFP as the gene name in TAIR (http://www.arabidopsis.org/servlets/Search?action=new_search&type=gene) and the OFP gene sequences and protein sequences of Arabidopsis (Arabidopsis thaliana) were obtained from GenBank.Conserved OVATE domains were identified in AtOFPs using the Conserved Domain Database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) with default parameters.
Further,to identify all potential OVATE domaincontaining proteins in Arabidopsis,we used DUF623 to query the Pfam Database (http://pfam.sanger.ac.uk;Finn et al.,2006).
Information about the position of the OFP genes on the chromosome was collected from TAIR (http://www.arabidopsis.org/jsp/ChromosomeMap/tool.jsp).The segmental genomic duplication information of Arabidopsis at Plant Genome Duplication Database (http://chibba.agtec.uga.edu/duplication/index/home;Tang,2008a;Tang,2008b) was used to determine the presence of AtOFP genes on duplicated chromosomal segments,with a maximum distance permitted between collinear gene pairs of 500 kb.
ClustalX (version 2.0) program (Thompson et al.,2002) was used to perform the multiple sequence alignments.Alignments were exported to the PHYLIP program 3.69 (Felsenstein,2005;Tuimala,2006) and used to generate unrooted neighbor joining trees and bootstrap values,using 1 000 resampled data matrixes.
Subcellular localization analysis of AtOFPs
Analysis of the subcellular localization of AtOFPs was performed by ProtComp Version 9.0 in infodb (http://linux1.softberry.com/berry.phtml?topic=protcomppl&group=programs&subgroup=proloc).
Identification of proteins interacting with AtOFPs
The protein TAIR loci of AtOFPs was used to search against the predicted and literature-curated Arabidopsis PPI data set (http://atpid.biosino.org/index.php;Cui et al.,2008),Arabidopsis thaliana Protein Interactome Database,to identify proteins that interact with AtOFPs.
Expression profiling of AtOFPs
Expression profile data of the AtOFPs in 47 tissues of Arabidopsis thaliana with three duplications was extracted from the BAR eFP browser (http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi;Winter et al.,2007),a database based on map of Arabidopsis development and the AtGenExpress Consortium Data,cell-type or seed-specific data and other data,and the data were normalized by the GCOS method with TGT value of 100.
Results
Analysis of AtoFPs amino acid sequence
Annotation of the TAIR Arabidopsis genome had identified 18 putative OFP genes (Table1) (http://www.arabidopsis.org/servlets/Search).However,taking advantage of the Conserved Domain Database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi),we found that the predicted translation products of two of these,AtOFP9 (AT4G04030) and AtOFP17 (AT2G30395),which were previously identified as members of the plant specific ovate protein family (Hackbusch et al.,2005),did not contain the OVATE domain (DUF623).In order to identify all putative OVATE domain-containing protein sequences in Arabidopsis,the OVATE domain,DUF623,was used as a query to search against the Pfam database (http://pfam.sanger.ac.uk/).This search resulted in 24 putative OVATE protein sequences (including the sequences corresponding to different gene models from the same locus and obsoleted protein sequences) in Arabidopsis.Then,by removing the redundant sequences identified from both methods,we finally identified 17 putative OVATE domain-containing protein sequences in Arabidopsis.The corresponding novel ovate gene (At2G36026) was named AtOFP19 by following the existing numbering system of the OVATE family in Arabidopsis (Table 1).The 17 AtOFP proteins were 159-349 amino acid in length and sequence alignment showed that they all contained a OVATE domain located in the C-terminus (Fig.1).

Table1 Members of the AtOFP gene family and their predicted protein structure
Distribution of 17 AtoFP homologs
Analyzing on the chromosomal distribution of the 17 AtOFP homologs by TAIR,we found that AtOFP12,AtOFP4 and AtOFP14 were distributed on both ends of chromosomes 1,AtOFP7,AtOFP2,AtOFP16,AtOFP19 and AtOFP15 were presented in the lower arm of chromosome 2,AtOFP6 and AtOFP18 were located on the lower arm of chromosome 3,AtOFP11 and AtOFP5 were on the lower arm of chromosome 4,and AtOFP1,AtOFP13,AtOFP8,AtOFP10 and AtOFP3 were distributed on chromosome 5 (Fig.2).AtOFP19 and AtOFP15 were tandemly arranged,same at AtOFP6 and AtOFP18.Interestingly,by analyzing the superimposition of segmental duplication on the dataset of chromosomal location of AtOFP genes in the Plant Genome Duplication Database (http://chibba.agtec.uga.edu/duplication/index/home),we found that the formation of four gene pairs,AtOFP2 and AtOFP4 (gene sequence identity 63.28%),AtOFP3 and AtOFP4 (gene sequence identity 51.81%),AtOFP12 and AtOFP16 (gene sequence identity 59.89%),and AtOFP15 and AtOFP18 (gene sequence identity 59.23%),may be due to the segmental duplication of the Arabidopsis genome.On the basis,these OFP genes,AtOFP2 and AtOFP4,AtOFP3 and AtOFP4,AtOFP12 and AtOFP16,and AtOFP15 and AtOFP18,may have similar function in Arabidopsis.
Phylogenetic tree of the AtOFP family
To further understand the evolutionary relationships of the 17 OFP proteins,a phylogenetic tree was generated based on the sequence alignments of 18 full-length OVATE domain-containing proteins (17 AtOFPs and one Slovate,tomato ovate protein ).
The unrooted neighbor joining phylogenetic tree generated by PHYLIP program 3.69 showed that the 17 AtOFP proteins can be classified into two major groups (ⅠandⅡ) (Fig.3).The OVATE proteins of groupⅠincluded the known Slovate which was responding to the pear-shaped tomato fruit (Liu et al.,2002),the active transcriptional repressor AtOFP1 and the putative plant-specific transcription factors AtOFP2 and AtOFP7 (Wang et al.,2007).Besides,AtOFP1 and AtOFP5,which were closely grouped in the phylogenetic tree,were shown to associate with the cytoskeleton and to regulate subcellular localization of TALE homeodomain proteins (Hackbusch et al.,2005).The AtOFP11,AtOFP12 and AtOFP16 which were in the groupⅡhad been identified as the most closely related proteins to AmOVA by Maximum likelihood analysis of full length amino acid sequences and distance and maximum parsimony analysis of the conserved DUF623 domain (Duttke,2010).The AmOVATE which mapped to LF2.2 was suggested to regulate leaf width (Duttke,2010).From this point of view,OFP proteins in groupⅡ may be associated with organ shape.Importantly,overexpression of AtOFP1,AtOFP2,AtOFP4,AtOFP5 and AtOFP7,groupⅠ genes but not of any other groupⅡAtOFP genes,resulted in characteristic kidney-shaped cotyledons in young seedlings (Wang et al.,2011),corresponding to our phylogenetic analysis.

Fig.1 Alignment of full length amino acid sequences of 17 AtOFP proteins by the CLUSTALX program

Fig.2 Distribution of the AtOFP genes on Arabidopsis chromosomes
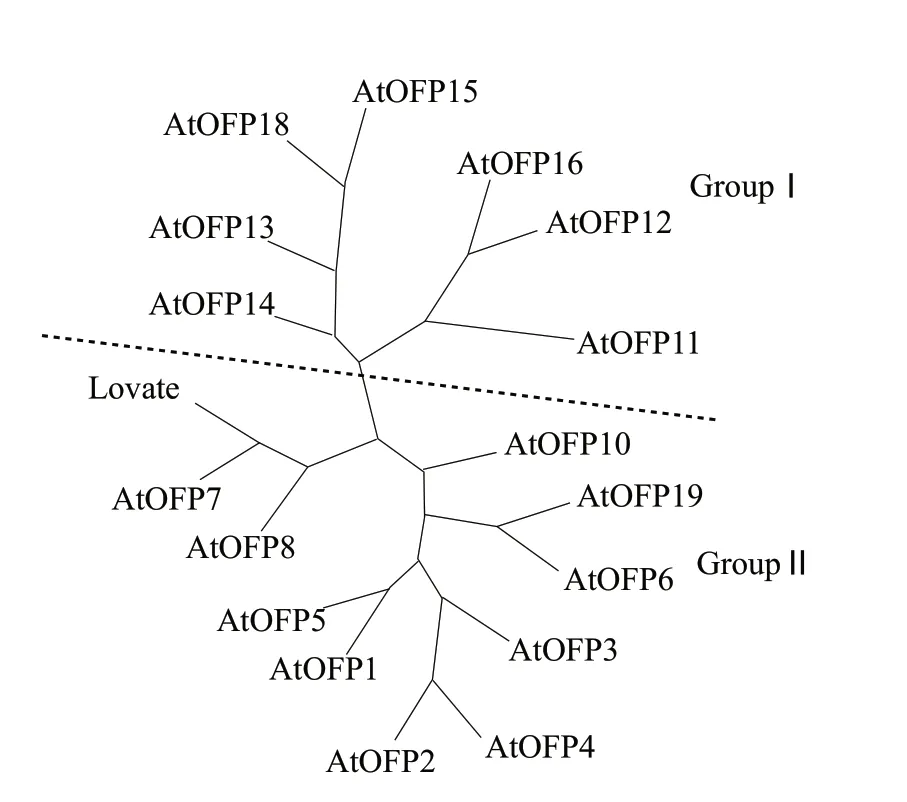
Fig.3 Phylogenetic analysis of Arabidopsis ovate family members and tomato ovate protein
Subcellular localization of AtOFPs
To provide a foundation for determining the function of the AtOFP proteins,ProtComp -Version 9 (http://linux1.softberry.com/berry.phtml? topic=protcomp pl&group=programs&subgroup=proloc) was used with the default parameters to identify the subcellular localization of AtOFPs.
It was previously reported that most members of the AtOFPs family contained a predicted nuclear localization signal (Hackbusch et al.,2005;Liu et al.,2002).Our results revealed that the majority of Arabidopsis OFP proteins (11 members) were located in nuclear,four members (AtOFP6,AtOFP13,AtOFP18 and AtOFP19) were presented in chloroplast,and two (AtOFP4 and AtOFP8) were distributed in cytoplasmic (Table 2).All the four AtOFPs predicted in chloroplast had chloroplast transit peptide predicted.The prediction of subcellular localization of AtOFP1,2 and 7 was consistent with the report that AtOFP1,AtOFP2 and AtOFP7 might represent a new class of plant-specific transcription factors (Wang et al.,2007).
Investigation of proteins interacting with AtOFPs
We used the TAIR loci of AtOFPs to searched against the Arabidopsis thaliana Protein Interactome Database (Version3.00;http://atpid.biosino.org/index.php;27) to investigate proteins interacting with AtOFPs.
The results are shown in Table 3.A total of 58 proteins (including the same proteins interacting with different AtOFPs) were involved in the interactions with AtOFPs,and they all came from text mining by using yeast two-hybrid technology (Hackbusch et al.,2005).After removing the redundant proteins,there were 16 proteins in total.The names,TAIR gene locus and interaction frequencies of the 16 proteins were as the followings: BLH3 (AT1G75410;6),BLH1(AT2G35940;6),KNAT7 (AT1G62990;5),KNAT1 (AT4G08150;5),KNAT3 (AT5G25220;5),BLH10 (AT1G19700;4),BLH2 (AT4G36870;4),KNAT4 (AT5G11060;4),KNAT5 (AT4G32040;4),BEL1 (AT5G41410;3),BLH4 (AT2G23760;3),BLH6 (AT4G34610;3),KNAT6 (AT1G23380;2),KNAT2 (AT1G70510;2),ATH1 (AT4G32980;1) and BLH5 (AT2G27220;1).Intriguingly,all the 16 proteins belonged to TALE homeodomain protein (Hackbusch et al.,2005).Actually,the AtOFPs were first characterized in a comprehensive survey of TALE protein interactions (Hackbusch et al.,2005 ).

Table2 Prediction of subcellular localization of AtOFPs by ProtComp-Version 9

Table3 Proteins interacting with AtOFPs in Arabidopsis thaliana Protein Interactome Database
Proteins with homology could function redundantly in regulating plant growth and development.Besides,the number of shared interactions might be taken as a crude measure of the overlap in two proteins functions (Hackbusch et al.,2005).In our study,we found eight AtOFPs with redundant TALE interaction proteins,and apart from AtOFPs/TALE interactions,there were TALE/TALE interactions at the same time (Hackbusch et al.,2005).These interactions above provided support for the possibility that some AtOFPs could function redundantly.For instance,most of the interaction proteins (11 of 13) of AtOFP1 were the interaction proteins of other AtOFPs at the same time.Thus,even there was no AtOFP1 in vivo,other AtOFPs could also interact with the related TALE proteins to play a corresponding function.In fact,a possible loss-of-function mutation in AtOFP1 really does not confer significant defects in morphology (Wang et al.,2007).TALE homeodomain proteins are fundamental regulators of plant meristem function and leaf development (Hackbusch et al.,2005).For instance,ATH11 (AT4G32980) is a positive mediator of a phyB-specific signal transduction cascade controlling GA levels by regulating the expression of GA20ox and GA3ox (García-Martinez et al.,2001);loss of BP/KNAT1 function results in reduced growth of floral internodes,pedicels and the style during reproductive growth (Scofield et al.,2008),while similar phenotypes were also observed among plants over-expressing AtOFP genes (Wang et al.,2007);suppression of ectopic activity of BELL-KNOX TALE complexes,which might be mediated by AtOFP5,is essential for normal development and cell specification in the Arabidopsis embryo sac (Gabriela et al.,2007);KNAT7 may be involved in secondary cell wall biosynthesis,whose mutants (SALK_002098) have moderately irregular xylem development (David et al.,2005).As AtOFPs interact with all these proteins,they may form functional complexes to realize the related regulation,together.
Hackbusch (2005) found that the single conserved domain of the AtOFPs-the OVATE domain-mediated the interaction with the homeodomains of both BELL and KNOX proteins (TALE proteins).As homeodomain transcription factors,KNOX and BELL proteins could bind to DNA.Thus,even there is no apparent DNA binding domain,AtOFPs could also regulate the expression of genes,and may play an important role in plant growth and development.
Expression profiles of AtOFP genes in different tissues of Arabidopsis
The spatial and temporal expression patterns of genes are often correlated with gene function.To investigate the AtOFPs expression pattern in Arabidopsis,an expression profile analysis was preformed on BAR eFP browser (http://bbc.botany.utoronto.ca/efp/cgibin/efpWeb.cgi;Winter et al.,2007),a microarraybased transcriptome profiling of 47 stages of vegetative and reproductive development.
A total of nine AtOFP encoding genes (AtOFP1,AtOFP2,AtOFP3,AtOFP7,AtOFP13,AtOFP14,AtOFP15,AtOFP16 and AtOFP18 ) were collected from the database and analysis based on the signal values showed that nine AtOFP genes were expressed at different levels and exhibited a variety of expression patterns (Fig.4).Overall,nine AtOFP genes were expressed in all of 47 Arabidopsis tissues investigated.AtOFP1 had a significantly higher expression in hypocotyl than in other tissues (Fig.4A),whereas AtOFP2 had a low expression in all of 47 tissues (Fig.4B),AtOFP3 had a slightly higher expression in stem,2nd internode (Fig.4C),AtOFP7 had higher expression in hypocotyl,shoot apex of inflorescence and transition (Fig.4D),AtOFP13 had high expression in all of 47 tissues,especially in flower (Fig.4E),AtOFP14 had a little higher expression in siliques (Fig.4F),AtOFP15 was first up-regulated and then downregulated during flower development (Fig.4G),AtOFP16 expressed at high levels in leaves (Fig.4H) and AtOFP18 was highly expressed in stamens and siliques (Fig.4I).However,none of the genes showed tissue-specific differential expression in Arabidopsis.

Fig.4 Expression patterns of nine AtOFP members in 47 different tissues of Arabidopsis(A–I).
Previousl studies have suggested that AtOFPs might have functions in gametophyte development (Gabriela et al.,2007;Skinner et al.,2009;Hackbusch et al.,2005).The expression patterns of AtOFPs from the database is coincident to this function presumption to some degree.Recently,Wang et al.(2007) found that AtOFP1 prom: GUS was expressed mainly in the roots and lower part of hypocotyls in 4-day-old Arabidopsis seedlings,in the roots including root hair,shoots,vasculatures and trichomes of 10-day-old seedlings,in the inflorescence,especially flowers with mature pollen,and the lower parts of young siliques in mature plants,which also supported the expression feature of the AtOFP1 revealed by microarray-based transcriptome profiling in the database.
Discussion
OVATE domain-containing proteins in Arabidopsis
Arabidopsis Ovate Family Proteins (AtOFPs) is a plant-specific protein family.Nine members of Arabidopsis thaliana ovate family proteins (AtOFPs) were indicated a close functional connection to TALE homeodomain proteins (Hackbusch et al.,2005).Furthermore,18 AtOFPs were characterized by the conserved C-terminal domain shared with the tomato OVATE protein (Slovate) by Hackbusch et al (2005).However,according to the newly protein sequences analysis,we found that the AtOFP9 and AtOFP17 did not contain the OVATE domain,while another protein which we named AtOFP19 had a OVATE domain.AtOFP19 was a hypothetical protein supported by ESTs with a OVATE domain (DUF623) located in 94-153 of 183 amino acids.AtOFP19 (TAIR:AT2G36026) was presented in the lower arm of chromosome 2 in Arabidopsis,and the most close related Arabidopsis protein of AtOFP19 was AtOFP6 (TAIR;sequence identity 60.11%).As a result,there were actually 17 AtOFPs in Arabidopsis.
Evolution of OVATE proteins in plant
By phylogenetic analysis on the unrooted neighbor joining phylogenetic tree about the 17 AtOFPs and one Slovate,we found that the AtOFPs could be classified into two major groups.And the tomato ovate protein (Slovate) was located in groupⅠ,the same to AtOFP1.Furthermore,the AtOFP1 over-expression phenotype resembled the over-expression phenotype of the Slovate gene.Over-expression of OVATE in tomato produced round fruit,exserted stigmas,repressed plant growth,and changed vegetative (leaf lets became rounder,and serration became less apparent) and floral (uneven suppression of different floral organ growths) architecture (Liu et al.,2002).Elevated expression of AtOFP1 transcript generated reduced lengths of aerial organs including hypocotyl,cotyledons,leaves (both rosette and cauline leaves changed to be round shape),leaf petioles,inflorescence stems,floral organs and siliques (Wang et al.,2007),and style and stigma protruded from the flower (Hackbusch et al.,2005).We all know that tomato (Solanum lycopersicum Mill.) is a plant of Solanaceae,while Arabidopsis (Arabidopsis thaliana L.Heynh) belongs to Cruciferae,the almostly same phenotypes of their OVATE over-expression mutant indicate that the OVATE protein may have a conserved function in plants over long evolutionary periods.Although we do not determine its function precisely right now,it can be concluded that the OVATE is a conserved function protein with a broad influence on plant growth and development.
Potential function of Arabidopsis AtOFPs genes
It has been showed that OVATE might represent a previously uncharacterized class of regulatory genes of plant growth (Liu et al.,2002).Besides,Wang et al.(2007) also found that AtOFP1 is an active transcriptional repressor that has a role in regulating cell elongation in plants.In our present study,11 of 17 AtOFPs were showed to be located in nuclear,four of 17 AtOFPs were presented in chloroplast with chloroplast transit peptide predicted,and the remained two were distributed in cytoplasmic.From this point,ovate proteins may represent a kind of pleiotropic developmental regulator controlling gene expression in nuclear and chloroplast.
It is evident that transcriptional regulation plays a pivotal role in the control of gene expression in plants,and four independent reports have shown that approximately 2 000 genes encode TFs in Arabidopsis (Mitsuda and Ohme-Takagi,2009).More importantly,it is emerged that the evolution of plant form will be most readily accomplished by changes in the cisregulatory regions of transcriptional regulators (Doebley and Lukens,1998).We performed a genomewide analysis of the Arabidopsis ovate protein family,and the results provided the evidence for a complex pattern of expression and regulation of this gene family,suggesting that ovate proteins may act as members of highly integrated networks in controlling plant growth and development.However,for identifying target genes and monitoring protein interactions at subcellular resolution,further experimental function studies of this multigene family is important.
Aoki K,Ogata Y,Shibata D.2007.Approaches for extracting practical information from gene co-expression networks in plant biology.Plant Cell Physio,148: 381-390.
Brewer M T,Moyseenko J B,Monforte A J,et al.2007.Morphological variation in tomato: a comprehensive study of quantitative trait loci controlling fruit shape and development.Journal of Experimental Botany,58: 1339-1349.
Cui J,Li P,Li G,et al.2008.AtPID: Arabidopsis thaliana protein interactome database-an integrative platform for plant systems biology.Nucleic Acids Res,36: 999-1008.
David MB,Leo A.H.Z,Joanne E,et al.2005.Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics.The Plant Cell,17: 2281-2295.
Doebley J,Lukens L.1998.Transcriptional regulation and the evolution of plant form.Plant Cell,10(7): 1075-1082.
Duttke Sascha H C.2010.Three-dimensional quantification of growth.John Innes Centre,Norwich.
Felsenstein J.2005.PHYLIP-Phylogeny Inference Package,version 3.69.University of Washington,Seattle,USA.
Finn R D,Mistry J,Tate1 J,et al.2010.The Pfam protein families database.Nucl Acids Res,38: 211-222.
Gabriela C P,Hee-Ju Y,Venkatesan S.2007.Cell-fate switch of synergid to egg cell in Arabidopsis eostre mutant embryo sacs arises from misexpression of the BEL1-Like homeodomain gene BLH1.The Plant Cell,19: 3578-3592.
García-Martinez J L,Gil J.2001.Light regulation of gibberellin biosynthesis and mode of action.Plant Growth Regul,20: 354-368.
Gonzalo MJ,Knaap E.2008.A comparative analysis into the genetic bases of morphology in tomato varieties exhibiting elongated fruit shape.Theor Appl Genet,116: 647-656.
Hackbusch J,Richter K,Müller J,et al.2005.A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins.Proc Natl Acad Sci USA,102: 4908-4912.
Hendrick U P,Booth N O.1907.Mendelian characters in tomato.Proc Am Soc Hort Sci,5: 19-24.
Knaap E,Lippman Z B,Tanksley S D.2002.Extremely elongated tomato fruit controlled by four quantitative trait loci with epistatic interactions.Theor Appl Genet,104: 241-247.
Ku H M,Doganlar S,Chen K Y,et al.1999.The genetic basis of pearshaped tomato fruit.Theor Appl Genet,9: 844-850.
Lindstrom E W.1925.Inheritance in tomatoes.Genetics,10: 305-317.
Lindstrom E W.1926.Linkage inheritance in tomatoes.Iowa State Col Jour Soc,1: 3-13.
Lindstrom E W.1927.The inheritance of ovate and related shapes of tomato fruits.Jour Agric Res,34: 961-985.
Liu J,Van Eck J,Cong B,et al.2002.A new class of regulatory genes underlying the cause of pear-shaped tomato fruit.Proc Natl Acad Sci USA,99: 13302-13306.
Macarthurn J W.1926.Linkage studies with the tomato.Genetics,11: 387-405.
Macarthurn J W.1928.Linkage studies with the tomato Ⅱ three linkage groupes.Genetics,13: 410-420.
Mitsuda N,Ohme-Takagi M.2009.Functional analysis of transcription factors in Arabidopsis.Plant Cell Physiol,50(7): 1232-1248.
Price H C,Drinkard A W.1908.Inheritance in tomato hybrids.Va Agric Exp Stn Bull,177: 17-53.
Scofield S,Dewitte W,Murray J A.2008.A model for Arabidopsis class-1 KNOX gene function.Plant Signaling Behav,3: 257-259.
Skinner D J,Gasser C S.2009.Expression-based discovery of candidate ovule development regulators through transcriptional profiling of ovule mutants.BMC Plant Biology,9: 29.
Tang H,Bowers J E,Wang X,et al.2008.Synteny and collinearity in plant genomes.Science,320: 486-488.
Tang H,Wang X,Bowers J E,et al.2008.Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps.Genome Research,18: 1944-1954.
Thompson J D,Gibson T J,Higgins D G.2002.Multiple sequence alignment using ClustalW and ClustalX.Curr Protoc Bioinformatics.
Toufighi K,Brady S M,Austin R,et al.2005.The botany array resource: e-northerns,expression angling,and promoter analyses.Plant J,43: 153-163.
Tuimala J.2006.A primer to phylogenetic analysis using the PHYLIP package.5th ed.Center for Scientific Computing Ltd,Espoo,Finland.
Wang S,Chang Y,Guo J,et al.2007.Arabidopsis ovate family protein 1 is a transcriptional repressor that suppresses cell elongation.Plant Journal,50: 858-872.
Wang S,Chang Y,Guo J,et al.2011.Arabidopsis ovate family proteins,a novel transcription factor family,control multiple aspects of plant growth and development.PLoS ONE,6(8): e23896.doi:10.1371/journal.pone.0023896.
Winter D,Vinegar B,Nahal H,et al.2007.An "electronic fluorescent pictograph" browser for exploring and analyzing large-scale biological data sets.PLoS ONE,2: e718.
 Journal of Northeast Agricultural University(English Edition)2012年3期
Journal of Northeast Agricultural University(English Edition)2012年3期
- Journal of Northeast Agricultural University(English Edition)的其它文章
- Regulatory Network of Transcription Factors in Response to Drought in Arabidopsis and Crops
- Comparison of Net Photosynthetic Rate in Leaves of Soybean with Different Yield Levels
- Multiplex PCR System Optimization with Potato SSR Markers
- Analysis on Combining Ability for Characters of Male Sterile Lines in Rapeseed (Brassica napus L.)
- Study on Mutant Induction of Gladiolus by in vitro Culture of Petals
- Research on Tobacco Transformation of Vacuolar H+-ATPase Subunit c Gene from Iris lacteal
