FE65: a hub for neurodevelopment
Yuqi Zhai, Laura Lok-Haang Ng, Dennis Dik-Long Chau, Kwok-Fai Lau
FE65, initially identified as a binding partner of amyloid precursor protein (APP), is an adaptor protein enriched in the brain and regulated during development.FE65 belongs to the FE65 protein family.This family is comprised of three members,FE65, FE65 like-1 (FE65L1), and FE65 like-2(FE65L2).The three members share a conserved structure: a tryptophan-tryptophan (WW) domain and two successive phosphotyrosine-binding (PTB)domains.Despite being structurally similar, the three proteins differ in their expression patterns.FE65 is brain-enriched while FE65L1 and FE65L2 are more ubiquitously expressed.Therefore, FE65 has drawn more research interest than the other two homologs.Insights into the potential functions of the FE65 family have been obtained through studies on its interaction partners.Recently,emerging evidence suggests multiple roles of FE65 and its interactors during neuronal developmental processes including neurogenesis, neuronal migration and positioning, neurite outgrowth, and synaptic plasticity (Figure 1).
Neurogenesis: Neurogenesis refers to the process of generating new neurons in the brain.It plays a crucial role in brain development, learning,and memory.While the exact role of FE65 in neurogenesis is still under investigation, it has been implicated in multiple neuronal developmental processes including the proliferation of neural stem or progenitor cells and neuronal cell fate decision.
FE65 has been shown to interact with signaling proteins and transcription factors that control cell proliferation such as Notch (Kim et al., 2012).The Notch signaling pathway is known to regulate the proliferation of neural progenitor cells.The binding of FE65 enhances the interaction of E3 ubiquitin ligase Itch and Notch1 to promote the proteasomal degradation of Notch1 (Kim et al., 2012).In the nucleus, FE65 also disrupts the formation of Notch intracellular domain-recombining binding protein suppressor of hairless transcriptional regulatory complex which suppresses the Notch intracellular domain-mediated expression of Hairy and enhancer of split 1, a transcription factor that plays a crucial role in maintaining and inhibiting the differentiation of neural progenitor cells (Kim et al., 2012).
The roles of FE65 in regulating neuronal cell differentiation have also been investigated.The abnormal dynamics of Gonadotropin-Releasing Hormone 1-mediated neurogenesis in a mouse model lacking the 97 kDa FE65 isoform suggest a role of FE65 in regulating the process (Forni et al.,2011).Notably, FE65 interacts with contactin-2/TAG1-APP signaling to suppress neurogenesis(Ma et al., 2008).Additionally, FE65 is suggested to modulate neurogenesis via regulating gene transcription.APP intracellular domain (AICD)-FE65 has been reported to form a transcriptional regulatory complex with Tat interactive protein 60 kDa (Tip60).Several genes that participate in neurogenesis have been reported to be regulated by the AICD-FE65-Tip60 complex, including stathmin, which is a highly conserved neuronal protein associated with neuronal migration and neurogenesis through regulating microtubule dynamics.Tetraspanin KAI1 is also an AICDFE65-Tip60 complex-controlled gene.However,controversial roles have been reported for the complex in regulating KAI1 expression.Moreover,AICD-FE65 is reported to interact with the nucleosome assembly factor SET through the FE65 WW domain to activate the transcription of KAI1.On the other hand, FE65 has been demonstrated to downregulate the expression of KAI1 by interacting with estrogen receptor alpha.While the aforementioned findings highlight the functional significance of FE65 interactions in neurogenesis and cell fate regulation, further investigations are still underway to determine the exact roles of FE65 in these processes.
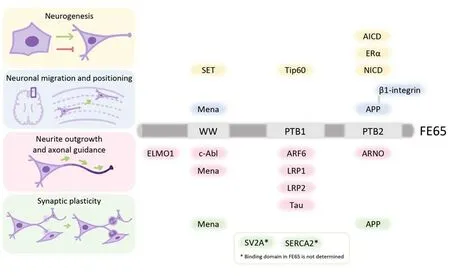
Figure 1 |Summary of FE65 structure and its interactors in neurodevelopment.The structure and the domains of FE65 responsible for the binding to its interacting partners are illustrated.FE65-interactors that are associated with neurogenesis (yellow), neuronal migration and positioning (blue), neurite outgrowth and axonal guidance (pink), and synaptic plasticity (green) are indicated.Created with Procreate and Microsoft PowerPoint.AICD: Amyloid precursor protein intracellular domain; APP: amyloid precursor protein; ARF6:ADP-ribosylation factor 6; ARNO: ADP-ribosylation factor nucleotide-binding site opener; c-Abl: c-Abl tyrosine kinase;ELMO1: engulfment and cell motility protein 1; ERα: estrogen receptor alpha; LRP1: low-density lipoprotein receptorrelated protein-1; LRP2: low-density lipoprotein receptor-related protein-2; NICD: notch intracellular domain; PTB:phosphotyrosine-binding; SERCA2: sarco/endoplasmatic reticulum calcium ATPase 2; SET: nucleosome assembly protein SET; SV2A: synaptic vesicle glycoprotein 2A; Tip60: tat interactive protein 60 kDa; WW: tryptophan-tryptophan.
Neuronal migration and positioning: Neuronal migration is essential for the establishment of the nervous system as the process allocates newly differentiated neurons to form functional neuronal layers and circuitry.FE65–/–/FE65L1–/–double knockout mice show cortical dysplasia,which indicates the role of FE65 in guiding neuronal migration and positioning during cortical development (Guenette et al., 2006).However, controversial roles have been reported for the complex in regulating KAI1 expression.Moreover, AICD-FE65 is reported to interact with the nucleosome assembly factor SET through the FE65 WW domain to activate the transcription of KAI1.On the other hand, FE65 has been demonstrated to downregulate the expression of KAI1 by interacting with estrogen receptor alpha.While the aforementioned findings highlight the functional significance of FE65 interactions in neurogenesis and cell fate regulation, further investigations are still underway to determine the exact roles of FE65 in these processes.
It is noteworthy that mice lacking both FE65 and FE65L1 exhibit a similar defect in neuronal positioning as inApp–/–/Aplp1-/-/Aplp2-/-tripleknockout mice (Guenette et al., 2006).Given that APP interacts with FE65, the above knockout mouse studies suggest the two proteins influence the same signaling pathway(s).FE65 has been shown to potentiate the stimulatory effect of APP on cell migration.This effect may be due to the interaction of FE65 and/or APP with β1-integrin, a regulator of extracellular matrix and cytoskeleton adhesion, or mammalian homolog of Drosophila Enabled (Mena), a mediator of actin dynamics(Sabo et al., 2003).
Neurite outgrowth and axon guidance:Intriguingly, growing evidence indicates FE65 positively regulates neurite outgrowth by interacting with proteins that have been identified to be associated with neurite outgrowth,including ADP-ribosylation factor 6 (ARF6) and Engulfment and Cell Motility 1 (ELMO1).It has been demonstrated that FE65 interacts with ARF6 and the Rac1-GEF complex ELMO1/Dedicator of cytokinesis 1 (ELMO1/DOCK1) simultaneously(Chan et al., 2020).Through such interactions,FE65 potentiates the plasma membrane targeting of ELMO1 via the ARF6-mediated endosomal recycling pathway.
There are some other FE65 interactors that have also been implicated in neurite outgrowth, such as Tau which is essential for growth cone motility(Nensa et al., 2014).Moreover, the WW domain of FE65 has been demonstrated to interact with the cytoskeleton regulator c-Abl (Perkinton et al.,2004).Nevertheless, the precise effects of the binding between FE65 and these interactors on neurite outgrowth remain to be determined.
As abnormal axonal projections are observed in the hippocampus of FE65-/-/ FE65L1-/-mice,the roles of FE65 in axonal guidance have been proposed.Of note, FE65 interacts with some regulators of axonal guidance such as Mena which mediates neuronal pathfinding downstream of the guidance molecule Netrin-1 (Sabo et al., 2003).Moreover, low-density lipoprotein receptor-related protein 1 (LRP1) and LRP2, which are FE65 PTB1 interactors (Pietrzik et al., 2004), function as axon guidance receptors.
Synaptic plasticity: FE65-/-/FE65L1-/-mice also develop long-term potentiation deficits that implicate their roles in synaptic plasticity (Strecker et al., 2016).It is noteworthy,App-/-/Aplp1-/-/Aplp2-/-triple-knockout mice show similar synaptic dysfunction.These findings suggest a functional connection between FE65 and APPs.Additionally,the FE65-APP complex may recruit other molecules to modulate synaptic plasticity including Mena (Sabo et al., 2003).FE65 may also influence synaptic plasticity via interactions with synaptic vesicle glycoprotein 2A, an essential component in synaptic vesicle fusion, and sarco/endoplasmatic reticulum calcium ATPase 2, a synaptic calcium channel (Nensa et al., 2014).
Conclusion: In the past two decades, the functions of FE65 have been unveiled through the identification of its interactors using different approaches.However, each identification method has its own limitations.Yeast two-hybrid screening is widely used for this purpose.It is known that yeast cells have limited capability to mimic the mammalian environment due to the absence of certain mammalian post-translational modifications.Affinity purification coupled with mass spectrometry has also been utilized to uncover the FE65 interactome.However, detecting membrane protein interactions and weak/transient interactions with this method is challenging.
Apparently, alternative/new methods should be considered for the identification of the full complement of FE65 interactors.For example,proximity labeling techniques like BioID can enhance the detection of subtle interactions in mammalian cells.Several interactors of FE65 have been implicated in various neurodevelopmental pathways.The association between FE65 and these interactors is expected to have significant roles in these processes.As a multi-interface hub,FE65 may mediate the crosstalk of different neurodevelopmental signaling pathways by facilitating the assembly of multimeric protein complexes.Further studies on the roles of these candidate interactions would uncover valuable mechanistic insights into the neuronal functions of FE65 during neurodevelopment.
Al though k noc kout m ice studies have demonstrated that FE65 family proteins have functional redundancy, FE65L1 and FE65L2 exhibit notable differences from FE65.According to the expression atlas, the distribution of FE65 and FE65L1 are similar in the brain.This is concordant with the notion that only double knockout, rather than FE65 or FE65L1 single knockout animals,develop evident phenotype.In contrast, FE65L2 has a higher expression level in the cerebral cortex and the hypothalamus.Further investigation is required to elucidate the roles of FE65L2 in these regions.
Yuqi Zhai, Laura Lok-Haang Ng,Dennis Dik-Long Chau*, Kwok-Fai Lau*
School of Life Sciences, Faculty of Science, The Chinese University of Hong Kong, Hong Kong Special Administrative Region, China
*Correspondence to:Dennis Dik-Long Chau, PhD,diklongchau@cuhk.edu.hk; Kwok-Fai Lau, PhD,kflau@cuhk.edu.hk.
https://orcid.org/0000-0002-1238-2101(Dennis Dik-Long Chau)
https://orcid.org/0000-0002-0193-1152(Kwok-Fai Lau)
Date of submission: August 31, 2023
Date of decision: November 3, 2023
Date of acceptance: November 21, 2023
Date of web publication: December 21, 2023
https://doi.org/10.4103/1673-5374.391188
How to cite this article:Zhai Y, Ng LLH, Chau DDL,Lau KF (2024) FE65: a hub for neurodevelopment.Neural Regen Res 19(9):1883-1884.
Open access statement:This is an open access journal, and articles are distributed under the terms of the Creative Commons AttributionNonCommercial-ShareAlike 4.0 License,which allows others to remix, tweak, and build upon the work non-commercially, as long asappropriate credit is given and the new creations are licensed under the identical terms.
- 中国神经再生研究(英文版)的其它文章
- NADPH oxidase 4 (NOX4) as a biomarker and therapeutic target in neurodegenerative diseases
- Circadian rhythm disruption and retinal dysfunction:a bidirectional link in Alzheimer’s disease?
- Interplay between the glymphatic system and neurotoxic proteins in Parkinson’s disease and related disorders: current knowledge and future directions
- Roles of neuronal lysosomes in the etiology of Parkinson’s disease
- Therapeutic advances in neural regeneration for Huntington’s disease
- The advantages of multi-level omics research on stem cell-based therapies for ischemic stroke

