Basal defense is enhanced in a wheat cultivar resistant to Fusarium head blight
Xinlong Gao,Fan Li,Yikun Sun,Jiaqi Jiang,Xiaolin Tian,Qingwen Li,Kaili Duan,Jie Lin,Huiquan Liu,Qinhu Wang
State Key Laboratory for Crop Stress Resistance and High-Efficiency Production,College of Plant Protection,Northwest A&F University,Yangling 712100,China
Abstract Fusarium head blight (FHB),mainly caused by the fungal pathogen Fusarium graminearum,is one of the most destructive wheat diseases.Besides directly affecting the yield,the mycotoxin residing in the kernel greatly threatens the health of humans and livestock.Xinong 979 (XN979) is a widely cultivated wheat elite with high yield and FHB resistance.However,its resistance mechanism remains unclear.In this study,we studied the expression of genes involved in plant defense in XN979 by comparative transcriptomics.We found that the FHB resistance in XN979 consists of two lines of defense.The first line of defense,which is constitutive,is knitted via the enhanced basal expression of lignin and jasmonic acid (JA) biosynthesis genes.The second line of defense,which is induced upon F.graminearum infection,is contributed by the limited suppression of photosynthesis and the struggle of biotic stress-responding genes.Meanwhile,the effective defense in XN979 leads to an inhibition of fungal gene expression,especially in the early infection stage.The formation of the FHB resistance in XN979 may coincide with the breeding strategies,such as selecting high grain yield and lodging resistance traits.This study will facilitate our understanding of wheat-F.graminearum interaction and is insightful for breeding FHB-resistant wheat.
Keywords: Fusarium head blight,Xinong 979,lignin,jasmonic acid,photosynthesis,Fusarium graminearum
1.Introduction
Wheat is the most widely cultivated crop that feeds 30% of humans on earth.However,its production is devastated by many plant diseases (Figueroaet al.2018;Richardet al.2021).Fusariumhead blight (FHB),which is mainly caused byFusarium graminearum,is one of the worst fungal diseases worldwide (Deanet al.2012;Xiaet al.2020).The prevalence and re-emergence of FHB are also increasing in recent years (Osborne and Stein 2007;Zhang Het al.2012;Maet al.2020).FHB severely affects the yield and quality of wheat (McMullenet al.2012;Salgadoet al.2015).In particular,the mycotoxins,such as deoxynivalenol (DON) and nivalenol,residing in the grains are severe threats to food safety (Chenet al.2019;Johnset al.2022).
Breeding FHB-resistant wheat is the most effective and environment-friendly approach to control this disease (Dwebaet al.2017;Esseet al.2019).Thousands of wheat and its relative accessions are surveyed for FHB resistance to achieve this purpose.Unfortunately,most of them are susceptible,and no immunity accession is found (Maet al.2020).At present,about 200 quantitative trait loci (QTLs) have been identified.Among these QTLs,seven are assigned as FHB resistance genes,namelyFhb1-Fhb7.OnlyFhb1andFhb7have been cloned (Liet al.2019;Suet al.2019;Wang Het al.2020).Fhb1,which is carried by the Sumai 3 and Wangshuibai,encodes a nuclear-localized,histidine-rich calcium-binding protein that reduces 20-50% FHB severity (Liet al.2019;Suet al.2019).Fhb7,which is carried byThinopyrum elongatum,encodes a glutathione-S-transferase (GST) that detoxifies DON to inhibit the blight spreading within the spike (Wang Het al.2020).Although the actual function of theseFhbgenes in host defense is not clear,these genes are regarded as promising candidates for breeding FHB-resistant wheat (Daiet al.2022).
For survival,plants have evolved a range of defense strategies to fight against pathogens.Generally,it could be divided into constitutive defense and induced defense (Andersonet al.2010;Doughari 2015).Constitutive or preformed defense is the first line of plant defense,including the physical and chemical barriers that prevent infection at the preinvasive stage (Singhet al.2021;Wanet al.2021).Induced defense is the last line of plant defense,and it requires plants to sense the presence of pathogens (de Wit 2007;Zhou and Zhang 2020;Ngouet al.2022).Plants can perceive pathogen infection by recognizing pathogen-associated molecular patterns (PAMPs) through cell surface pattern recognition receptors,leading to PAMP-triggered immunity (PTI) (Jones and Dangl 2006;Monaghan and Zipfel 2012).During the co-evolution of plant and pathogen,PTI is frequently interfered by pathogen effectors,leading to effector-triggered susceptibility (Jones and Dangl 2006;Wang Qet al.2020).Some of the effectors are recognized by intracellular nucleotide-binding and leucine-rich repeat (NLR) receptors,leading to effector-triggered immunity (ETI) (Jones and Dangl 2006;Eitas and Dangl 2010).Recently studies suggest that some PAMPs and effectors are blurred (Thommaet al.2011),and PTI and ETI could collaborate to ensure robust defenses (Yuanet al.2021).
Like other plants,wheat also has multiple lines of defense againstF.graminearum(Walteret al.2010).For successful colonization,F.graminearummust overcome the first line of wheat defense,such as wax,cuticle,and cell wall (Kikotet al.2009).The wheat genome encodes a large number of receptor-like kinases,receptor-like proteins,or NLR proteins (IWGSC 2018).Some of them are effective against wheat pathogens (Figueroaet al.2018).For example,severalNLRgenes can recognize rust effectors,leading to ETI (Mapurangaet al.2022).This is a major type of resistance in wheat against these biotrophic pathogens (Cuiet al.2015;Gebrie 2016;Mapurangaet al.2022).Hence,wheat also has a strong immune system.Because no wheat accession immune to FHB is known yet,ETI againstF.graminearumis probably not present or has not evolved (Maet al.2020).Therefore,our current knowledge on wheat-induced resistance against FHB shall all be in the branch of PTI.The preformed defenses and PTI are often called basal defenses,which play essential roles in the fight against FHB.Except for these preformed physical and chemical barriers,PTI typically triggers a unique set of physiological responses,such as reactive oxygen species (ROS) production,callose deposition,biosynthesis of antimicrobial metabolites and defense hormones,and so on,to deal with the pathogen attacks (Zhang and Zhou 2010;Bigeardet al.2015).At present,five types of basal FHB resistance (type I to V) are recognized.Wheat resistance to initial infection,blight spreading,or mycotoxin accumulation is defined as type I,II,and III resistance,respectively (Mesterhazy 1995;Bai and Shaner 2004).
The breeding of FHB-resistant wheat is hampered by the quantitative nature of the known QTLs and limited knowledge of wheat-F.graminearuminteraction (Dwebaet al.2017;Xuet al.2022).Studying the defense mechanisms can provide alternative approaches for utilizing and improving the FHB resistance,in addition to the application ofFhbgenes themselves.Due to the conservation of PTI processes (Han 2019;Ngouet al.2022),many defense marker genes are employed to monitor different types of PTI.With the development of technologies,transcriptomics,proteomics,and metabolomics have been conducted on a few FHBresistant accessions to dissect the defense mechanisms of different FHB-resistant varieties.In the FHB-resistant landrace Wangshuibai,proteomic profiling suggests the ordered activation of Ca2+,salicylic acid (SA),and jasmonic acid (JA)/ethylene (ET) pathways are critical to its FHB resistance (Dinget al.2011);and a transcriptomics study reveals thatPR5,PR14,and JA signaling were crucial for its FHB resistance mediated byFhb1(Xiaoet al.2013).In the FHB-resistant Sumai 3,a previous metabolomics study suggests that its resistance is mainly associated with phenylpropanoid and flavonoid metabolites (Gunnaiah and Kushalappa 2014);the later transcriptomic analysis reveals that plant hormones,such as SA and JA,have a predominant role in orchestrating its FHB resistance (Wanget al.2018).RNA-seq analysis on FHB-resistant Nyubai,Wuhan 1,and HC374 suggests that the defense mechanisms are genotype-specific;and some GST,membrane proteins,and receptor kinases are likely the common components involved in FHB resistance (Panet al.2018).
Recently,efforts in breeding have invented a lot of excellent wheat varieties,and some of them exhibit better FHB resistance in agricultural practice.Xinong 979 (XN979),which is bred at Northwest A&F University,is one of those with moderate FHB resistance.It shows stable and lasting resistance against FHB in the ten years of cultivation in China.In addition to FHB,XN979 exhibits excellent resistance to wheat stripe rust.With high quality,high yield,and early maturity,XN979 has become one of the main varieties in Henan,Shaanxi,Hubei,and the rice-wheat rotation areas of Jiangsu and Anhui provinces (Shiet al.2019).However,the defense mechanisms of XN979 remain unclear.
Here,to study the defense mechanisms of XN979,we compared the transcriptomic differences between the FHB-resistant XN979 and the FHB-susceptible Xiaoyan 22(XY22),two genotypes with a pedigree of Xiaoyan 6.By studying their differences in the absence and presence ofF.graminearum,we found that the FHB resistance in XN979 consists of two lines of defense.Lignin pathway genes and JA biosynthesis genes likely contribute to the first line of defense.The second line of defense,which is induced uponF.graminearuminfection,is associated with the photosynthesis genes and biotic stress-responding genes.These findings facilitate our understanding of wheat-F.graminearuminteraction and may give insights into breeding FHB-resistant wheat.
2.Materials and methods
2.1.Plant inoculation
Fusarium graminearumstrain PH-1 was used for inoculation.The fungus was routinely cultured on potato dextrose agar plates at 25°C.The freshly harvested conidia,which grew for four days in carboxymethyl cellulose liquid medium with shaking at 175 r min-1,were suspended in ddH2O at a final concentration of 105conidia mL-1.The FHB-resistant cultivar XN979 and FHB-susceptible cultivar XY22 were sown in the field under natural conditions.At the anthesis stage,the spike of XN979 and XY22 were inoculated with 10 μL of conidial suspensions.Inoculated spikes were covered with a transparent bag for two days to maintain moisture.The development of the disease was assayed at 14 days post-inoculation.The first symptomless spikelet adjacent to the first weak bleaching spikelet was regarded as the early infection sample,while the bleached spikelet was regarded as the late infection sample.
2.2.RNA-seq analysis and identification of DEGs
Total RNAs were isolated with TRIZOL reagent.The library construction and RNA sequencing were performed at Novogene Bioinformatics Technology (Beijing,China),using paired-ends sequencing technology with Illumina HiSeq 2500.The quality of sequencing data was evaluated with FastQC.Only clean reads were used for analysis.
The sequencing reads (NCBI SRA database,PRJNA960823) from XN979 and XY22 were mapped to the wheat genome (IWGSC 2018),with hisat2 (Kimet al.2019),using the default parameters.The alignments were sorted and converted to bam files with SAMtools (Daneceket al.2021).Transcript quantification was performed with featureCounts (Liaoet al.2014),based on wheat genome annotation v1.1.
To evaluate the biological variability among samples,the multidimensional scaling cluster analysis was carried out with the count value of the digital gene expression profile,using the plotMDS function of edgeR (Robinsonet al.2010).The differentially expressed genes (DEGs) were also identified with this package.In brief,the “Trimmed Mean of M-values” method was used to normalize gene expression between samples.Genes with |log2Fold change (FC)|≥1,and false discovery rate (FDR)<0.05 were regarded as significantly differentially expressed.The multiple tests correction were performed with Benjamini-Hochberg (BH) algorithm.
2.3.Gene ontology enrichment analysis
To perform gene ontology (GO) annotation,three sources of GO terms were used.The first source of GO terms of wheat genes was assigned using the annotation from InterProScan (Joneset al.2014).The second source of GO annotation was retrieved on wheat genes using the MAPPER program of EggNOG (Cantalapiedraet al.2021).And the third wheat GO annotations were downloaded from the agriGO database (Tianet al.2017).After removing redundancy,the GO annotations from the three sources were integrated to obtain the final GO annotation of wheat.
The GO enrichment analysis was performed on the DEGs using the Parent-Child-Union method developed in Ontologizer (Baueret al.2008).The BH algorithm was used to correct the FDR (<0.05) of multiple tests for overexpressed GO terms.Revigo (Supeket al.2011) was used to reduce and visualize enriched GO terms.
2.4.MapMan analysis
The differences between the gene expression in XN979 and XY22 were studied with MapMan (Thimmet al.2004).The wheat pathway annotation files were generated with Mercator deposited in the plaBi database,with default parameters.Gene expression level was calculated with the transcripts per millions (TPM) method.The log2FC in the mock plant were calculated directly.To calculate the log2FC upon infection,the expression levels were first normalized by the corresponding mock expression.Pathway enrichment analysis was performed using the Wilcoxon test with an FDR below 0.05.Heatmaps were generated with the pheatmap package.
2.5.Gene expression analysis of F.graminearum genes
To compare the difference ofF.graminearumgenes when infecting XN979 and XY22,the conidia used for inoculation were sequenced using the same method of wheat sequencing.The genome and latest annotation ofF.graminearumwere downloaded from FgBase (Luet al.2022).The other downstream analyses,such as DEG identification and GO enrichment analysis were similar to that in wheat.Carbohydrate-active enzyme (CAZY) genes were predicted with the dbCAN3 server (Zhenget al.2023).Candidate secreted effector protein (CSEP) genes were identified as follows: First,secreted proteins (SP) with N-terminal signal peptides were precited with SignalP 6.0 (Teufelet al.2022);second,SP with transmembrane domains predicted by DeepTMHMM (Hallgrenet al.2022) were excluded;third,the resulted proteins were used to feed EffectorP 3.0 (Sperschneider and Dodds 2022) to obtain the CSEP.The secondary metabolite (SM) genes were generated previously by Sieberet al.(2014).
2.6.Lignin staining and JA contents measurement
Lignin staining was performed with phloroglucinol.For JA content measurement,GC-MS was employed.Briefly,the lemmas were grounded in liquid nitrogen and dissolved in ethyl acetate with an ultrasonic bath for 30 min.After drying with a vacuum thickener,the extracts were derivatized withN-methyl-N-(trimethylsilyl)trifluoroacetamide (MSTFA) and dissolved inn-hexane.Finally,1 μL sample was injected with splitless mode to the GC-MS device (GCMS-QP2010,Shimadzu,Japan) with a Rxi-5ms column (30 m×0.25 mm×0.25 μm).Helium (>99.999%) was used as the carrier gas at a constant flow rate of 1 mL min-1.The oven program was set as follows: 80°C for 10 min and then 2°C min-1to 150°C,holding for 1 min,and finally to 270°C at a rate of 25°C min-1and maintained for 9 min.Injection,ion source,and interface temperatures were kept at 200,300,and 280°C,respectively.The parental ion ofm/z282 [M-SiC3H9]-with a retention time of 48.82 min was used for JA quantitation.
3.Results
3.1.Limited blight spreading and infection-responsive genes in XN979
To confirm the FHB resistance of XN979 observed in natural fields,we inoculated the heads of XN979 (FHBresistant) and XY22 (FHB-susceptible),and compared the severity of FHB disease on these two cultivars.After a two-week infection,about 10 spikelets were bleached in XY22.In comparison,only 1-2 spikelets were bleached in XN979 (Fig.1-A).The significantly slower spreading ofF.graminearumin XN979 (Fig.1-B) indicates that XN979 has a stronger type II resistance against FHB.
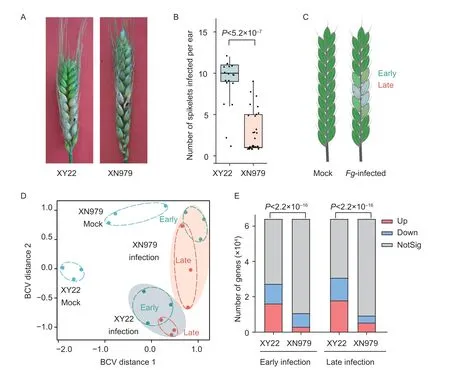
Fig.1 XN979 is resistant to Fusarium graminearum infection.A,the phenotype of wheat cultivar XY22 and cultivar XN979 inoculated with F.graminearum.The black point indicates the inoculation site.Fusarium graminearum strain PH-1 was used for inoculation,and photos were taken 14 days post-inoculation.B,statistics on the number of infected spikelets on the XY22 and XN979.Each point represents a spike.Student’s t-test was used to evaluate the significance.C,a schematic diagram shows the sampling of the RNA-seq.Each infected spike actually has both early (the frontiers of infection) and late (the heavily infected spikelets) infected tissues.The symptomless and bleached spikelets,representing different degrees of F.graminearum (Fg)-infection,were taken as early and late infected tissues,respectively.D,the multidimensional scaling plot illustrates the biological variation among XN979 and XY22 samples.Each dot represents a biological replicate of the RNA-seq.BCV,biological coefficient of variation.E,statistics on the number of differentially expressed genes between infected and mock samples.The upregulated (up),downregulated (down),and not significant (NotSig) genes were shown in red,blue,and grey,respectively.The significances are calculated based on X2 tests.
To dissect the difference between XN979 and XY22 at the transcriptomic level,we sampled the early and late infected spikelets,as well as mock spikelets,for RNA-seq analysis (Fig.1-C).In all the samples,about 85% of the clean reads were aligned to wheat genome (Appendix A).Multidimensional scaling plot (Fig.1-D) showed that the samples in XN979 and XY22 obviously differ,whenever in the uninfected or infected samples.This data indicates the distinct transcriptomic responses to FHB in these two cultivars.
To compare the FHB-responsive genes in these two cultivars,we identified the DEGs of XN979 and XY22 uponF.graminearuminfection,respectively (Appendices B-E).Among the 63,334 genes expressed,only 14-17% were differentially expressed in XN979.In contrast,43-48% of them in XY22 were identified as DEGs (Fig.1-E).The fewer DEGs in XN979 may result from the resistant cultivar being less affected byF.graminearum.Interestingly,this phenomenon was observed even in the late infection,suggesting that the limited FHB-responsive genes are the nature of FHB-resistant XN979.
3.2.XN979-specific FHB-responsive genes are associated with plant defense
To find out whether the FHB-responsive genes in XN979 contributes to the FHB resistance,we identified XN979-specificF.graminearum-induced genes (Fig.2-A-C) and performed GO enrichment analysis (Fig.2-D-E;Appendices F-H).Interestingly,we found that the GO term “response to vitamin B1” was significantly enriched in early infection (Fig.2-D).Close examination showed that the genes associated with this GO term arePR1,which is recognized as a marker gene for plant defense.The data suggest that effective early defense has been activated during the infection of XN979.

Fig.2 Identification and GO enrichment analysis of the Fusarium head blight (FHB)-responsive genes in XN979.A,Venn diagram shows the XN979-specific upregulated genes in the early infection.B,Venn diagram shows the XN979-specific upregulated genes in the late infection.C,Venn diagram shows the XN979-specific upregulated genes in both early and late infection.“↑” and “↓” represent the upregulated and downregulated genes,respectively.The area of the Venn diagram highlighted in red indicates the XN979-specific responsive genes against FHB.D,GO enrichment analysis of XN979-specific upregulated genes in the early infection.E,GO enrichment analysis of XN979-specific upregulated genes common in early and late infection.Representative GO terms were plotted in a two-dimensional semantic space by REVIGO.The bubble color indicates the adjusted P-value;the bubble size indicates the frequency of the GO term in the underlying GO annotation database.
Among the 828 XN979-specificF.graminearuminduced genes in the early infection (Fig.2-A) and 1,622 XN979-specificF.graminearum-induced genes in the late infection (Fig.2-B),246 of them were commonly induced in both stages (Fig.2-C).GO enrichment analysis on these commonly induced genes showed that genes associated with photosynthesis and polyamine metabolism are significantly enriched (Fig.2-E).Polyamines are known to induce DON production inF.graminearum(Gardineret al.2010;Rochaet al.2020;Tanget al.2021).The upregulation of these polyaminedegrading genes may reduce the accumulation of polyamines in XN979,result in less toxin induction,and prevent the spreading of FHB blight.In addition,the upregulation of photosystem-related genes may provide XN979 with more energy and ROS to further reinforce the defense againstF.graminearuminfection.These results suggest that the XN979-specific FHB-responsive genes are associated with plant defense.
3.3.FHB resistance in XN979 consists of two lines of plant defense
To systematically compare the responses of these two cultivars challenged toF.graminearum,we performed MapMan analysis (Appendix I).Multiple Wilcoxon tests corrected by the Benjamini-Hochberg procedure were employed to assess the significance of pathway expression differences between XN979 and XY22.Surprisingly,we found that their differences in FHB resistance may have two layers of source (Fig.3-A).One layer of the difference lies in the basal expression of mock plants in the absence of pathogen (basal difference),such secondary metabolism,signaling,hormone metabolism,and lipid metabolism pathways.Another layer of the difference lies in the FHB-responsive genes that are specifically affected duringF.graminearuminfection (infection-induced difference),such as photosystem,tetrapyrrole synthesis,redox,and mitochondrial electron transport pathways.These two layers of difference may form two lines of defense that contribute to the FHB resistance in XN979.

Fig.3 Overview of the differentially expressed pathway and the expression of lignin biosynthesis genes in XN979 and XY22.A,a summary of the MapMan analysis.Adjusted P-values of Wilcoxon tests were mapped from red (low) to white (high).B,the expression of lignin biosynthesis genes in XN979 and XY22.Each heatmap illustrates the gene expression of the designated pathway/genes.Gene expression levels,as measured by transcripts per million (TPM),were mapped from black to red to represent lower to higher expression.The six columns of the heatmap are the mock,early,and late infection of XN979 and XY22,respectively.A green dot in the left bottom corner of the heatmap represents the gene expression is significantly different in XN979 and XY22 in the labeled samples.Light green represents adjusted P<0.05,and the dark green represents adjusted P<10-5.PAL,phenylalanine ammonia-lyase;C4H,cinnamate-4-hydroxylase;4CL,4-coumarate-CoA ligase;HCT,hydroxycinnamoyl transferase;C3H,coumarate-3-hydroxylase;CCoAOMT,caffeoyl-CoA 3-O-methyltransferase;CAD,cinnamyl alcohol dehydrogenase;CCR,cinnamoyl-CoA reductase;COMT,caffeoyl O-methyltransferase;F5H,ferulate 5-hydroxylase.
3.4.Enhanced expression of basal defense genes in XN979 occurs in the absence of pathogen
To study the first layer of difference,we firstly investigated the secondary metabolism bin that significantly differs in XN979 and XY22 mock plants (Fig.3-A).Close examination showed that lignin biosynthesis is the most significant pathway under this bin (Appendix I;Fig.3-B).Lignin is important basal defense compound that acts as a physical barrier of pathogen attack.Interestingly,we found that thePHENYLALANINE AMMONIA-LYASE(PAL) andCINNAMATE-4-HYDROXYLASE(C4H) genes,which are required for the first two steps of lignin biosynthesis,exhibit a higher basal expression level in XN979 than that in XY22 (Fig.3-B).TwoArabidopsiscinnamyl alcohol dehydrogenase (CAD),CAD5 and CAD4,has the highest level of catalytic activity and is required for the final step of lignin monomers biosynthesis.Its ortholog genes in XN979 also exhibited high basal expression levels (Fig.3-B).The enhanced expression of these lignin biosynthesis genes may lead to a more robust basal defense,thus reduce the early colonization of pathogens and spreading of the blight.
Secondly,among the bins of hormone metabolism,JA metabolism is the most significant pathway in XN979 and XY22 mock plants (Appendix I).To investigate which branches were differentially expressed between these two cultivars,we examined the gene expression in JA biosynthesis,conjugation,transport,and signaling pathways (Fig.4).Only JA biosynthesis genes showed a significant difference in the XN979 and XY22 mock plants.Expanded analysis showed that the first two genes,LIPOXYGENASE(LOX) andALLENE OXIDASE SYNTHASE(AOS),which are involved in JA biosynthesis in the chloroplast,exhibited higher basal expression levels in XN979.The enhanced expression of these JA biosynthesis genes may contribute to the enhanced basal defense in XN979.

Fig.4 The expression of jasmonic acid (JA) pathway genes in XN979 and XY22.Each heatmap illustrates the gene expression of the designated pathway/genes.Gene expression levels,as measured by transcripts per million (TPM),were mapped from black to red to represent lower to higher expression.The six columns of the heatmap are the mock,early,and late infection of XN979 and XY22,respectively.A green dot in the left bottom corner of the heatmap represents the gene expression is significantly different in XN979 and XY22 in the labeled samples.Light green represents adjusted P<0.05,and the dark green represents adjusted P<10-5.Chemical abbreviations: α-LeA,α-linolenic acid;13-HPOT,(13S)-hydroperoxy octadecatrienoic acid;12,13-EOT,12,13(S)-epoxy-(9Z,11E,15Z)-octadecatrienoic acid;OPDA,12-oxo-phytodienoic acid;OPC8,3-oxo-2(2´[Z]-pentenyl)-cyclopentane-1-octanoic acid;JA,jasmonic acid;JA-Ile,jasmonoyl-L-isoleucine.Protein abbreviations: LOX,lipoxygenase;AOS,allene oxidase synthase;AOC,allene oxidase cyclase;JASSY,oxophytodienoate export protein;ACS,OPC8-CoA synthetase;PXA1,peroxisomal ABCtransporter 1;OPR3,oxophytodienoate reductase;ACX1,OPC8-CoA oxidase;ACH,acyl-CoA thioesterase;JOX/JAO,jasmonic acid oxidase;JAR1,jasmonoyl-amino acid synthetase 1;JAT,jasmonic acid transporter;COI1,coronatine insensitive 1;JAZ,jasmonate ZIM-domain protein;CYP94B,jasmonoyl-amino acid hydroxylase;CYP94C,jasmonoyl-amino acid carboxylase.
To verify whether the constitute enhancement of lignin biosynthesis genes in XN979 resulted in more lignin accumulation,we performed Weisner staining assays on the lemma of XN979 and XY22 withoutF.graminearuminfection.Consistently,the stained reddish area in XN979 is much more than that in XY22 (Fig.5-A).To examine if the enhanced expression of JA biosynthesis genes in XN979 leads to an elevation of JA content,we employed gas chromatography-mass spectrometry (GC-MS) to measure the content of JA in the lemma of both cultivars absentF.graminearuminfection.Again,we found that the JA level is significantly higher in XN979 than in XY22 (Fig.5-B and C).These data further support that XN979 has a constitutively enhanced basal defense.

Fig.5 XN979 exhibits a constitutively enhanced basal defense.A,Weisner staining shows the lignin content of the lemma of XN979 and XY22.B,representative chromatograms show the jasmonic acid (JA) contents of the lemma of XN979 and XY22.The parental ion of JA (m/z 282 [M-SiC3H9]-) with a retention time (RT) at 48.82 min is indicated.C,relative JA level in the lemma of XN979 and XY22.Bars represent the mean±SD of five biological replicates (n=5),and the mean of JA content in XY22 was set to 1.**,P-value<0.01 (Student’s t-test).
3.5.Infection-induced expression of chloroplast pathways may reinforce FHB resistance in XN979
To investigate the secondary layer of difference,we analyzed the photosystems (PS),which is the most significant bin in infection-induced resistance (Fig.3-A).PS is the functional unit for photosynthesis.It converts light into chemical energy,acts as a major source of ROS inside plant cells,and positively regulates the plant defense (Padmanabhan and Dinesh-Kumar 2010;Suet al.2018).Analysis of the gene expression in lightreaction (Fig.6) showed that they are comparable between XN979 and XY22 in the absence ofF.graminearum.After infection,the majority of lightreaction genes were suppressed.This is consistent with previous studies (Yanget al.2013;Yang and Luo 2021) showing the general suppression of photosynthesis in plants challenged withF.graminearum.However,the suppression of some of the lightreaction genes,such asPSB(core of PS II),LHCA(light-harvesting complex I),PSA(core of PS I),andFd(ferredoxin) genes,were significantly weaker in XN979 than that in XY22 (Fig.6),suggesting that the photosystems in XN979 may still work better underF.graminearumattack.Similar hypo suppression was also observed in Calvin cycle genes (Fig.6).

Fig.6 The expression of photosystem genes in XN979 and XY22.Each heatmap illustrates the gene expression of the designated pathway/genes.Gene expression levels,as measured by transcripts per million (TPM),were mapped from black to red to represent lower to higher expression.The six columns of the heatmap are the mock,early,and late infection of XN979 and XY22,respectively.A green dot in the left bottom corner of the heatmap represents the gene expression is significantly different in XN979 and XY22 in the labeled samples.Light green represents adjusted P<0.05,and the dark green represents adjusted P<10-5.LHCB,lightharvesting complex II;PSB,core protein of PSII;Cyt b6/f,cytochrome b6f complex;LHCA,light-harvesting complex I;PSA,core protein of PSI;PC,plastocyanin;Fd,ferredoxin;FNR,ferredoxin-NADP+ oxidoreductase;OEC,oxygen-evolving complex;PQ,plastoquinone.
Interestingly,theLHCB(light-harvesting complex II) genes,which had a relatively poor expression in the mock plants,were upregulated in response toF.graminearuminfection (Fig.6).LHCB binds chlorophyll directly and acts as the antenna to absorb photons to drive photosynthesis.The heavy induction of theseLHCBgenes in XN979 may further support the running of the photosystem machinery,and thus provide sufficient infection-induced resistance against FHB.We noticed that tetrapyrrole synthesis,which leads to the synthesis of chlorophyll,was also a component of infection-induced resistance (Fig.3-A).Similar to the bin of PS,most genes involved in this pathway were generally suppressed in infection,but to a less extent in XN979 (Fig.7).Some of thePROTOCHLOROPHYLLIDE REDUCTASEgenes,which make the precursor chlorophyll directly,exhibited a consistent upregulation in the early and late infection of XN979 (Fig.7).Therefore,it seems there is much more chlorophyll available during the infection of XN979.The synergy of LHCB and chlorophyll may reinforce FHB resistance in XN979 in the chloroplast.

Fig.7 The expression of tetrapyrrole biosynthesis genes in XN979 and XY22.Each heatmap illustrates the gene expression of the designated pathway/genes.Gene expression levels,as measured by transcripts per million (TPM),were mapped from black to red to represent lower to higher expression.The six columns of the heatmap are the mock,early,and late infection of XN979 and XY22,respectively.A green dot in the left bottom corner of the heatmap represents the gene expression is significantly different in XN979 and XY22 in the labeled samples.Light green represents adjusted P<0.05,and the dark green represents adjusted P<10-5.GluRS,glutamyl-tRNA synthetase;GluTR,glutamyl-tRNA reductase;GSA-AT,glutamate-1-semialdehyde-2,1-aminomutase;ALA dehydratase,5-aminolaevulinic acid dehydratase;PBG deaminase,porphobilinogen deaminase;Mg-proto IX MT,magnesium protoporphyrin IX methyltransferase;Mg-proto IX ME cyclase,magnesium-protoporphyrin IX monomethyl ester (oxidative) cyclase;PΦB synthase,phytochromobilin synthase.
3.6.Struggles of XN979 against F.graminearum infection in the biotic stress pathway
To detect whether the genes related to biotic stress were differentially expressed in XN979 and XY22 afterF.graminearuminfection,we further analyzed the biotic stress pathway (Fig.8).Although the basal expression of these biotic stress genes was somewhat different,they were generally suppressed uponF.graminearuminfection,even in the FHB-resistant XN979.This is consistent with that both XN979 and XY22 are finally infected after inoculation.

Fig.8 The expression of biotic pathway genes in XN979 and XY22.Each heatmap illustrates the gene expression of the designated pathway/genes.Gene expression levels,as measured by transcripts per million (TPM),were mapped from black to red to represent lower to higher expression.The six columns of the heatmap are the mock,early,and late infection of XN979 and XY22,respectively.A green dot in the left bottom corner of the heatmap represents the gene expression is significantly different in XN979 and XY22 in the labeled samples.Light green represents adjusted P<0.05,and the dark green represents adjusted P<10-5.BR,brassinolide;ABA,abscisic acid;ET,ethylene;SA,salicylic acid;JA,jasmonic acid;GST,glutathione-S-transferase;NLR,nucleotide-binding,leucine-rich repeat proteins;MAPK,mitogen-activated protein kinases;PR proteins,pathogenesis-related proteins;ERF,ethylene response factors;bZIP,basic leucine zipper;DOF,DNA binding with one finger.
However,some of the biotic stress genes in XN979 still seemed to struggle withF.graminearumin the early or late infection.For example,some of theNLRgenes,which are essential for recognizing pathogen avirulent effectors that lead to plant immunity,were less suppressed in XN979 (Fig.8).This is reminiscent of RNA-seq analysis of infection byosp24mutant.OSP24encodes an effector that is required for the full virulence ofF.graminearuminfection.When knockout,collective upregulation of wheatNLRgenes also occurred upon its infection (Jianget al.2020).Currently,no FHB immunity germplasm is available.Whether the collective lower suppression ofNLRgenes,or their concerted expression,could crosstalk with PTI (Yuanet al.2021) to fight againstF.graminearum,remains unclear.
Although the battlefield of XN979 seems a mess (Fig.8),some of the effective defenses may have occurred.For example,consistent with our analysis above,some of thePRgenes,as well as secondary metabolism (e.g.,lignin) genes,were all upregulated in XN979 during infection (Fig.8).The genes related to defense hormones are also likely to combat withF.graminearum(Fig.8).Unlike JA-and BR-related genes that had basal differences,a few of ET-related genes,and someERFtranscript factor genes,were likely to try to change the interaction of wheat-F.graminearum.In addition,some genes involved in oxidative defense,heat shock,proteolysis,and cell wall defense could also help the struggles of XN979 againstF.graminearuminfection.
3.7.Many infection-related genes of F.graminearum were compromised in the early infection of XN979
Examinations of the blight development and gene expression in these two cultivars consistently indicate that XN979 has an effective defense againstF.graminearuminfection.To find if there is evidence on the pathogen side supporting these observations,we identifiedF.graminearumgenes upregulated during infection,by comparison to the conidia used for inoculation (Fig.9-A).Interestingly,we found that the number ofF.graminearumupregulated genes is significantly lower in XN979 than that in XY22,especially in the early infection (Fig.9-A).This suggests thatF.graminearumfailed to,or delayed to,express the infection-related genes,possibly due to the pathogen had encountered a harsh interacting environment from the host plant.

Fig.9 Fusarium graminearum gene expression during infection of XN979 and XY22.A,the proportion of upregulated (up) and downregulated (down) F.graminearum genes in the colonization of XN979 and XY22.The significances are calculated with X2 tests.B,GO enrichment analysis of the upregulated fungal genes during wheat infection.Only the GO terms with an enrichment fold>2 are shown.BP,biological process;CC,cellular component;MF,molecular function.C,the expression of F.graminearum CAZY genes in different samples.D,the expression of F.graminearum CSEP genes in different samples.E,the expression of F.graminearum SM genes in different samples.TRI genes and fusaoctaxin A biosynthesis genes,encoding two known chemical virulence factors for plant infection,are shown separately.The dendrogram highlighted in red indicates that the expression pattern is consistent with cluster IV.C-E,the color key from green to purple represents the gene expression levels (in transcripts per millions (TPM)) from low to high.
GO enrichment analysis revealed that the products of these upregulated genes were mainly located in the extracellular region,or involved in the carbohydrate metabolic process (Fig.9-B).We,therefore,hypothesized that some of the extracellular proteins,such as CAZY and CSEP which important forF.graminearuminfection (Wanget al.2017),may compromise in expression when infecting XN979.Subsequent analyzing the expression patterns of the CAZY and CSEP genes showed that both of them formed five distinct clusters (Fig.9-C and D).As we hypothesized,a substantial of them in cluster IV (93 of the 547 expressed CAZY genes and 35 of the 365 expressed CSEP genes) failed to upregulation when infecting XN979.Secondary metabolites,such as the mycotoxins ofF.graminearum,are also important extracellular virulence factors.Analyzing the expression patterns revealed that 128 (cluster IV) of the 738 expressed SM genes failed to upregulation in the early infection of XN979 (Fig.9-E).DON mycotoxin is important for FHB spreading in rachis (Baiet al.2002),and fusaoctaxin A is required for the cell-to-cell invasion (Jiaet al.2019).Examinations of the expression of trichothecene (TRI) and fusaoctaxin A biosynthesis genes confirmed that these secondary metabolites genes explicitly required forF.graminearuminfection were compromised in the early infection of XN979 (Fig.9-E).
4.Discussion
Breeding FHB-resistant wheat is the persistent pursuit of green,sustainable agriculture.Due to the limited FHB resistance resource,studying and utilizing the defense mechanisms is critical for improving FHB-resistant traits.In this study,we compared the blight development and transcriptomic response of XN979 and XY22 challenged withF.graminearum,and found that XN979 does have a slower FHB spreading in spike and fewer DEGs.Functional enrichment analysis on the XN979-specific FHB-responsive genes showed they are associated with plant defense.Therefore,XN979 is FHB-resistant in terms of blight spreading and transcriptomic response.Pathway-to-pathway comparison of the transcriptomic difference revealed that two lines of defense were enhanced in XN979 (Fig.10).

Fig.10 A proposed model for Fusarium head blight (FHB) resistance in XN979.The FHB resistance in XN979 consists of two lines of defense.The first line of defense is constitutive that preformed in the uninfected XN979.It mainly comprises lignin (thickened cell wall) and JA-based defense.The second line of defense is induced upon F.graminearum infection.In susceptible XY22,suppression of wheat chloroplast-based defense,possibly by fungal effectors,leads to the bleaching of the spikelet,and finally formed head blight of wheat.However,in resistant XN979,the hypo suppression of photosynthesis genes formed an infection-specific defense.These two lines of defense enhanced the type II resistance in XN979 and made the failure induction of F.graminearum infection-related genes,especially in the early infection stage.DON,deoxynivalenol.
4.1.Lignin-and JA-based constitutive defenses are enhanced in XN979 without infection
Lignin is an essential component of the cell wall that acts as a physical barrier preventing the spread of pathogens (Sattler and Funnell-Harris 2013;Leeet al.2019).The expression of lignin biosynthesis genes,such asPAL,C4H,COUMARATE-3-HYDROXYLASE(C3H),andCAD4/5,were constitutively higher in resistant XN979 than that in the susceptible XY22.PAL is the first ratelimiting enzyme in the lignin synthesis pathway (Hahlbrock and Scheel 1989) and plays a role in both type I and type II resistances (Wuet al.2022).CAD4 and CAD5 have been shown to play major roles in the lignin biosynthesis (Kimet al.2004).These higher levels of lignin biosynthesis genes may strengthen the basal lignin level before infection,and promote efficient lignin reinforcement during infection,to enhance FHB resistance in XN979.
Studies on Sumai 3 showed that its FHB resistance is mainly associated with phenylpropanoid metabolites (Paranidharanet al.2008;Gunnaiah and Kushalappa 2014).In Sumai 3 derived FHB-resistant CM82036,which carriesFhb1andQfhs.ifa-5A,PALis strongly upregulated at 72 h post-infection (Steineret al.2009).Microarray analysis also revealedPALis upregulated in FHB-resistant double haploid line GS-1-EM0040 at 8 h post-infection (Foroudet al.2012).Immunogold labeling of lignin showed that,after infection,the lignin content in FHB-resistant cultivars Arina and Frontana is significantly higher than that in FHB-susceptible cultivar Agent (Kang and Buchenauer 2000).These studies consistently indicate that the lignin biosynthesis pathway is important for defending FHB,similar to what happened in XN979.
It should be noted that,in XN979,the higher expression ofPALhas already occurred in the absence of the pathogen.According to the expression ofPALgenes in XN979 and XY22,lignin biosynthesis is constantly enhanced in uninfected,early-infected,and late-infected samples of XN979 (Fig.10).The enhanced ligninbased physical barrier may be the major defense that hinders pathogen spreading in the spike of XN979.This constitutive defense may be the nature of XN979,and its rust resistance may also be contributed by the ligninbased physical defense.The expression elevation of multiple genes in lignin biosynthesis is unlikely to be random.Its formation probably coincided with the breeding selected for lodging resistance trait positively associated with lignin level.
Like in the lignin pathway,the first two genes required for JA biosynthesis,LOXandAOS,were simultaneously higher in FHB-resistant XN979.Traditionally,JA is believed to be a defense hormone against necrotrophic pathogens (Bari and Jones 2009).However,increasing evidence suggests that it is also involved in defending against biotrophic and hemibiotrophic pathogens (Anticoet al.2012;Guptaet al.2020).For example,in rice,JA is required to protect against hemibiotrophicMagnaporthe oryzae(Riemannet al.2013).JA can directly inhibit the growth ofF.graminearum,and in particular,in wheat,the application of JA could reduce the spreading of FHB (Qiet al.2016).Hence JA is most likely to positively regulate wheat defense against this hemibiotrophic pathogen (Trail 2009;Zhang X Wet al.2012;Qiuet al.2019).In line with this evidence,microarray and qRT-PCR analysis in Sumai 3 found that bothLOXandAOSgenes were FHBresponsive (Li and Yen 2008).Therefore,the constitutive enhanced JA biosynthesis in the mock plant of XN979 may also be necessary for the FHB resistance (Fig.10).
During infection,the constitute expression of some JA biosynthesis genes seems to be suppressed,similar to the JA biosynthesis suppression observed inBrachypodium distachyon-F.graminearuminteraction (Zhuet al.2021).The hormone pathways,including the JA pathway,are frequently manipulated by pathogen effectors (Patkar and Naqvi 2017;Han and Kahmann 2019).Thus,it is possible thatF.graminearumhad changed the behaviors of host JA defense during infection,which is not overcome in XN979.Therefore,although XN979 may have a richer constitutive JA level,it may only be effective at the initial infection,and its effects may be partially masked afterF.graminearuminfection.
4.2.Chloroplast-based inducible defense is enhanced in XN979 during infection
Chloroplast is an organelle responsible for photosynthesis,which captures light energy to make sugars.It is also crucial for plant defense,since it is the place for synthesizing defense phytohormone SA and JA (Kangasjarviet al.2012;Medina-Pucheet al.2020).The lightreaction of photosynthesis,specifically photosystem II,is a primary ROS production site in green plants and is a critical component of early defense during infection (Göhre 2015;de Torres Zabalaet al.2015;Xuet al.2019).Some pathogens have evolved effectors to target chloroplast defense (de Torres Zabalaet al.2015;Xuet al.2019;Medina-Pucheet al.2020).Therefore,chloroplast is now recognized as a battlefield for plant-pathogen interaction (Littlejohnet al.2020;Medina-Pucheet al.2020;Kachrooet al.2021;Yanget al.2021).Generally,the maintenance of normal photosynthesis is crucial for plant defense (Huotet al.2014).
Analysis of the gene expression revealed that the photosynthesis was overall inhibited after infection of XN979 and XY22,suggesting the photosynthesis is successfully inhibited byF.graminearuminfection,whether through active suppression coordinated by growth-defense trade-off (Huotet al.2014),or passive suppression by pathogen effectors (de Torres Zabalaet al.2015;Xuet al.2019).The overall suppression of lightreaction,Calvin cycle,and chlorophyll biosynthesis are highly consistent with the hallmark of FHB -the bleaching of infected spikelets (Fig.10).It is also compatible with the fact that FHB is more severe in rainy conditions where photosynthesis may be interrupted in nature.Notably,our study found that the suppression of photosynthesis is limited in XN979.This is supported again by the GO enrichment analysis of the FHBresponsive genes,which showed that XN979 might have more robust photosynthesis in the early infection of XN979.Therefore,our study is consistent with the fact that photosynthesis has a role in defending against pathogen infection,and the limited photosynthesis suppression in infection of XN979 contributed to the FHB resistance (Fig.10).
Both GO enrichment and MapMan analyses suggested that the light-harvest process might be enhanced in XN979.AfterF.graminearuminfection,theLHCBgene was dramatically upregulated in XN979.The LHCB binds chlorophyll and is the antenna in lightreaction,directly capturing photons to drive photosynthesis.In rice,light-induced phosphorylation of LHCB5 contributed to broad-spectrum resistance againstMagnaporthe oryzaeinfection,viaROS accumulation (Liuet al.2019).Therefore,the lightreaction is likely to be boosted byLHCBduring the infection of XN979,and it then enhances photosynthesis or generates more ROS,to form the inducible defense againstF.graminearuminfection.On rainy days which FHB favors,this chloroplast baseddefense is especially pronounced,since clouds mask the sunlight required for photosynthesis.The formation of the enhanced photosynthesis gene expression in XN979 probably coincided with the breeding selected for highyield traits.
4.3.Hints from XN979 for FHB-resistance breeding
Our findings are insightful into the breeding of FHBresistant wheat.First,breeding wheat with both high yield and FHB resistance is possible.The formation of the FHB resistance in XN979 might coincide with the breeding strategy,such as selecting high grain yield and lodging resistance traits.In breeding practices,selecting wheat lines with higherPALandLHCBexpression may result in higher yields and well FHB-resistance.Second,breeding wheat with DON resistance is possible.Polyamines are inducers of DON production inF.graminearum(Gardineret al.2010;Rochaet al.2020;Tanget al.2021).The early infected XN979 were enriched in genes related to polyamine catabolism,and consistently,we found thatF.graminearumshowed a failure induction of DON biosynthesis genes.Therefore,selecting wheat lines with high polyamine degradation rates may enable the breeding of mycotoxin-free wheat.
5.Conclusion
XN979 is a widely cultivated wheat elite with high yield and FHB resistance.This study showed that its FHB resistance consists of two lines of defense.The first line of defense is constitutive,knitted by higher expression of lignin-and JA-related genes.The second line of defense is inducible,mainly contributed by limited photosynthesis suppression.These defenses lead to a compromised expression ofF.graminearuminfection-related genes,such as SM,CAZY,and CSEP genes.This study will facilitate the understanding of wheat-F.graminearuminteraction and is insightful for breeding FHB-resistant wheat.
Acknowledgements
We thank Ping Xiang (Northwest A&F University,China) for help with the analysis of JA contents,and Drs.Cong Jiang,Guanghui Wang,and Ming Xu (Northwest A&F University) for fruitful discussions.This work was supported by the grants from the National Key R&D Program of China (2022YFD1400100),the National Natural Science Foundation of China (32072505 and 31701747),the Chinese Universities Scientific Fund (2452020222),and the National Innovation and Entrepreneurship Training Program for College Students,China (202110712255).
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendicesassociated with this paper are available on https://doi.org/10.1016/j.jia.2023.06.014
 Journal of Integrative Agriculture2024年4期
Journal of Integrative Agriculture2024年4期
- Journal of Integrative Agriculture的其它文章
- OsNPF3.1,a nitrate,abscisic acid and gibberellin transporter gene,is essential for rice tillering and nitrogen utilization efficiency
- Fine mapping and cloning of the sterility gene Bra2Ms in nonheading Chinese cabbage (Brassica rapa ssp.chinensis)
- Optimized tillage methods increase mechanically transplanted rice yield and reduce the greenhouse gas emissions
- A phenology-based vegetation index for improving ratoon rice mapping using harmonized Landsat and Sentinel-2 data
- Combined application of organic fertilizer and chemical fertilizer alleviates the kernel position effect in summer maize by promoting post-silking nitrogen uptake and dry matter accumulation
- miR-24-3p promotes proliferation and inhibits apoptosis of porcine granulosa cells by targeting P27
