一株多药耐药Comamonas kerstersii细菌的基因组分析(英文)
王慧 明德松 王明席
Genomic Insights of A Multi-Drug Resistant Comamonas kerstersii Isolate
WANG Hui1, MING Desong2, WANG Mingxi1
( 1. School of Medicine, Huaqiao University, Xiamen 361021, China;
2. Quanzhou First Hospital Affiliated to Fujian Medical University, Quanzhou 362000, China )
Abstract: A multi-drug resistant C. kerstersii 121606 was isolated from a patient with a urinary tract infection, and the antimicrobial susceptibility testing (AST) and whole genome sequencing were performed, then comparative genomic analyses were conducted with seven representative Comamonas species strains and Acidovorax species by average nucleotide identity (ANI) analysis using OrthoANI and single nucleotide polymorphism (SNP) analysis by snpTree web server. Finally, genomic sequence annotation was conducted with RAST server, as well as functional annotation of orthologous clusters by OrthoVenn, prediction of antibiotic resistance genes (ARGs) by CARD, CRISPRs by CRISPR recognition tool, and prophages by PHAST. These data revealed that C. kerstersii 121606 was a multi-drug resistant bacterium with similar genetic components to other seven Comamonas species and Acidovorax species. The presence of ARGs in its genome helped to explain its multi-drug resistance mechanisms. These findings provide valuable insights for the study of novel antibiotics to manage multi-drug resistant C. kerstersii infections.
Keywords:Comamonas kerstersii; multi-drug resistance; comparative genomic analysis; antibiotic resistance gene; CRISPR; prophage
CLC Number: 中圖分类号:Q 939.48; R 378文献标志码:Document Code: A Article Number: 文章编号:1000-5013(2024)01-0035-12
摘要:从1名尿路感染患者中分离出了1株多药耐药的Comamonas kerstersii (C. kerstersii)菌株121606,对其进行了抗微生物药敏试验(AST)和全基因组测序;然后将其与7个具有代表性的Comamonas菌株和Acidovorax菌种进行基因组比较分析,包括使用OrthoANI分析平均核苷酸同一性(ANI),以及通过snpTree网络服务器进行单核苷酸多态性(SNP)分析。最后,使用RAST服务器进行基因组序列,使用OrthoVenn软件对同源簇进行功能注释,通过CARD数据库对抗生素耐药基因(ARGs)进行预测,并利用CRISPR识别工具预测CRISPR,以及利用PHAST软件预测前噬菌体。结果表明:C. kerstersii 121606是一种多药耐药细菌,其遗传成分与其他7个Comamonas和Acidovorax菌种相似;其基因组中存在的ARGs有助于解释其多药耐药机制。这些发现为研究新型抗生素来控制多药耐药C. kerstersii感染提供了有价值的见解。
关键词:Comamonas kerstersii; 多药耐药性; 比较基因组分析; 抗生素耐药性基因; CRISPR; 前噬菌体中图分类号:
Comamonas kerstersii (C. kerstersii) is a gram-negative, aerobic, motile rod bacterium[1] that mainly caused the gastrointestinal tract infections[2-4]. It is rarely resistant to antibiotics[2, 5]. However, we isolated a multi-drug resistant strain, 121606, from the urine of an 82-year-old male patient with a urinary tract infection secondary to prostatic hyperplasia. This strain was resistant to 18 out of 21 antibiotics, including β-lactams, aminoglycosides, quinolones, sulfonamide and colistin, indicating that it was a multi-drug resistant strain. The property of multi-drug resistance of C. kerstersii 121606 has not been not reported. As the infections of C. kerstersii are rapidly increasing, it is imperative to understand the genomic feactures and the antibiotic resistance mechanisms of C. kerstersii strain 121606 for better management of the increasing C. kerstersii infections. To achieve these purposes, in this paper, we performed genomic sequencing for strain 121606, conducted comparative genomic analyses, and correlated the antibiotic resistance genes (ARGs) in the genome with its multi-drug phenotype.
1 Materials and Methods
1.1 Bacterium Identification and Sntimicrobial Susceptibility Testing
Bacterium identification and antimicrobial susceptibility testing (AST) of C. kerstersii 121606 were performed by BD PhoenixTM automated identification and antibiotic susceptibility testing system with the NMIC/ID-4 panel (Becton, Dickinson and Company, Franklin Lakes, NJ, USA), according to CLSI categories.
1.2 DNA Extraction and 16S rRNA Gene Sequencing
DNA extraction and 16S rRNA gene sequencing of C. kerstersii 121606 were performed as described previously[6]. It was deposited into GenBank under genus Acidovorax (1 460 bp, GenBank No. KY014106).
1.3 Genome Sequencing, Assembly, Annotation and Analysis of General Components in C. kerstersii 121606 Genome
Sequencing of the genomic DNA of C. kerstersii 121606 was completed with Illumina HiSeqTM 2000 (Illumina, San Diego, CA, USA) with 500 bp library preparations. The clean data were obtained (476 Mb). Genome assembly was carried out by SOAP de novo (version 2.04), and the genomic size was 3.34 Mb (196 contigs, GenBank assembly accession No. GCA_002002445.1, ASM200244v1).
The genomic sequences were annotated with RAST server[6]. The identification of rRNA genes, genomic GC content, insertion sequence (IS) elements was completed with RNAmmer 1.2 server[7].
1.4 Comparative Genomic Analyses of C. kerstersii Strains
To understand the origin, genomic compositions of C. kerstersii 21606, five representative Comamonas species and two Acidovorax species were comparatively analysed (GenBank accession numbers: C. kerstersii 8943, CP020121.1; C. kerstersii J29, NZ_LFYP00000000.1; C. kerstersii UBA11446, DOQQ00000000.1; C. testosterone TK102, CP006704.1; C. aquatica CJG, CP016603.1; Acidovorax sp. RAC01, NZ_CP016447.1; Acidovorax sp. 1608163, NZ_CP033069.1). Both C. kerstersii 8943 and C. kerstersii J29 were isolated from the same source-dialysis effluent of patients with peritonitis caused by continuous ambulatory peritoneal dialysis (CAPD) by the same team from Zhejiang Province, China[8].
Average nucleotide identity (ANI) analysis was operated by OrthoANI[9]. The single nucleotide polymorphism (SNP) differences between each C. kerstersii genomes were calculated using the snpTree web server for the assembled genomes with minimum coverage and minimum distance between SNPs[10]. Functional annotation of orthologous clusters among the C. kerstersii strains was conducted by OrthoVenn[11]. The prediction of CRISPRs (clustered regularly interspaced short palindromic repeats), prophages in our C. kerstersii 121606 were carried out with CRISPR recognition tool (CRT, version CRT1.1)[12], PHAST[13] respectively.
1.5 Analysis of Antibiotic Resistance Mechanisms of C. kerstersii 121606 at Genomic Level
The ARGs in C. kerstersii 121606 was predicted through CARD database, then filtered with more stringent cut-off parameters (identity>50%, query coverage>50%, subject coverage>50%, match length>100 amino acids, identical amino acids>100).
2 Experimental Results and Analysis
2.1 Identification and Antibiotic Susceptibilities Profiles of C. kerstersii 121606
With BD PhoenixTM automated identification and AST system, C. kerstersii 121606 was initially misidentified as Sphingobacterium multivorum (92% confidence). Moreover, because its 16S rRNA gene sequence displayed high identity with Acidovorax sp. 98-63833 (GenBank No. AY258065.1), it was again misidentified as Acidovorax sp. and deposited as Acidovorax sp. in GenBank after BlastN analysis.
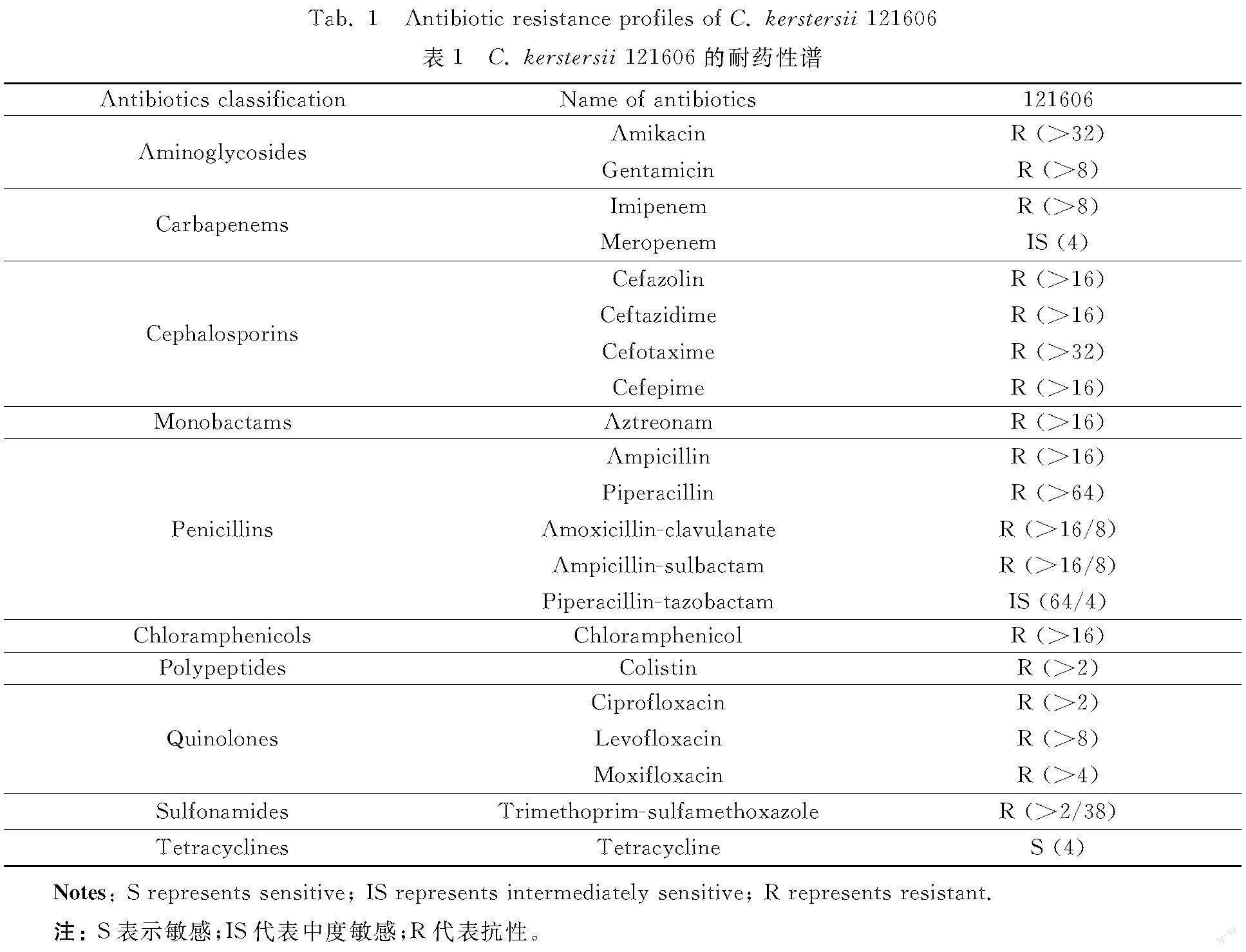
The AST (table 1) displayed that C. kerstersii 121606 was resistant to 18 out of 21 tested antibiotics, including aminoglycosides (amikacin, gentamicin), nearly all β-lactams {penicillins (ampicillin,piperacillin, amoxicillin-clavulanic acid, ampicillin-sulbactam, piperacillin-tazobactam), cephalosporins (cefazolin, ceftazidime, cefotaxime, cefepime), carbapenems (imipenem), monobactams (aztreonam)}, chloramphenicols (chloramphenicol), polypeptides (colistin), quinolones (ciprofloxacin, levofloxacin, moxifloxacin), sulfonamides (trimethoprim/sulfamethoxazole), intermediately susceptible to meropenem, piperacillin-tazobactam, and sensitive to tetracycline. These data showed that C. kerstersii 121606 possessed a multi-drug resistant phenotype.
2.2 General Genomic Components of C. kerstersii 121606
The assembledgenome was composed by 196 contigs and 53 scaffolds, the repeat accounted for 31.2%. The genome size was 3 419 381 bp, and GC content was 59.68%. It had 3 291 coding sequences (CDS), 3 128 insertion sequences (ISs), 17 tRNAs, 3 rRNAs.
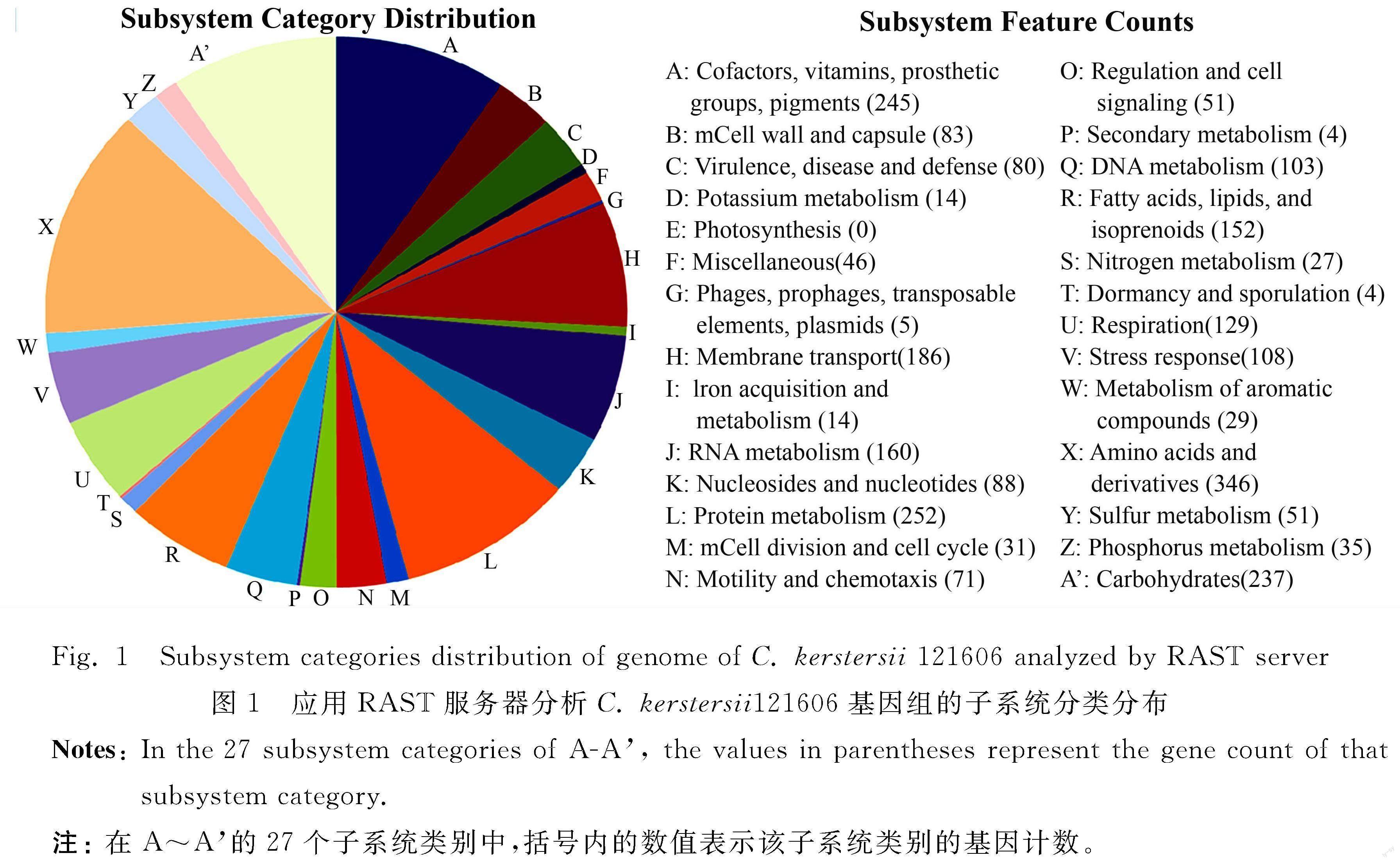
2.3 Subsystem Features of C. kerstersii 121606 and Orthologues in Four Representative Comamonas Species and Two Acidovorax Species
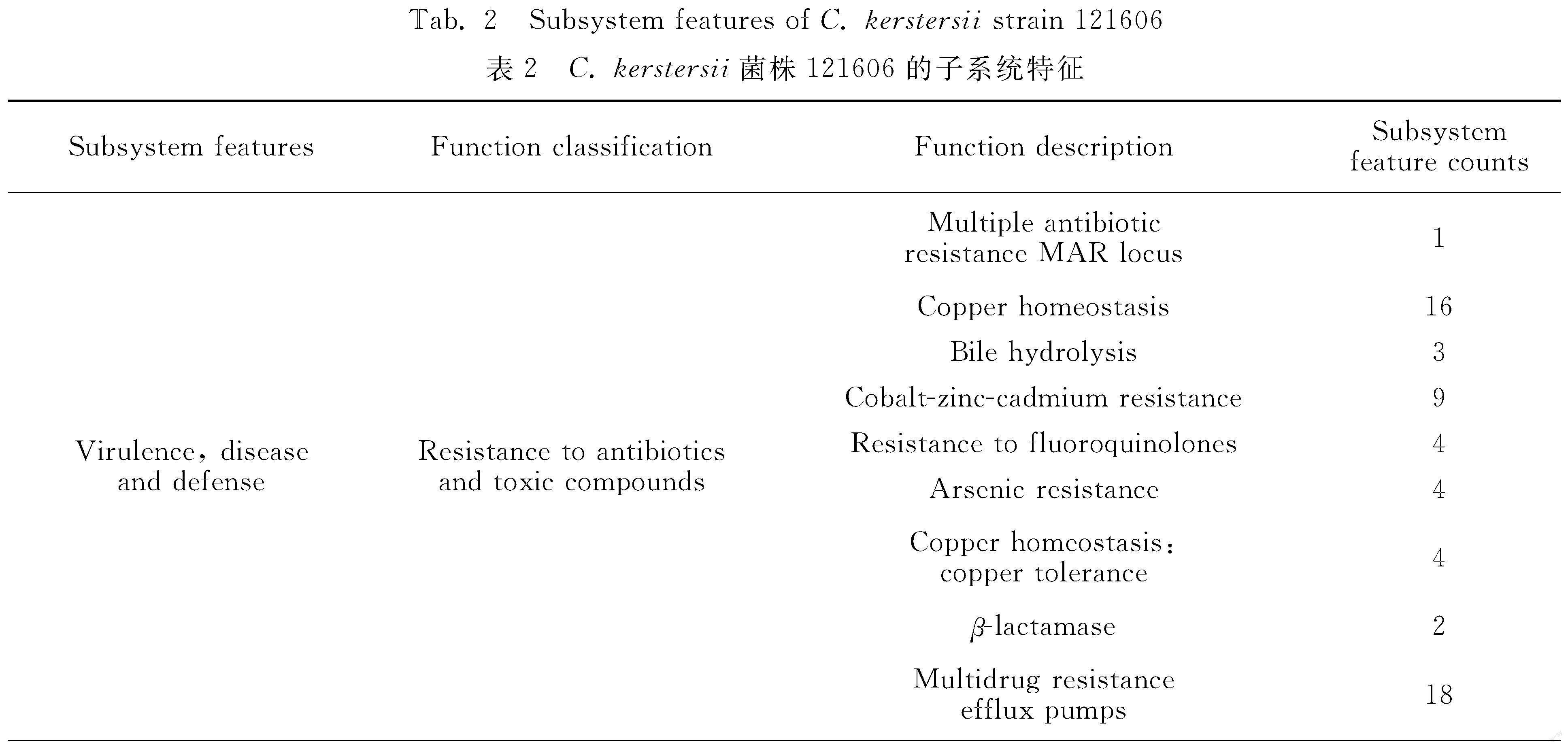
The subsystem features of C. kerstersii 121606 generated by RAST server revealed that it contained 27 subsystem categories (A-A′ in figure 1). For example, it contained 80 genes related to virulence, disease and defense, 6 related to phage, prophage (table 2). Further subsystem category analysis revealed that C. kerstersii 121606 had two phages, which was consistent to the number predicted by PHAST, as described below.
2.4 ANI Analysis and Phylogenic Tree of C. kerstersii 121606 With Five Representative Comamonas Species and Two Acidovorax Species
To reconfirm this C. kerstersii isolate 121606, we calculated the ANI values with other five representative Comamonas species and two closely related Acidovorax species. C. kerstersii 121606 showed high identities with C. kerstersii 8943, J29, UBA11446 (99.39%, 99.40%, 99.08%, respectively), and its identities with C. testosterone TK102, C. aquatica CJG, Acidovorax sp. 1608163, and Acidovorax sp. RAC01, were low (76.71%, 81.34%, 74.95%, 75.07%, respectively). According to the microbial taxonomy for species delineation ((95±0.5)% cut-off for ANI)[14], these data suggested that C. kerstersii 121606 really belonged to the species C. kerstersii, the identification as S. multivorum by the BD PhoenixTM automated identification method or as Acidovorax sp. by 16S rRNA gene sequence was not correct.
The phylogenetic tree based on SNPs showed thatC. kerstersii 121606, 8943, J29 and UBA11446 grouped together and other bacteria constituted a monophyletic cluster (figure 2). In figure 2, the tree was built using snpTree server with default parameters; the maximum likelihood tree was based on SNPs alignment; tree was shown in the rectangular cladoGram format in which branches were drawn with reference to branch lengths. The tree topology evaluation was based on percentage of concordanceTheir distance to C. testosterone TK102, C. aquatica CJG, Acidovorax sp. 1608163 and Acidovorax sp. RAC01 was the same as the ANI-based phylogenetic tree (not shown).
The orthologoues of C. kerstersii 121606 were compared with other three C. kerstersii genomes (strain 8943, J29 and UBA11446). It showed that strain 121606 had 2 955 proteins, 2 943 clusters, 11 singletons. 2 095 clusters were shared by these four C. kerstersii strains. Further, the gene family clusters of these five Comamonas species and two Acidovorax species were presented in figure 3. In agreement with above statements, C. kerstersii strain 121606, 8943, J29 and UBA11446 exhibited a closely related similarity in single- or multiple-copy orthologs.
2.5 Prediction of CRISPR in C. kerstersii 121606
CRISPRs are vital constituents of RNA-based viral defense systems possessed by bacteria and archaea[15]. CRISPR loci are composed by leader sequence, repeats, spacers and cas gene cassette[15]. C. kerstersii 121606 was predicted to contain 10 nearly identical 29-nucleotide CRISPR repeats (GGTTGCCCCGCACACGCGGGGATAGGCCC) (Range: 2657125-2657702 in genome), which separated by 32 nucleotides spacers in average length. The constituent characteristics of these CRISPRs were same as those originally found in Escherichia coli genome in 1987[16].
2.6 Prediction of Prophages in C. kerstersii 121606
The C. kerstersii 121606 genome harbored an incomplete (a) and an intact prophage region (b) (figure 4). The intact prophage region b was predicted to extend from 1 799 206 bp to 1 836 855 bp (37 650 bp in length) and it carried 35 CDSs. Significantly, one integrase used as the marker of mobile DNA elements[17] was identified in b, and the att sites used to determine the extent of the prophage[18] were also found in b, meaning that the intact prophage region b is disseminatable? This assumption requires laboratory verification.
2.7 Resistome of C. kerstersii 121606
C. kerstersii 121606 (table 3) harbored a number of ARGs contributing to the resistance to β-lactams (blaKPC-2, blaSPM-1), fluoroquinolone (AANT(4′)-Ib, apmA, mfd, opmH, oqxB), polymyxin (basR), elfamycin (basS, lpxC), fosfomycin (pvrR), aminocoumarin, elfamycin, fluoroquinolone, isoniazid, lipopeptide antibiotics, and rifamycin[tet(35)], or encoding efflux pump complex subunits or regulators (acrB, acrD, acrF; mexA, mexB, mexQ, oprM, mexR; smeB, smeD, smeE; adeJ; mdtF). The presence of these collective ARGs was consistent with the AST profiles of C. kerstersii 121606.
3 Discussion
3.1 Underestimated C. kerstersii Infections in Clinical Diagnosis
As mentioned above, during our laboratory diagnosis, C. kerstersii 121606 was initially misidentified as S. multivorum. Later, it was misidentified as Acidovorax sp. even by the gold standard diagnosis method, 16S rRNA gene sequencing. In fact, we had the same misidentification experience with other two C. kerstersii, 202149 and 12322-1 (unpublished). Their identities as C. kerstersii were not determined until their genomic sequences were obtained for ANI analysis. There are increasing reports of C. kerstersii misidentification due to unreliable phenotypic identification tests prior to the application of MALDI-TOF[4,19-20]. As a result, actual C. kerstersii infections might be seriously underestimated.
3.2 Origin of C. kerstersii 121606 Revealed by Comparative Genomic Analyses
The phylogenetic trees based on both ANI values and SNPs confirmed that C. kerstersii 121606 is a strain of the species kerstersii and is closely related to C. kerstersii 8943 and C. kerstersii J29. Additionally, the orthologous analysis revealed that strains C. kerstersii 121606, 8943, and J29 contained highly similar single- or multiple-copy orthologs. It is known that C. kerstersii 8943 and C. kerstersii J29 were isolated from the same source in Zhejiang Province, China[8], which is not far from Quanzhou City, the isolation site of C. kerstersii 121606. Therefore, C. kerstersii 121606 might have the same ancestor as C. kerstersii 8943 and J29.
3.3 Multi-Drug Phenotype of C. kerstersii 121606 Explained by Presence of Number of Antibiotic Resistance Genes in Genome
According to table 1, C. kerstersii 121606 exhibited the multi-drug phenotype of C. kerstersii, which had not been previously reported. For instance, it was noted that nine of the antibiotics to which C. kerstersii 121606 was resistant (amikacin, gentamicin, imipenem, cefepime, cefotaxime, ciprofloxacin, ampicillin-sulbactam, piperacillin-tazobactam, trimethoprim-sulfamethoxazole) were sensitive in 14 clinical C. kerstersii strains[21]. Additionally, 11 of the antibiotics to which C. kerstersii 121606 was resistant (ampicillin, ampicillin-sulbactam, cefotaxime, ceftazidime, cefepime, imipenem, gentamicin, amikacin, ciprofloxacin, colistin, trimethoprim-sulfamethoxazole) were susceptible in four clinical C. kerstersii isolates[5]. The AST profiles of strain 121606 (table 1) could be explained by the corresponding ARGs in its genome (table 3).
β-lactams are the primary antibiotics used to treat bacterial infections. The rapid increase in bacterial resistance to β-lactams posed a serious health threat. The mechanisms of β-lactam resistance include inactivation by β-lactamases, efflux pumps, reduced permeability, and altered transpeptidases[22]. C. kerstersii 121606 possessed at least the former two mechanisms.
Firstly, the presence of the β-lactamase encoding genes blaSPM-1 and blaKPC-2 in C. kerstersii 121606 explained its resistance to the tested β-lactams, including penicillins (ampicillin, piperacillin, amoxicillin-clavulanic acid, piperacillin-tazobactam), cephalosporins (ceftazidime, cefotaxime, cefepime), monobactams (aztreonam), and carbapenems (imipenem) (table 1).
SPM-1 (Sao Paulo metallo-β-lactamases) is a Class B metallo-β-lactamase, first identified in Pseudomonas aeruginosa strain 48-1997A isolated in Sao Paulo, Brazil[23]. It shows hydrolytic activity against penicillins (benzylpenicillin, ampicillin, carbenicillin, azlocillin, piperacillin), with the highest activity for benzylpenicillin, ampicillin, and piperacillin, and preferential hydrolyzing activity against cephalosporins (cephalothin, cefuroxime, cefotaxime, cefoxitin, cefepime), carbapenems (imipenem, meropenem), the serine β-lactamase inhibitor tazobactam, but not against the monobactam aztreonam or the serine β-lactamase inhibitor clavulanic acid. The latter two are competitive inhibitors of SPM-1[23]. The presence of blaSPM-1 was consistent with the resistance of C. kerstersii 121606 to ampicillin, piperacillin, cefotaxime, cefepime, and imipenem.
KPC-2 is a homolog of various Klebsiella pneumoniae carbapenemases (KPCs), the most typical example of a class A carbapenemase, which also confers resistance to most β-lactams. KPC-1 was first found in a K. pneumoniae isolate in 1996 in the United State, and showed hydrolytic activity against penicillin (benzylpenicillin, ampicillin, cloxacillin, piperacillin), cephalosporins (cefotaxime), carbapenem (imipenem and meropenem), and monobactams (aztreonam)[24]. The KPC-1 transforming expressing bacteria showed resistance to penicillins (ampicillin, amoxicillin-clavulanic acid, piperacillin-tazobactam), cephalosporins (ceftazidime, cefoxitin, cefpodoxime, cefotaxime, ceftriaxone), carbapenems (imipenem, meropenem), and monobactams (aztreonam)[24]. Initially identified KPC-2 also has hydrolytic activity against penicillins and cephalosporins but not cephamycins and ceftazidime. Recently emerged KPC-2 variants evolved by single and double amino acid substitution among clinical isolates showed increased hydrolytic acitivity against ceftazidime and consequently increased resistance to ceftazidime, while maintaining the resistance capacity to penicillins and carbapenems[25]. The presence of blaKPC-2 was closely related to the resistance of C. kerstersii 121606 to ampicillin, amoxicillin-clavulanic acid, piperacillin-tazobactam, ceftazidime, cefotaxime, imipenem, and aztreonam.
Secondly, for theefflux pumps, the C. kerstersii 121606 genome possessed several type of multidrug efflux systems, which would futher enhance resistance to β-lactams and other antibiotics listed in table 1.
The genes acrD, acrB, and acrF encode three types of RND (Resistance Nodulation and cell Division) efflux pumps that confer resistance to monobactams (aztreonam), cephalosporins (cefazolin), and aminoglycosides (amikacin, gentamicin) and quinolones (ciprofloxacin, levofloxacin, moxifloxacin). For example, the AcrD (encoded by acrD) containing AcrAD-TolC efflux pump confers resistance to aztreonam, cefazolin in Salmonella typhimurium[26], and aminoglycosides (amikacin, gentamicin, neomycin, kanamycin, and tobramycin) in E. coli[27]. AcrAB-TolC, a major and clinically important efflux pump in Gram-negative bacteria, and another type of RND efflux pumps, AcrEF-TolC, contribute to resistance to fluoroquinolones, which are often used as therapeutic alternatives in infections[28].
For the component and structure of RND efflux pumps in Gram-negative bacteria, for example, the AcrAB-TolC multidrug efflux pump complex is formed by assembling the homotrimeric inner membrane protein (IMP) AcrB, which recognizes, binds and translocates substrates, and transduces the energy (proton motive force, PMF) for the drug/proton antiport process, the hexameric membrane fusion protein (MFP) AcrA, which forms a membrane fused tubular structure inside the periplasmic space, and the homotrimeric outer membrane factor (OMF) TolC[29], constituting a main means of intrinsic resistance against cytotoxic substances[30].
The TolC encoding gene tolC, and the AcrEF efflux pump encoding genes acrE and acrF were predicted in C. kerstersii 121606, but not acrA. acrE was redicted to encode a 399 amino acid-AcrE protein in C. kerstersii 121606 through CARD database, but it was filtered out and not listed in table 3 because its identity with the reference gene was below 50%. However, AcrE and AcrD/AcrF are homologues of AcrA and AcrB, and AcrE can substitute AcrA to form a functional complex with AcrD by associating with the same residues identified in AcrB binding[31]. So, we supposed that C. kerstersii 121606 possessed functional AcrEF, AcrED, or AcrEB-TolC multidrug efflux pumps, resulting in resistance to monobactams (aztreonam), cephalosporins (cefazolin), aminoglycosides (amikacin, gentamicin) and quinolones (ciprofloxacin, levofloxacin, moxifloxacin) (table 1).
In addition, C. kerstersii 121606 may have MexAB-OprM, which is found in P. aeruginosa, as it harbored their encoding genes mexA, mexB, oprM and one of the repressor gene, mexR. Overexpression of MexAB-OprM induced by mexR mutations can export various antibiotics, including penicillins (penicillin G cloxacillin, nafcillin, amoxicillin, piperacillin, carbenicillin, sulbenicillin), cephems (cefamandole, cefuroxime, cefoperazone, cefotaxime, ceftizoxime, ceftriaxone, ceftazidime, cefpirome, cefepime, cefozopran, cefoselis, cefoxitin), oxacephems (moxalactam, flomoxef), monobactams (aztreonam), quinolones (nalidixic acid, piromidic acid, pipemidic acid, cinoxacin, norfloxacin, ofloxacin, enoxacin, ciprofloxacin, tosufloxacin, sparfloxacin), macrolides (erythromycin, oleandomycin, spiramycin), lincomycins (lincomycin), chloramphenicols (chloramphenicol), and novobiocin (novobiocin)[32]. If MexAB-OprM really conferred the resistance of C. kerstersii 121606 to penicillins (amoxicillin, piperacillin), cephalosporins (cefotaxime, ceftazidime, cefepime), monobactams (aztreonam), quinolones (ciprofloxacin), chloramphenicol (chloramphenicol)] (table 1), sequencing of the full-length mexR gene in C. kerstersii 121606 should be performed to verify whether it is a loss-of-function mutant[33], leading to overexpression of MexAB-OprM and contributing to the resistance. In fact, C. kerstersii 121606 may express a series of Mex efflux pumps as it contained the genes mexA-N, mexP-R , mexV-Y, mexA-N, mexP-R, mexV-Y, oprA, oprJ, oprM, oprN, oprZ[34], most of which were not listed in table 3 due to the stringent filtering parameters mentioned above.
C. kerstersii 121606 might also possess anthor two homologues of the mexAB-oprM efflux pumps,SmeABC and SmeDE (a SmeB homologue) F (a SmeC homologue) multidrug efflux pump, as their encoding genes smeA (encoding the inner-membrane fusion lipoprotein), smeB (encoding the RND transporter), smeC (encoding the outer-membrane efflux lipoprotein), smeD, smeE, smeF (encoding the outer membrane protein), and the regulation genes smeR (encoding the two-component system sensory kinase which positively regulates expression of the smeA,smeB,smeC genes), smeS (encoding the two-component system response regulator) were predicted through CARD database, and smeS and smeR were located upstream of the smeABC genes and the smeDEF genes as reported[35]. We should mention that, smeA, smeC, smeF, smeR, and smeS were not put into table 3 because their identities with the reference genes in CARD database were below 50%. The SmeABC efflux pump confers the resistance to aminoglycosides (amikacin, gentamicin, kanamycin, streptomycin, tobramycin), penicillins (penicillin, carbenicillin, ampicillin), (cefsulodin, cefotaxime, cefoperazone, cefepime, cefpirome), quinolones (nalidixic acid, ciprofloxacin,norfloxacin,trovafloxacin)[35]. The efflux pump SmeDEF has broad substrate specificity and contributes to the resistance to a variety of antimicrobials, including quinolones, macrolides, chloramphenicol, and novobiocin[34], trimethoprim-sulfamethoxazole in clinical isolates of Stenotrophomonas maltophilia [36]. Aside from conferring the resistance to β-lactams (listed in table 3), the probable expression of AcrEF, AcrED, AcrEB-TolC, MexAB-OprM, smeABC, SmeDEF multidrug efflux pump also contributed to the resistance to aminoglycosides (amikacin, gentamicin), quinolones (ciprofloxacin, levofloxacin, moxifloxacin), chloramphenicol (chloramphenicol), sulfonamides (trimethoprim-sulfamethoxazole).
Overall, these ARGs contained inC. kerstersii 121606 genome explained its multi-drug resistance.
4 Conclusion
Comparative genomic analyses revealed that our clinical C. kerstersii isolate 121606 belonged to the species C. kerstersii and shared similar genetic components with five other Comamonas species and the closely related Acidovorax species. The ARGs in the genome were consistent with its multi-drug resistance phenotype. All the data needs to be verified by laboratory experiments and could help to find new drug to combate infections caused by multi-drug resistant C. kerstersii.
Ethics Statement: This article does not contain any studies withhumans or animals performed by any of the authors. This work was approved by the Ethics Committee of Quanzhou First Hospital.
References:
[1] WAUTERS G,DE BAERE T,WILLEMS A,et al.Description of Comamonas aquatica comb. nov. and Comamonas kerstersii sp. nov. for two subgroups of Comamonas terrigena and emended description of Comamonas terrigena[J].Int J Syst Evol Microbiol,2003,53(3):859-862.DOI:10.1099/ijs.0.02450-0
[2] LIU Xuejiao,QIAO Xiwen,HUANG Tianmin,et al.Comamonas kerstersii bacteremia[J].Med Mal Infect,2020,50(3):288-290.DOI:10.1016/j.medmal.2019.12.005.
[3] BENNANI H,HANCHI AL,SORAA N.A young child with acute perforated appendicitis due to Comamonas kerstersii: A rare case report[J].Pan Afr Med J,2022,41:186-193.DOI:10.11604/pamj.2022.41.186.29615.
[4] BISWAS J S,FITCHETT J,O′HARA G.Comamonas kerstersii and the perforated appendix[J].J Clin Microbiol,2014,52(8):3134.DOI:10.1128/JCM.00909-14
[5] ALMUZARA MN,VERA OCAMPO C,BAKAI R,et al.Intra-abdominal infections due to Comamonas kerstersii[J].J Clin Microbiol,2013,51(6):1998-2000.DOI:10.1128/JCM.00659-13.
[6] WANG M,LIN N,ZHANG Y,et al.The antibiotic resistance and pathogenicity of a multidrug-resistant Elizabethkingia anophelis isolate[J].Microbiologyopen,2019,8(11):e804.DOI:10.1002/mbo3.804.
[7] HU S,WU Y,ZHOU Y,et al.Comparative genomic analysis of Myroides odoratimimus isolates[J].Microbiologyopen,2019,8(2):e00634.DOI:10.1002/mbo3.634.
[8] JIANG Xiawei,LIU Wenhong,ZHENG Beiwen.Complete genome sequencing of Comamonas kerstersii 8943,a causative agent for peritonitis[J].Sci Data,2018,5(1):180222.DOI:10.1038/sdata.2018.222.
[9] LEE I,KIM Y O,PARK S C,et al.OrthoANI: An improved algorithm and software for calculating average nucleotide identity[J].Int J Syst Evol Microbiol,2016,66(2):1100-1103.DOI:10.1099/ijsem.0.000760.
[10] LEEKITCHAROENPHON P,KAAS RS,THOMSEN MC,et al.snpTree: A web-server to identify and construct SNP trees from whole genome sequence data[J].BMC Genomics,2012,13(Suppl 7):S6(1-8).DOI:10.1186/1471-2164-13-S7-S6.
[11] WANG Y,COLEMAN-DERR D,CHEN G.OrthoVenn: A web server for genome wide comparison and annotation of orthologous clusters across multiple species[J].Nucleic Acids Res,2015,43(W1):W78-W84.DOI:10.1093/nar/gkv487.
[12] BLAND C RT,SABREE F,LOWE M,et al.CRISPR recognition tool (CRT): A tool for automatic detection of clustered regularly interspaced palindromic repeats[J].BMC Bioinformatics,2007,8:209-216.DOI:10.1186/1471-2105-8-209.
[13] ZHOU You,LYNCH K H,DENNIS J J,et al.PHAST: A fast phage search tool[J].Nucleic Acids Res,2011,39:W347-W352.DOI:10.1093/nar/gkr485.
[14] GORIS J,KLAPPENBACH J A,COENYE T, et al.DNA-DNA hybridization values and their relationship to whole-genome sequence similarities[J].Int J Syst Evol Microbiol,2007,57(1):81-91.DOI:10.1099/ijs.0.64483-0.
[15] WIEDENHEFT B,DOUDNA J A.RNA-guided genetic silencing systems in bacteria and archaea[J].Nature,2012,482(7385):331-338.DOI:10.1038/nature10886.
[16] ISHINO Y,MAKINO K,AMEMURA M,et al.Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product[J].J Bacteriol,1987,169(12):5429-5433.DOI:10.1128/jb.169.12.5429-5433.1987.
[17] BENNETZEN JL.Transposable element contributions to plant gene and genome evolution[J].Plant Mol Biol,2000,42(1):251-269.
[18] VENTURA M,KLEEREBEZEM M,DE VOS WM,et al.The prophage sequences of Lactobacillus plantarum strain WCFS1[J].Virology,2003,316(2):245-255.DOI:10.1016/j.virol.2003.08.019.
[19] OPOTA O,ZANETTI G,JATON K,et al.Bacteremia caused by Comamonas kerstersii in a patient with diverticulosis[J].J Clin Microbiol,2014,52(3):1009-1012.DOI:10.1128/JCM.02942-13.
[20] RONG K,ALMUTAWA F.Comamonas kerstersii Bacteremia of Unknown Origin[J].Case Rep Infect Dis,2022,2022:1129832.DOI:10.1155/2022/1129832.
[21] ALMUZARA M,ESTRAVIZ ML,ELLIS A,et al.First report of Comamonas kerstersii causing urinary tract infection[J].New Microbes New Infect,2018,24:4-7.DOI:10.1016/j.nmni.2018.03.003.
[22] SAWA T,KOOGUCHI K,MORIYAMA K.Molecular diversity of extended-spectrum β-lactamases and carbapenemases, and antimicrobial resistance[J].J Intensive Care,2020,8:13.DOI:10.1186/s40560-020-0429-6.
[23] MURPHY T A,SIMM A M,TOLEMAN M A,et al.Biochemical characterization of the acquired metallo-beta-lactamase SPM-1 from Pseudomonas aeruginosa[J].Antimicrob Agents Chemother,2003,47(2):582-587.DOI:10.1128/AAC.47.2.582-587.2003.
[24] YIGIT H,QUEENAN A M,ANDERSON G J,et al.Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae[J].Antimicrob Agents Chemother,2001,45(4):1151-1161.DOI:10.1128/AAC.45.4.1151-1161.2001.
[25] MEHTA SC,RICE K,PALZKILL T.Natural variants of the KPC-2 carbapenemase have evolved increased catalytic efficiency for ceftazidime hydrolysis at the cost of enzyme stability[J].PLoS Pathog,2015,11(6):e1004949.DOI:10.1371/journal.ppat.1004949.
[26] CUESTA BERNAL J,EL-DELIK J,et al.Characterization and molecular determinants for β-lactam specificity of the multidrug efflux pump AcrD from Salmonella typhimurium[J].Antibiotics (Basel),2021,10(12):1494.DOI:10.3390/antibiotics10121494.
[27] ROSENBERG EY,MA D,NIKAIDO H.AcrD (acriflavine resistance D) of Escherichia coli is an aminoglycoside efflux pump[J].J Bacteriol,2000,182(6):1754-1756.DOI:10.1128/JB.182.6.1754-1756.2000.
[28] CHETRI S,DOLLEY A,BHOWMIK D,et al.Transcriptional response of AcrEF-TolC against fluoroquinolone and carbapenem in Escherichia coli of clinical origin[J].Indian J Med Microbiol,2018,36(4):537-540.DOI:10.4103/ijmm.IJMM_18_308.
[29] KOBYLKA J,KUTH MS,MLLER RT,et al.AcrB: A mean, keen, drug efflux machine[J].Ann N Y Acad Sci,2020,1459(1):38-68.DOI:10.1111/nyas.14239.
[30] PUGH HL,CONNOR C,SIASAT P,et al.E. coli ST11 (O157:H7) does not encode a functional AcrF efflux pump[J].Microbiology (Reading),2023,169(4):001324.DOI:10.1099/mic.0.001324.
[31] ALAV I,BAVRO V N,BLAIR JMA.Interchangeability of periplasmic adaptor proteins AcrA and AcrE in forming functional efflux pumps with AcrD in Salmonella enterica serovar Typhimurium[J].J Antimicrob Chemother,2021,76(10):2558-2564.DOI:10.1093/jac/dkab237.
[32] TSUJIMOTO H,NISHINO T.Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-oprM efflux pumps in Pseudomonas aeruginosa[J].Antimicrob Agents Chemother,2000,44(12):3322-7.DOI:10.1128/AAC.44.12.3322-3327.2000.
[33] SRIKUMAR R,PAUL CJ,POOLE K.Influence of mutations in the mexR repressor gene on expression of the MexA-MexB-oprM multidrug efflux system of Pseudomonas aeruginosa[J].J Bacteriol,2000,182(5):1410-1414.DOI:10.1128/JB.182.5.1410-1414.2000.
[34] ZHANG Li,LI Xianzhi,POOLE K.SmeDEF multidrug efflux pump contributes to intrinsic multidrug resistance in Stenotrophomonas maltophilia[J].Antimicrob Agents Chemother,2001,45(12):3497-3503.DOI:10.1128/AAC.45.12.3497-3503.2001.
[35] LI Xianzhi,ZHANG Li,POOLE K.SmeC, an outer membrane multidrug efflux protein of Stenotrophomonas maltophilia[J].Antimicrob Agents Chemother,2002,46(2):333-343.DOI:10.1128/AAC.46.2.333-343.2002.
[36] SNCHEZ MB,MARTNEZ JL.The efflux pump SmeDEF contributes to trimethoprim-sulfamethoxazole resistance in Stenotrophomonas maltophilia[J].Antimicrob Agents Chemother,2015,59(7):4347-4348.DOI:10.1128/AAC.00714-15.

