Regulatory Network of Transcription Factors in Response to Drought in Arabidopsis and Crops
Chen Li-miao ,Li Wen-bin,and Zhou Xin-an
1 Key Laboratory of Oil Crop Biology,Ministry of Agriculture,Oil Crops Research Institute,CAAS,Wuhan 430062,China
2 Key Laboratory of Soybean Biology,Ministry of Education,Soybean Research Institute,Northeast Agricultural University,Harbin 150030,China
Introduction
Transcription factors (TFs),which are known as trans-acting elements,can bind to cis-acting elements located in the promoter of stress-inducible genes,and regulate their expressions.Stress-inducible gene expressions are regulated by some signal pathways,such as AREB/ABF,DREB,NAC,MYB/ MYC,WRKY,NFYA,HD-ZIP etc (Table 1).AREB/ABF is ABA-dependent ,DREB is ABA-independent,while NAC,MYB/MYC,WRKY,NFYA,and HD-ZIP families include several subfamilies,in which some are ABA-dependent,the other are ABA-independent,even different members in the same subfamily involve in different signal pathways.But they are not totally isolated.The crosstalk of multiple stress signal pathways puts these TFs together.These TFs show differential transcript regulations in response to different stresses (Table 2).
Plants have adapted to respond to various environment stresses,such as drought,high-salinity,extreme temperature etc.,through a series of stress stimuli,signal perception,signal transduction,stressresponsive gene expression,appropriate morphological and physiological,molecular and cellular level changes occurred in plants,they protect themself from the damage of biotic and abiotic stresses.TFs play an important role in signal transduction.(Fig.1).

Table1 Sorts of transcription factors in different families
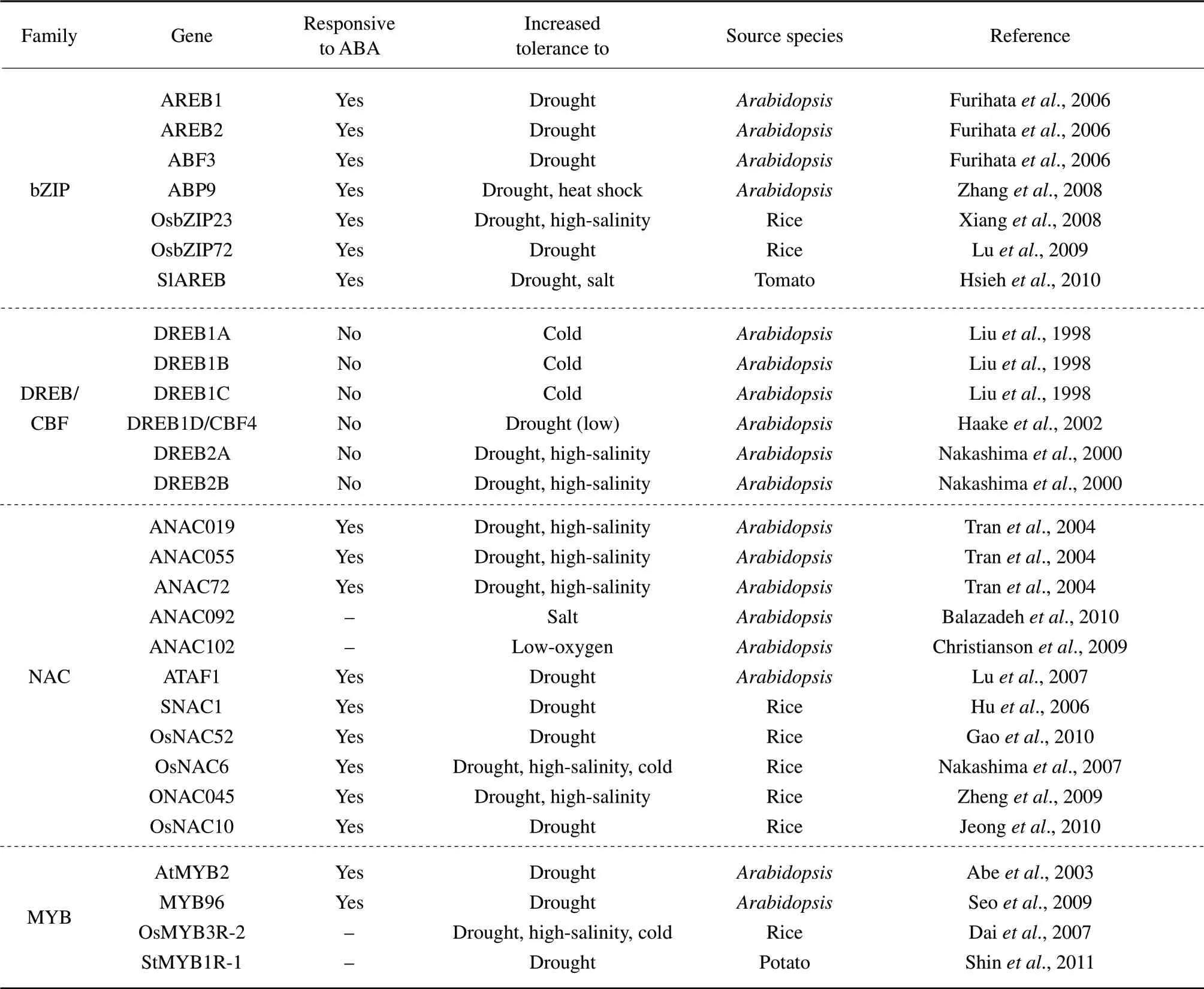
Table2 Abiotic stress tolerance of transgenic plant over-expressing transcription factors in different species

Continued

Fig.1 Transcriptional regulatory networks of TFs involved in abiotic stress-responsive gene expressions
Study of Drought-inducible Genes in Arabidopsis
Microarray analyses in Arabidopsis identify the products of the drought-inducible genes,which are divided into two types (Shinozaki et al.,2003).One is functional protein,such as water channel,detoxification enzyme,late embryogenesis abundant (LEA) protein,key enzyme for osmolyte biosynthesis (praline,sugar),and protease.Another is regulatory protein,including transcription factor,protein kinase,phospholipid metabolism,ABA biosynthesis and so on.In which,transcription factors play an important role in regulating drought-inducible gene expression.
AREB/ABFs regulated pathway
Some TFs,such as basic leucine zippers (bZIPs),regulate stress-responsive gene expressions through ABA signal pathway.There are some cis-elements,termed Abscisic Acid-Responsive Element Binding Proteins (ABREs),binding to bZIP-type AREB/ABFs in the promoter of ABA-regulated genes.AREB1,AREB2,and ABF3 in the AREB/ABFs subfamily need to be activated by ABA and can be induced under drought.These three transcription factors could form dimers in nucleus,and could interact with an SNF1-related protein kinase 2 (SnRK2) protein kinase designated SRK2D/SnRK2.2,which phosphorylated the AREB1 polypeptide (Furihata et al.,2006).To further study AREB1,AREB2,and ABF3 function,an areb1 areb2 abf3 triple mutant was constructed.The mutant was less sensitive to ABA and had weaker drought tolerance.Therefore,AREB1,AREB2,and ABF3 as transcription factors,collaboratively regulated ABA-signaling gene expression under drought (Yoshida et al.,2010).In Arabidopsis,a bZIP transcription factor,ABP9 (ABA-responsive-element (ABRE) binding protein 9) constitutive expression could improve the photo-synthetic capacity of plants under stress,including drought and heat shock (Zhang et al.,2008).There are some bZIP TFs responding to drought and other stress in crop plants.Expression of OsbZIP23 could be induced by most stresses,such as drought,salt,abscisic acid (ABA),and polyethylene glycol (PEG) treatments.Trans-activation assay in yeast suggested that OsbZIP23 was a transcriptional activator.Transient expression in onion cells revealed OsbZIP23 protein located in nucleus.Over-expressing OsbZIP23 in rice significantly improved tolerance to drought and high-salinity stresses and sensitivity to ABA.On the contrary,a null mutant of this gene showed significantly decreased sensitivity to a high concentration of ABA and decreased tolerance to drought stress and high-salinity,and this phenotype could be complemented by transforming the OsbZIP23 back into the mutant.These demonstrated adequately that OsbZIP23 was an important regulator in ABAdependent drought and high-salinity signal pathway (Xiang et al.,2008).OsbZIP72 was another positive transcription factor.It not only showed a hypersensitivity to ABA and a higher ability of drought tolerance,but also activated expression of ABA response genes,such as LEAs (Lu et al.,2009).The rice bZIP TF OsABI5 was also isolated from rice panicles.Expression of OsABI5 was induced by ABA and high salinity,but was down-regulated by drought and cold (4℃) stress in seedlings.Over-expression of OsABI5 in rice conferred high sensitivity to salt stress.In contrast,downregulation of OsABI5 improved stress tolerance,but decreased fertility of rice.These results demonstrated that OsABI5 might regulate stress response and plant fertility (Zou et al.,2008).In addition,a tomato bZIP transcription factor,SlAREB in Arabidopsis thaliana and tomato plants regulated stress-related genes including AtRD29A,AtCOR47,and SlCI7-like dehydrin under ABA and abiotic stress treatments.Taken together,these results showed that SlAREB might regulate some stress-responsive genes and that its over-production improved plant tolerance to water deficit and salt stress (Hsieh et al.,2010).
DREBs regulated pathway
Other TFs are in response to drought stress,but no respond to ABA,such as DREBs.The dehydration responsive element binding proteins (DREBs) are important transcription factors that regulate the abiotic stress-related genes and further improve stress tolerance to plants.DREBs were divided into DREB1/CBF and DREB2 (Yamaguchi-Shinozaki and Shinozaki,2005) which responsed to different stress signals as cold and drought.They both belong to the EREBP family of transcription factors,and bind to DRE (dehydration-responsive element)/CRT (C-RepeaT),both of which were cis-acting elements in the promoters of stress-inducible genes (Yamaguchi-Shinozaki and Shinozaki,1994).Their conserved DNA-binding motifs were CCGAC.The Arabidopsis genome contained six DREB1/CBF genes and eight DREB2 genes (Sakuma et al.,2002).DREB1A,DREB1B,and DREB1C were strongly induced by cold,but not by drought and high-salinity stress (Liu et al.,1998).However,DREB2A and DREB2B genes were on the contrary (Nakashima et al.,2000).Other DREB1 and DREB2 genes were weakly induced (Sakuma et al.,2002),and some DREB1/CBF genes as DREB1D/CBF4 also expressed low under drought stress,indicating crosstalk between the DREB1/CBF and the DREB2 pathways under drought stresses (Haake et al.,2002;Magome et al.,2004).DREB1/CBF and DREB2-homologous genes were identified in rice,named 10 OsDREB1s and four OsDREB2s,respectively.They had a similar function with these in Arabidopsis to abiotic stress.
NAC regulated pathway
NAC (NAM,ATAF and CUC) transcription factors (TFs),which are one kind of plant-specific TFs families,have been reported to enhance different stress tolerance such as drought,high salinity and cold.Many NAC TFs have been identified in model Arabidopsis and crops.More than 105 putative NAC TFs existed in Arabidopsis,140 in rice (Ooka et al.,2003),101 in soybean (Pinheiro et al.,2009) and 152 in tobacco (Rushton et al.,2008),respectively.The core motif CACG was identified,which was NAC recognition sequence (NACRS),by analyzing the promoter of the drought inducible EARLY RESPONSE TO DEHYDRATION1 (ERD1) gene.The expression of ERD1 under drought depended on both the "CACTAAATTGTCAC" ZFHDRS and the"ANNNNNTCNNNNNNNACACGCATGT" NACRS sequences (Simpson et al.,2003).They both acted as transcription activators in response to drought stress.In Arabidopsis,some NAC TFs could be induced by drought,high-salinity,low temperature or other stress.It was reported that ANAC019,ANAC055 or ANAC72 over-expression could improve some stressinducible genes up-regulated and increase drought tolerance in transgenic plants (Fujita et al.,2004;Tran et al.,2004).They also regulated jasmonic acidsignaled defense response (Ooka et al.,2003;Bu et al.,2008).RD26 gene encoding a NAC transcription factor was responsive not only to dehydration,but also to NaCl,ABA and jasmonic acid treatments.The transgenic plants over-expressing RD26 cDNA were hypersensitive to ABA,and inversely,the transgenic plants with RD26 repressed were insensitive to ABA.The expressions of many ABA-and stress-induced genes including RD20 and GLY genes were up-regulated in plants over-expressing RD26 and repressed in plants with RD26 repressed.In Arabidopsis protoplasts,RD26 activated a promoter of the GLY gene that was up-regulated in plants over-expressing RD26.Thus,GLY was the target gene of RD26.These results demonstrated that RD26,as a transcription activator,involved in stress-responsive ABA-dependent signal transduction pathway (Fujita et al.,2004).ANAC092/ AtNAC2/ORE1 was found to act during senescence and respond to salt stress (Balazadeh et al.,2010).ANAC102 was reported to be induced by low-oxygen stress,and decreasing ANAC102 expression would reduce seed germination efficiency under a 0.1% oxygen treatment,but increasing expression had no effect on seed germination.Indicating ANAC102 as an important regulator of seed germination under waterlogging (Christianson et al.,2009).ATAF1/ ATAF2 both negatively regulated stress-responsive gene expressions,the difference was that ATAF1 was strongly induced by dehydration and abscisic acid (ABA) treatment,indicating it regulated droughtresponsive pathway (Lu et al.,2007),ATAF2 repressed the expression of pathogenesis-related genes in Arabidopsis (Delessert et al.,2005).In rice,there are also some NAC TFs involving in drought,highsalinity,low temperature etc.stress.SNAC1 (STRESSRESPON-SIVE NAC 1) over-expression significantly increased drought tolerance in transgenic rice,whose seed setting was 22%-34% higher than that of control during drought stress.The transgenic rice was more sensitive to ABA treatment and losed water more slowly than WT by closing more stomatal pores,yet showed no significant difference in the rate of photosynthesis.Therefore,the yield had not been affected (Hu et al.,2006).OsNAC52,a rice NAC transcription factor,could respond to ABA.OsNAC52 over-expression activated the expression of downstream genes in transgenic Arabidopsis and enhanced tolerance to drought stresses but not growth retardation (Gao et al.,2010).Similarly,a rice NAC gene,ONAC045,was also induced by drought,high salt,and low temperature stress and ABA treatment in leaves and roots.Over-expressing ONAC045 in rice enhanced tolerance to drought and salt treatments (Zheng et al.,2009).OsNAC10 expressed in roots significantly,and be induced by drought,high salinity,and ABA.In transgenic rice,OsNAC10 over-expression increased drought tolerance.Under the control of root-specific promoter RCc3,root diameter of OsNAC10 plants was thicker by 1.25-fold than that of the constitutive promoter GOS2 and non-transgenic plants grain yield added by 17% in drought stress condition,9% in normal condition (Jeong et al.,2010).In soybean,101 NAC domain-containing proteins were divided into 15 different subgroups,in which six previously described GmNAC proteins (GmNAC1 to GmNAC6) were located in the nucleus and responded to various stress.GmNAC 2-5 acted as transactivators.GmNAC2-4 were significantly induced by osmotic stress.GmNAC3-4 were also induced by ABA,JA and salinity,but had not different effects on low temperature.The transient expression of GmNAC1,GmNAC5 and GmNAC6 in tobacco leaves promoted senescence and further resulted in cell death.The function and participating in stress-responsive pathways NAC TFs in soybean would be further studied.
MYB regulated pathway
The MYB family is a large family and exists in all eukaryotes.MYB proteins are main regulators in plant development,metabolism and response to biotic and abiotic stresses.AtMYB2 and AtMYC2 both bound cis-elements in the RD22 promoter and cooperatively activated the RD22.Microarray analysis suggested that target genes of MYC/MYB overexpression in transgenic plants might be alcohol dehydrogenase and ABA-or jasmonic acid (JA)-inducible genes (Abe et al.,2003).Over-expression of AtMYB2 and AtMYC2 showed hypersensitivity to ABA and improved drought tolerance of the transgenic plants.A R2R3-type MYB transcription factor,MYB96,regulated drought stress response by integrating ABA and auxin signals.The MYB96-mediated ABA signals were integrated into an auxin signaling pathway that involved a subset of GH3 genes encoding auxin-conjugating enzymes.MYB96 over-expression in Arabidopsis exhibited enhanced drought resistance with reduced lateral roots.On the other hand,a T-DNA insertional knockout mutant was more susceptible to drought.Taken together,MYB96 was a molecular link that mediated ABA-auxin cross talk in drought stress response and lateral root growth (Seo et al.,2009).By a cDNA microarray approach to monitor the expression profile of rice under cold stress,OsMYB3R-2 was identified.Unlike most plant R2R3 MYB transcription factors,OsMYB3R-2 has three imperfect repeats in the DNA-binding domain.Expression of OsMYB3R-2 was induced by cold,drought,and salt stress.Over-expressing OsMYB3R-2 in Arabidopsis showed increased tolerance to cold,drought,and salt stress (Dai et al.,2007).A putative R1-type MYB-like transcription factor,StMYB1R-1,was also isolated as a putative stress-response gene using reverse northern-blot analysis under abiotic environmental stress conditions.StMYB1R-1 located to the nucleus and bound to the DNA sequence /AGATAA.StMYB1R-1 over-expression in potato plants improved plant tolerance to drought stress while having no significant effects on other agricultural traits.In addition,over-expression of StMYB1R-1 enhanced the expression of drought-regulated genes such as AtHB-7,RD28,ALDH22a1,and ERD1-like.These results demonstrated that StMYB1R-1 functions as a transcription factor involved in the activation of drought-related genes (Shin et al.,2011).
WRKY regulated pathway
Like MYB family,the WRKY gene family also encoded a large group of TFs.Proteins of this family contained one or two highly conserved WRKY domains and a zinc finger motif in the C-terminal region (Eulgem et al.,2000).The WRKY domain bound to the W box or SURE (sugar-responsive ciselement) found in promoters of target genes and regulated its transcription (Rushton et al.,1995;Sun et al.,2003).WRKY transcription factors were involved in plant responses to both biotic and abiotic stresses such as WRKY18,WRKY40,and WRKY60.WRKY18 and WRKY60 had a positive effect on plant ABA sensitivity for inhibition of seed germination and root growth.And the two WRKY genes also enhanced plant sensitivity to salt and drought stress.In contrast,WRKY40 repressed WRKY18 and WRKY60 in the effect on plant sensitivity to ABA and abiotic stress in germination and growth.Both WRKY18 and WRKY40 were rapidly induced by ABA,while induction of WRKY60 by ABA was delayed.ABA-inducible expression of was almost completely abolished in the WRKY18 and WRKY40 mutants.Thus,WRKY60 might be a direct target gene of WRKY18 and WRKY40 in ABA signaling.Taken together,these three related WRKY transcription factors formed a highly interacting regulatory network that regulated gene expression in both plant defense and stress responses by acting as either transcription activator or repressor (Chen et al.,2010).The ABO3,a WRKY transcription factor,regulated plant responses to abscisic acid and drought tolerance in Arabidopsis.The ABA sensitive mutant,abo3,was hypersensitive to ABA in both seedling establishment and seedling growth.However,stomatal closure was less sensitive to ABA,and the abo3 mutant was less drought tolerant than the wild type.Northern blot analysis showed that the expression of the ABA-responsive transcription factor ABF2/AREB1 was markedly lower in the abo3 mutant than in the wild type.The abo3 mutation also reduced the expression of stress-inducible genes RD29A and COR47,especially early during the ABA treatment.The ABO3 was able to bind the W-box in the promoter of ABF2 in vitro.These results indicated that ABO3 played an important role as a WRKY transcription factor in plant responses to ABA and drought stress (Ren et al.,2010).In addition,the expression of OsWRKY45 found in rice markedly induced by abscisic acid (ABA) and various stress factors in Arabidopsis,including NaCl,PEG,mannitol or dehydration,treatment with 0℃ and 42℃ as well as infection by Pyricularia oryzae Cav.and Xanthomonas oryzae pv.oryzae.Together,these results indicated that the OsWRKY45 may be involved in the signal pathways of both biotic and abiotic stress responses (Qiu and Yu,2009).Besides,in other plants,including soybean and barely,there were also some WRKY TFs responding to drought and other stresses.Soybean WRKY-type transcription factor,GmWRKY13,GmWRKY21,and GmWRKY54,conferred differential tolerance to abiotic stresses in transgenic Arabidopsis plants.GmWRKY21-transgenic Arabidopsis plants were tolerant to cold stress,whereas GmWRKY54 conferred salt and drought tolerance,possibly through the regulation of DREB2A and STZ/Zat10.Transgenic plants over-expressing GmWRKY13 showed increased sensitivity to salt and mannitol stress,but decreased sensitivity to ABA,when compared with wild-type plants.In addition,GmWRKY13-transgenic plants showed an increase in lateral roots.These results indicated that these three GmWRKY genes played differential roles in abiotic stress tolerance (Zhou et al.,2008).Constitutive expression of the barley HvWRKY38 transcription factor also enhanced drought tolerance in turf and forage grass (Paspalum notatum Flugge).Transgenic plants retained water better during dehydration,recovered faster and produced more biomass following rehydration and survived severe dehydration stress under controlled environment conditions in contrast to non-transgenic plants.They indicated that HvWRKY38 played the regulatory role in dehydration tolerance (Xiong et al.,2010).
NF-Y regulated pathway
NF-Y transcription factors existd ubiquitous in all eukaryotes and have roles in the regulation of various genes (McNabb et al.,1995;Edwards et al.,1998;Maity and Crombrugghe,1998;Mantovani,1999).The NF-Y transcription factor complex was composed of three unique subunits: NF-YA,NF-YB,and NFYC.The subunits NF-YB and NF-YC formed a heterodimer in the cytoplasm,and then translocated to the nucleus,where they combined with the third subunit,NF-YA,heterotrimeric NF-Y transcription factor (Frontini et al.,2004;Kahle et al.,2005).NF-Y transcription factor combined to CCAAT box,the core sequence in the promoter of regulative genes.Plant NF-Y TFs function appears to be important for responses to drought stress.Although a specific mechanism remains unknown.Over-expression of AtNF-YB1 and its ortholog in maize (Zea mays),ZmNF-YB2 improved drought tolerance (Nelson and Repetti,2007).However,no loss-of-function data were provided to study and indicated the function of drought in Arabidopsis.On the other hand,a publication provided both over-expression and loss-of-function data for NFYA5 (Li et al.,2008).The Arabidopsis thaliana NFYA5 transcript was strongly induced by drought stress in an abscisic acid (ABA)-dependent signal pathway.NFYA5 was highly expressed in vascular tissues and guard cells,and NFYA5 contained a target site for miR169,which was down-regulated by drought stress also by an ABA-dependent pathway.Analysis of the expression of miR169 precursors showed that miR169a and miR169c were substantially down-regulated by drought stress.Co-expression of miR169 and NFYA5 suggested that miR169a was more efficient than miR169c at repressing the NFYA5 mRNA level.nfya5 knockout plants and plants overexpressing miR169a showed enhanced leaf water loss and were more sensitive to drought stress than wild-type plants.By contrast,transgenic Arabidopsis plants over-expressing NFYA5 displayed reduced leaf water loss and were more resistant to drought stress than the wild type.Microarray analysis indicated that NFYA5 was an important regulator for the expression of a number of drought stress-responsive genes.Therefore,NFYA5 was crucial for drought tolerance,and its induction by drought stress occurred at both the transcriptional and posttranscriptional levels.
HD-Zip regulated pathway
The HD-Zip (Homeodomain-Leucine Zipper) family of transcription factors is unique to the plant kingdom.They are made up of four subfamilies in Arabidopsis,including HD-ZipⅠ,Ⅱ,Ⅲ,and Ⅳ.Some HD-Zip proteins participate in plant growth and development,while,others involve the action of hormones or are apt to respond to environmental stress.Here,we mainly reviewed recent studies for transcription factors of this family playing crucial role in environment stress,especially in drought.In 1999,ATHB6 (HD-ZipⅠ) was reported to be induced by water deficit,osmotic stress or exogenous treatment with abscisic acid (ABA),and the ATHB6 induction was impaired in the two ABA-insensitive mutants,abi1 and abi2.It demonstrated that ATHB6 might act downstream to both ABI1 and ABI2 in a signal transduction pathway mediating a drought stress response (Söderman et al.,1999).In contrast,the Arabidopsis thaliana homeodomain leucinezipper gene ATHB7 and ATHB12 (HD-ZipⅠ) acted to mediate a response to water deficit as a negative regulator of growth (Söderman et al.,1996;Hjellstrom et al.,2003;Olsson et al.,2004).The dehydrationinducible expression of the Arabidopsis thaliana EARLY RESPONSIVE TO DEHYDRATION STRESS 1 (ERD1) gene depended on the action together of zinc finger homeodomain ZFHD1 and NAC transcription factors.Using the yeast one-hybrid system,ZFHD1 transcriptional activator was isolated.The expression of ZFHD1 was induced by drought,high salinity and abscisic acid.The DNA-binding and activation domains of ZFHD1 were localized on the C-terminal homeodomain and N-terminal zinc finger domain,respectively.Microarray analysis of transgenic plants over-expressing ZFHD1 revealed that several stress-inducible genes were up-regulated in the transgenic plants.Using the yeast two-hybrid system,both ZFHD1 and NAC proteins were detected.Moreover,co-overexpression of the ZFHD1 and NAC genes restored the morphological phenotype of the transgenic plants to a near wild-type state and increase expression of ERD1 in transgenic Arabidopsis plants (Tran et al.,2006).Activated expression of an Arabidopsis HD-START Protein,HDG11(HDZip Ⅳ) enhanced drought tolerance with improved root system and reduced stomatal density (Yu et al.,2008).And HDG11 over-expression in transgenic Arabidopsis,tobacco and tall fescue all increased drought tolerance (Yu et al.,2008;Cao et al.,2009).HIPP26 from Arabidopsis thaliana belonged to the HIPP family of plant proteins,characterized by a heavy metal associated domain and an additional isoprenylation motif.It was induced during cold,salt and drought stress.By a yeast-two-hybrid approach,a strong interaction of HIPP26 with the zinc finger homeodomain transcription factor ATHB29 existed,which was known to play a role in dehydration stress response could be detected (Barth et al.,2009).A dehydration responsive nuclear-targeted HD-ZIP transcriptional regulator,CpHB-7 could combine with its target gene CDeT6-19,a known ABA and dehydration responsive dehydrin gene.CpHB-7 over-expression in transgenic desiccation-tolerant plant Craterostigma plantagineum reduced sensitivity towards ABA during seed germination and stomatal closure.CpHB-7 regulated ABA-responsive gene expression as a negative regulator,which was functionally similar to the Arabidopsis transcription factor,ATHB-6 (Deng et al.,2006).Brassica napus L.Homeodomain Leucine-Zipper Gene BnHB6 was reported to be induced by several stress and phytohormones including mannitol,NaCl,cold treatment,anaerobic culture,wounding,H2O2,abscisic acid (ABA),and salicylic acid (SA) treatments,but not by ultraviolet treatment (Yu et al.,2005).Hahb-4,a sunflower homeobox-leucine zipper gene,can be induced by drought and ABA.Its over-expression also increased drought tolerance.It showed that it might participate in the regulation of the expression of genes involved in developmental responses of plants to desiccation (Dezar et al.,2005).Besides,the TF was a new component of ethylene signaling pathways,and that it induced a marked delay in senescence.Plants overexpressing Hahb-4 were less sensitive to external ethylene,entered the senescence pathway later.Expression of this TF had a major repressive effect on genes related to ethylene synthesis,such as the ACO and the SAM,and on genes related to ethylene signaling,such as ERF2 and ERF5.Taken together,we proposed that Hahb-4 was involved in a novel conserved mechanism related to ethylene-mediated senescence that improved desiccation tolerance (Manavella et al.,2006).Nicotiana attenuata NaHD20 enhanced leaf ABA accumulation during water stress,and NaHD20 played a positive role in the expression of some dehydration-responsive genes including ABA biosynthetic genes (Re et al.,2011).
Crosstalk of Stress-responsive Gene Regulatory Network
The crosstalk exists not only in different regulatory systems,such as ABA-dependent or not,but also in different stresses-responsive gene expressions.Most drought-inducible genes are also induced by highsalinity stress,but only a few are cold-inducible.Stress-responsive gene regulatory is related to the cis-acting elements in the promoter of genes.The promoter of RD29A included both DRE and ABRE motifs,indicating that DRE/CRT functioned cooperatively with ABRE as a coupling element in ABA-responsive gene expression in response to drought stress (Narusaka et al.,2003).It was reported that genes of CBF/DREB1 family were ABA-independent and mainly induced by cold stress,but the drought-inducible gene CBF4 functioned to provide crosstalk between the DREB1/CBF and the DREB2 pathways under drought stress.A maize DRE-binding protein,DBF1,activated of the rab17 promoter by ABA (Kizis and Pages,2002).This also suggested the existence in some plants of an ABA-dependent pathway for the regulation of stress-inducible genes that involved in DRE/CRT.Drought-induced gene expressions such as ATMYB2 and ATMYC2 were also greatly enhanced by a jasmonic acid stress signal,which indicated crosstalk between abiotic-stress and biotic-stress pathways.
Conclusions and Perspectives
Molecular and genetic studies have identified many stress-inducible genes and their regulators,in the model plant,basic regulatory mechanism of genes expression also has been revealed,but it is only tip of the iceberg,we need fill up and perfect the network of stress-responsive gene expressions to understand the whole molecular basis.And the most important is digging the key factors at the crossing among different abiotic stress responses and phytohormone signal pathways of the same abiotic stress response.Then plant stress tolerance would have been improved by gene transfer.The constitutive promoters had better be replaced by inducible promoters to avoid the negative effects on plant growth.Ultimately,it would have been applied to crops and vegetable plants.
Abe H,Urao T,Ito T,et al.2003.Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcription activators in abscisic acid signaling.The Plant Cell,15(1): 63-78.
Balazadeh S,Siddiqui H,Allu A D,et al.2010.A gene regulatory network controlled by the NAC transcription factor ANAC092/ AtNAC2/ORE1 during salt-promoted senescence.The Plant Journal,62(2): 250-264.
Barth O,Vogt S,Uhlemann R,et al.2009.Stress induced and nuclear localized HIPP26 from Arabidopsis thaliana interacts via its heavy metal associated domain with the drought stress related zinc finger transcription factor ATHB29.Plant Mol Biology,69(1): 213-226.
Bu Q Y,Jiang H L,Li C B,et al.2008.Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses.Cell Research,18(7): 756-767.
Cao Y J,Wei Q,Liao Y,et al.2009.Ectopic overexpression of AtHDG11 in tall fescue resulted in enhanced tolerance to drought and salt stress.Plant Cell Report,28(4): 579-588.
Chen H,Lai Z B,Shi J W,et al.2010.Roles of Arabidopsis WRKY18,WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress.BMC Plant Biology,10(1): 281-326.
Christianson J A,Wilson I W,Llewellyn D J,et al.2009.The low-oxygen-induced NAC domain transcription factor ANAC102 affects viability of Arabidopsis seeds following low-oxygen treatment.Plant Physiology,149(4): 1724-1738.
Dai X Y,Xu Y Y,Ma Q B,et al.2007.Over-expression of an R1R2R3 MYB gene,OsMYB3R-2,increases tolerance to freezing,drought,and salt stress in transgenic Arabidopsis.Plant Physiology,143(4): 1739-1751.
Deng X,Phillips J,Brautigam A,et al.2006.A homeodomain leucine zipper gene from Craterostigma plantagineum regulates abscisic acid responsive gene expression and physiological responses.Plant Molecular Biology,61(3): 469-489.
Dezar C A,Gago G M,Gonzalez D H,et al.2005.Hahb-4,a sunflower homeobox-leucine zipper gene,is a developmental regulator and confers drought tolerance to Arabidopsis thaliana.Plants Transgenic Research,14(4): 429-440.
Delessert C,Kazan K,Wilson I W,et al.2005.The transcription factor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis.The Plant Journal,43(5): 745-757.
Edwards D,Murray J A,Smith A G.1998.Multiple genes encoding the conserved CCAAT-box transcription factor complex are expressed in Arabidopsis.Plant Physiology,117(3): 1015-1022.
Eulgem T,Rushton P J,Robatzek S,et al.2000.The WRKY superfamily of plant transcription factors.Trends Plant Science,5(5): 199-206.
Fujita M,Fujita Y,Maruyama K,et al.2004.A dehydration induced NAC protein,RD26,is involved in a novel ABA-dependent stresssignaling pathway.Plant Journal,39(6): 863-876.
Furihata T,Maruyama K,Fujita Y,et al.2006.ABA-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1.Proceedings of the National Academy of Sciences,103(6): 1988-1993.
Gao F,Xiong A S,Peng R H,et al.2010.OsNAC52,a rice NAC transcription factor,potentially responds to ABA and confers drought tolerance in transgenic plants.Plant Cell Tissue Organ Culture,100(3): 255-262.
Haake V,Cook D,Riechmann J L,et al.2002.Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis.Plant Physiology,130(2): 639-648.
Hjellstrom M.,Olsson A.S.B,Engstrom P,et al.2003.Constitutive expression of the water deficit-inducible homeobox gene ATHB7 in transgenic Arabidopsis causes a suppression of stem elongation growth.Plant Cell Environment,26(7): 1127-1136.
Hsieh T H,Li C W,Su R C,et al.2010.A tomato bZIP transcription factor,SlAREB,is involved in water deficit and salt stress response.Planta,231(6): 1459-1473.
Hu H H,Dai M Q,Yao J L,et al.2006.Over-expressing a NAM,ATAF,and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice.Proceedings of the National Academy of Sciences,103(35): 12987-12992.
Jeong J S,Kim Y S,Baek K H,et al.2010.Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions.Plant Physiology,153(1): 185-197.
Kahle J,Baake M,Doenecke D,et al.2005.Subunits of the heterotrimeric transcription factor NF-Y are imported into the nucleus by distinct pathways involving importin beta and importin13.Molecular Cell Biology,25(13): 5339-5354.
Kizis D,Pages M.2002.Maize DRE-binding proteins DBF1 and DBF2 are involved in rab17 regulation through the drought responsive element in an ABA-dependent pathway.The Plant Journal,30(6): 679-689.
Liu Q,Sakuma Y,Abe H,et al.1998.Two transcription factors,DREB1 and DREB2,with an EREBP/AP2 DNA binding domain,separate two cellular signal transduction pathways in drought-and low temperature-responsive gene expression,respectively,in Arabidopsis.The Plant Cell,10(8): 1491-1406.
Li W X,OonoY,Zhu J,et al.2008.The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and post-transcriptionally to promote drought resistance.The Plant Cell,20(8): 2238-2251.
Lu P L ,Chen N Z,An R,et al.2007.A novel drought-inducible gene,ATAF1,encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis.Plant Molecular Biology,63(2): 289-305.
Lu G J,Gao C X,Zheng X N,et al.2009.Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice.Planta,229(3): 605-615.
Magome H,Yamaguchi S,Hanada A,et al.2004.Dwarf and delayed-flowering 1,a novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor.The Plant Journal,37(5): 720-729.
Maity S N,Crombrugghe B.1998.Role of the CCAAT-binding protein CBF/NF-Y in transcription.Trends Biochemistry Science,23(5): 174-178.
Mantovani R.1999.The molecular biology of the CCAAT-binding factor NF-Y.Gene,239(1): 15-27.
Frontini M,Imbriano C,Manni I,et al.2004.Cell cycle regulation of NF-YC nuclear localization.Cell Cycle,3(2): 217-222.
McNabb D S,Xing Y,Guarente L.1995.Cloning of yeast HAP5: a novel subunit of a heterotrimeric complex required for CCAAT binding.Genes &Development,9(1): 47-58.
Nakashima K,Shinwari Z K,Sakuma Y,et al.2000.Organization and expression of two Arabidopsis DREB2 genes encoding DRE-binding proteins involved in dehydration-and high-salinity-responsive gene expression.Plant Molecular Biology,42(4): 657-665.
Narusaka Y,Nakashima K,Shinwari Z K,et al.2003.Interaction between two cis-acting elements,ABRE and DRE,in ABAdependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses.The Plant Journal,34(2): 137-148.
Nelson D E,Repetti P P,Adams T R,et al.2007.Plant nuclear factor
Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres.Proceedings of the National Academy of Sciences,104(42): 16450-16455.
Olsson A S B,Engstrom P,Soderman E.2004.The homeobox genes ATHB12 and ATHB7 encode potential regulators of growth in response to water deficit in Arabidopsis.Plant Molecular Biology,55(5): 663-677.
Ooka H,Satoh K,Doi K,et al.Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana.DNA Research,2003,10(6): 239-47.
Pinheiro G L,Marques C S,Costa M D B L,et al.2009.Complete inventory of soybean NAC transcription factors: sequence conservation and expression analysis uncover their distinct roles in stress response.Gene,444(1): 10-23.
Qiu Y P,Yu D Q.2009.Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis.Environmental and Experimental Botany,65(1): 35-47.
Re D A,Dezar C A,Chan R L,et al.2011.Nicotiana attenuata NaHD20 plays a role in leaf ABA accumulation during water stress,benzylacetone emission from flowers,and the timing of bolting and flower transitions.Journal of Experimental Botany,62(1): 155-166.
Ren X Z,Chen Z Z,Liu Y,et al.2010.ABO3,a WRKY transcription factor,mediates plant responses to abscisic acid and drought tolerance in Arabidopsis.The Plant Journal,63(3): 417-429.
Rushton P J,Macdonald H,Huttly A K,et al.1995.Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of alpha-Amy2 genes.Plant Molecular Biology,29(4): 691-702.
Rushton P J,Bokowiec M T,Han S,et al.2008.Tobacco transcription factors: novel insights into transcriptional regulation in the Solanaceae.Plant Physiology.147(1): 280-95.
Sakuma Y,Liu Q,Dubouzet J G,et al.2002.DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs,transcription factors involved in dehydration-and cold-inducible gene expression.Biochemical and Biophysical Research Communications,290(3): 998-1009.
Seo P J,Xiang F N,Qiao M,et al.2009.The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis.Plant Physiology,151(1): 275-289.
Shinozaki K,Yamaguchi-Shinozakiy K,Sekiz M.2003.Regulatory network of gene expression in the drought and cold stress responses.Current Opinion in Plant Biology,6(5): 410-417.
Shin D J,Moon S J,Han S,et al.2011.Expression of StMYB1R-1,a novel potato single MYB-Like domain transcription factor,increases drought tolerance.Plant Physiology,155(1): 421-432.
Söderman E,Hjellström M,Fahleson J,et al.1999.The HD-Zip gene ATHB6 in Arabidopsis is expressed in developing leaves,roots and carpels and up-regulated by water deficit conditions.Plant Molecular Biology,40(6): 1073-1083.
Söderman E,Mattsson J,Engstrom P.1996.The Arabidopsis homeobox gene ATHB-7 is induced by water deficit and by abscisic acid.The Plant Journal,10(2): 375-381.
Sun C,Palmqvist S,Olsson H,et al.2003.A novel WRKY transcription factor,SUSIBA2,participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter.The Plant Cell,15(9): 2076-2092.
Tran L S,Nakashima K,Sakuma Y,et al.2004.Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter.The Plant Cell,16(9): 2481-2498.
Tran L S P,Nakashima K,SakumaY,et al.2007.Co-expression of the stress-inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis.The Plant Journal,49(1): 46-63.
Xiang Y,Tang N,Du H,et al.2008.Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice.Plant Physiology,148(4): 1938-1952.
Xiong X,James V,Zhang H N,et al.2010.Constitutive expression of the barley HvWRKY38 transcription factor enhances drought tolerance in turf and forage grass (Paspalum notatum Flugge).Molecular Breeding,25(3): 419-432.
Yamaguchi-Shinozaki K,Shinozaki K.2005.Organization of cis-acting regulatory elements in osmotic-and cold-stress-responsive promoters.Trends in Plant Science,10(2): 88-94.
Yamaguchi-Shinozaki K,Shinozaki K.1994.A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought,low temperature,or high-salt stress.The Plant Cell,6(2): 251-264.
Yoshida T,Fujita Y,Sayama1 H,et al.2010.AREB1,AREB2,and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation.The Plant Journal,61(4): 672-685.
Yu H,Chen X,Hong Y Y,et al.2008.Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density.The Plant Cell,20(4): 1134-1151.
Yu S W,Zhang L D,Zuo K J,et al.2005.Brassica napus L.homeodomain leucine-zipper gene BnHB6 responds to abiotic and biotic stresses.Journal of Integrative Plant Biology,47(10): 1236-1248.
Zhang X,Wollenweber B,Jiang D,et al.2008.Water deficits and heat shock effects on photosynthesis of a transgenic Arabidopsis thaliana constitutively expressing ABP9,a bZIP transcription factor.Journal of Experimental Botany,59(4): 839-848.
Zheng X N ,Chen B ,Lu G J,et al.2009.Over-expression of a NAC transcription factor enhances rice drought and salt tolerance.Biochemical and Biophysical Research Communications,379(4): 985-989.
Zou M J,Guan Y C,Ren H B,et al.2008.A bZIP transcription factor,OsABI5,is involved in rice fertility and stress tolerance.Plant Molecular Biology,66(6): 675-683.
Zhou Q Y,Tian A G,Zou H F,et al.2008.Soybean WRKY-type transcription factor genes,GmWRKY13,GmWRKY21,and GmWRKY54,confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants.Plant Biotechnology Journal,6(5): 486-503.
 Journal of Northeast Agricultural University(English Edition)2012年3期
Journal of Northeast Agricultural University(English Edition)2012年3期
- Journal of Northeast Agricultural University(English Edition)的其它文章
- Comparison of Net Photosynthetic Rate in Leaves of Soybean with Different Yield Levels
- Multiplex PCR System Optimization with Potato SSR Markers
- Analysis on Combining Ability for Characters of Male Sterile Lines in Rapeseed (Brassica napus L.)
- Study on Mutant Induction of Gladiolus by in vitro Culture of Petals
- Research on Tobacco Transformation of Vacuolar H+-ATPase Subunit c Gene from Iris lacteal
- Genome-wide Analysis of Ovate Family Proteins in Arabidopsis
