桃PpSGR基因功能鉴定及其对乙烯合成的调控
邵允 张蒙蒙 陈云 王晓菲 董康 刘宁 张郎郎 谭彬 王伟 程钧 冯建灿

摘 要:【目的】乙烯合成及果肉褪綠是桃果实成熟过程中相伴出现的两个生理事件。STAY-GREEN(SGR)是参与植物叶片和果实褪绿的重要基因。然而,桃SGR基因在果实成熟及褪绿过程中的功能尚不清晰,旨在初步探究PpSGR基因在桃果实成熟及褪绿过程中的功能。【方法】以秋蜜红为试验材料,对PpSGR基因进行克隆,对PpSGR的核苷酸及氨基酸序列进行分析,对不同发育时期桃果肉中PpSGR的转录水平进行检测,并对PpSGR基因调控叶绿素降解及乙烯合成的功能进行研究。【结果】PpSGR编码区全长为831 bp;该基因编码的蛋白序列含有1个高度保守的SGR域。PpSGR基因的表达水平随着果肉逐渐褪绿呈现上升的趋势。瞬时过表达PpSGR基因后,桃叶片颜色明显褪绿,并且在300 mmol·L-1 NaCl的盐胁迫下,过表达PpSGR的叶片褪绿更加明显。此外,过表达PpSGR后,桃苗乙烯合成的限速基因PpACS1、PpACS4及PpACS6均表达上调,且内源乙烯释放量显著增多。【结论】对PpSGR的基因功能进行鉴定和研究,并分析了其对乙烯的调控作用,为进一步解析桃果实成熟及果肉褪绿提供了新的思路,也为不同成熟期桃品种的选育提供了理论基础。
关键词:桃;STAY-GREEN (SGR);褪绿基因;叶绿素降解;乙烯
中图分类号:S662.1 文献标志码:A 文章编号:1009-9980(2023)12-2513-11
收稿日期:2023-07-28 接受日期:2023-11-02
基金项目:河南省杰出外籍科学家工作室(GZS2020007);国家河南省联合项目(U1804114)
作者简介:邵允,女,在读硕士研究生,研究方向为果树分子生物学。E-mail:15937171226@163.com
*通信作者 Author for correspondence. E-mail:jcheng2007@163.com;E-mail:jcfeng@henau.edu.cn
Function identification of PpSGR gene and its regulation of ethylene synthesis in peach
SHAO Yun, ZHANG Mengmeng, CHEN Yun, WANG Xiaofei, DONG Kang, LIU Ning, ZHANG Langlang, TAN Bin, WANG Wei, CHENG Jun*, FENG Jiancan*
(College of Horticulture, Henan Agricultural University, Zhengzhou 450002, Henan, China)
Abstract: 【Objective】 Ethylene synthesis and disappearance of peel green color are two physiological events associated with fruit ripening of peach. STAY-GREEN (SGR) is a crucial gene that promotes disappearance of leaf and fruit green color in the plant kingdom. In various plant species, SGR genes exhibit similar functions in regulating chlorophyll. Peach (Prunus persica) is a significant fruit globally. However, the function of the PpSGR gene in the peach genome has not been identified, and its potential impact on green color disappearance and ripening of peach fruit remains unclear. This study aims to identify the SGR gene in the peach genome, analyze the expression level of the PpSGR gene in different fruit maturity stages, and preliminarily investigate the function of the PpSGR gene. 【Methods】The nucleotide sequences of the PpSGR gene were cloned from the Qiumihong cultivar. Bioinformatics analysis was conducted to analyze the gene structure of PpSGR in peach, the secondary and tertiary structure of the PpSGR protein was predict, the amino acid sequences of SGR in different species were compared, and a phylogenetic tree was constructed. Subsequently, the transcriptional level of the PpSGR gene in peach fruits at different maturity stages was analyzed using Quantitative Real-time PCR (qRT-PCR). The function of the PpSGR gene in chlorophyll degradation was preliminarily verified by transient overexpression in peach seedlings. Numerous studies have reported that the SGR gene can promote chlorophyll degradation under salt stress, leading to leaf yellowing. To verify the function of SGR under salt stress, the Fv/Fm values were measured and analyzed using a chlorophyll fluorescence analyzer after dark treatment of the whole peach seedlings transiently infected with the PpSGR gene. Additionally, there is evidence suggesting that ethylene can promote the expression of the SGR gene, but whether the SGR gene has a direct effect on ethylene synthesis remains to be studied. Therefore, we measured ethylene production and the transcriptional level of PpACSs genes involved in ethylene synthesis in the peach seedlings transiently overexpressing the PpSGR gene. 【Results】 The results of the bioinformatics analysis revealed that the length of the PpSGR gene sequencewas 1115 bp, consisting of 4 exons and 3 introns. The coding region of PpSGR spanned a total length of 831 bp, encoding 277 amino acids. Comparative analysis of SGR proteins from different species demonstrated that the PpSGR protein shared high homology with SGR proteins from other species. The middle region of the SGR protein contained a highly conserved SGR domain, while the N-terminal region contained a conserved chloroplast transit peptide. The C-terminal region of the SGR protein exhibited variability across different species. The amino acid sequence similarities between PpSGR and MdSGR, FvSGR or CsSGR were found to be 77.63%, 69.83% and 63.05%, respectively. A phylogenetic tree was constructed using amino acid sequences of SGR proteins from 13 different plant species, revealing that the PpSGR protein from peach was closely related to apples and strawberries, which belonged to the same Rosaceae family. The predicted tertiary structure of the PpSGR protein consisted mainly of α-helices and random graph curls, which was consistent with the predicted secondary structure. qRT-PCR results indicated that the transcription level of the PpSGR gene was lower during the early stage of peach fruit development when the flesh was still green. As the fruit matured gradually, the flesh turned green, and the expression level of PpSGR increased continuously. At 120 days after the full-bloom stage, the expression of PpSGR increased rapidly, reaching a level of 16 times higher than that at 80 days after the full-bloom stage, with the highest expression observed at 180 days after the full-bloom stage. This suggested that the expression level of the PpSGR gene gradually increased with the decolorization of peach fruit. Phenotypic analysis of PpSGR transgenic plants revealed that the leaves of transient PpSGR plants were light green, whereas the control plants had dark green leaves. This suggested that transient overexpression of the PpSGR gene led to chlorosis in peach seedlings. Under salt stress conditions (300 mmol·L-1 NaCl), the leaves of peach plants transiently overexpressing PpSGR exhibited a slower rate of yellowing, compared to control plants. The Fv/Fm value of the leaves in PpSGR-overexpressing plants was significantly lower than that of the control plants, indicating that the PpSGR gene may play an important role in leaf yellowing induced by the abiotic stress. Furthermore, the expression levels of genes involved in ethylene production were analyzed in peach seedlings transiently overexpressing PpSGR. The results demonstrated that the ethylene synthesis genes PpACS1, PpACS4 and PpACS6 were significantly induced and up-regulated in these seedlings. The expression of PpACS1 in peach seedlings with PpSGR overexpression was more than 32 times higher than that in control seedlings. Additionally, the ethylene production of peach seedlings with PpSGR overexpression showed a gradual increase. 【Conclusion】 Based on the aforementioned findings, it can be concluded that the PpSGR gene plays a role in promoting chlorosis in peach leaves. Moreover, our preliminary analysis suggests that PpSGR may enhance the function of ethylene synthesis in peach seedlings. This finding holds great significance for understanding the molecular mechanism of SGR in regulating ethylene synthesis. Additionally, it also provides some theoretical references for SGR gene function research, peach fruit ripening and post-harvest preservation.
Key words: Peach; STAY-GREEN (SGR); Degreening gene; Chlorophyll degradation; Ethylene
叶绿素降解是叶片衰老、果实成熟进程中发生的一系列重要的生化反应,由6种叶绿素代谢酶(chlorophyll catabolic enzymes,CCE)共同催化完成[1]。除了这6种CCE之外,STAY GREEN(SGR)在叶绿素降解过程中也发挥着非常重要的作用。SGR可以和6个CCE、光系统Ⅱ形成动态的蛋白复合体,从而形成叶绿素代谢通道,最大程度上减少代谢中间产物对细胞的毒害。有研究表明,SGR既不具有CCE活性,也不能直接结合叶绿素,这表明SGR可能不是一种直接参与叶绿素代谢的酶[2]。SGR参与叶绿素降解的功能在大豆[3]、水稻[4]、柑橘[5]、拟南芥[6]、甘蓝型油菜[7]等多个物种中都得到了验证。例如,Wang等[3]对大豆颜色变化的研究表明,GmSGR在控制种子黄化方面具有重要作用;Peng等[4]研究发现,敲除水稻OsSGR后植株能保持绿色;Zhu等[5]研究显示,CsSGRa能使烟草叶片中叶绿素含量显著降低,而突变CsSGRa后,棕橙果实和叶片中叶绿素的降解受到抑制,叶绿素含量显著增加。同样,在拟南芥中,AtSGR1被报道具有诱导叶绿素和叶绿素结合蛋白降解的作用,AtSGR1过表达后诱导拟南芥叶片逐渐褪绿[8]。此外,在番茄、辣椒、香蕉等植物中,发现SGR1同源基因突变后可延缓叶绿素的降解速度[9-12]。SGR基因具有响应非生物胁迫的功能,在盐胁迫下会表现出早期衰老和叶片褪绿速度加快的现象,而sgr突变体在非生物胁迫条件下表现出不褪绿的现象[9-10,13]。许多研究表明,在植物发育和衰老过程中叶绿素的分解代谢受到遗传程序的高度调控,因此,在非生物胁迫下对SGR的基因功能进行研究,对进一步阐明叶绿素降解的未知调节机制也具有重要作用。
不同物种中SGR基因在调控叶绿素方面表现出的类似现象,可能是由于不同物种中SGR蛋白结构较为保守(在其N-端含有一个叶绿体的信号肽,C端存在一个多变区域,两者之间存在一个高度保守的SGR保守域,且在C端多变区域中含有一个保守的富含半胱氨酸的基序)。
桃(Prunus persica)是广受消费者喜爱的重要水果之一。PpSGR基因在桃中的功能尚未鉴定,其对桃果实成熟可能存在的影响也未得到深入研究。笔者在本研究中对其编码的蛋白结构特征进行了生物信息学分析,并对PpSGR的基因功能进行鉴定和研究,这对桃成熟及采后保鲜等方面的研究具有一定的启发意义,也为进一步研究PpSGR基因在桃中的生物学功能奠定理论基础。
1 材料和方法
1.1 试验材料
以新疆毛桃实生苗为试验材料。将毛桃种子置于4 ℃低温贮存1个月,温水浸种后去除1/3种皮进行催芽5~7 d,将种胚单独种植在营养钵中,放置于温度为25 ℃、光周期为长日照条件(16 h光照/8 h黑暗)、光照为白光(140 μmol·m-2·s-1)的温室培养至幼苗,待试验使用[14]。
1.2 桃PpSGR过表达载体的构建及转化
根据PpSGR目的基因编码区(CDS)的全长序列,结合pSAK277载体上的EcoRⅠ和XhoⅠ为酶切位点,进行同源重组引物的设计,由上海生工公司进行引物的合成。以秋蜜红茎尖的cDNA为模板,利用合成的全长引物(pSAK277-PpSGR-F:AAAGAATTCGGTACCATGGGTACTTTGACTGCTGCTTC;pSAK277-PpSGR-R:TTTGTAATCCTCGAGGTTTGTTTCTTGGGTTTGGC)扩出PpSGR目的基因CDS区的全长序列,用限制性内切酶EcoRⅠ和XhoⅠ对空载体进行双酶切以获得线性化载体,于水浴锅中37 ℃酶切2~3 h。待PCR扩增反应及酶切反应结束后,用200 V的电压,经1.0%琼脂糖凝胶电泳检测,切下带有目的基因条带的凝胶,利用DNA凝胶回收试剂盒(庄盟生物)进行目的DNA片段的回收及纯化,将所得到的DNA溶液进行浓度及质量检测后用于后续的载体连接或置于-20 ℃冰箱中保存备用。利用SE无缝克隆和组装试剂盒SE Seamless Cloning and Assembly Kit(ZC231,北京庄盟生物)对PpSGR基因的纯化产物与线性化载体进行重组连接反应,以构建35S启动子驱动的pSAK277-PpSGR的过表达载体。步骤为:在冰上进行无缝克隆的连接反应体系的配制,轻轻将其混匀后,放于PCR仪上,37 ℃条件下反应30 min。
将构建好的载体质粒使用冻融法转入根癌农杆菌菌株GV3101中,利用真空渗透的方法瞬时侵染桃幼苗。
1.3 叶片颜色观察及叶绿素含量分析
将GV3101::SAK277-PpSGR基因侵染桃苗后,对整株植物进行黑暗处理,黑暗孵育后对叶片进行取样,使用叶绿素荧光分析仪(捷克PSI Flu)进行分析。如前所述[15]進行盐胁迫分析,将3周龄植物的离体叶片背面朝上漂浮在300 mmol·L-1 NaCl的3 mmol·L-1 MES 缓冲液(pH 5.8)中孵育数天。
1.4 总RNA的提取、cDNA的合成和实时定量PCR
提取转PpSGR基因秋蜜红桃的叶片总RNA,反转录第一链cDNA。实时定量PCR鉴定乙烯合成相关基因PpACS1、PpACS4、PpACS5和PpACS6的表达水平。利用SPSS Statistics 22对数据进行方差分析,用GraphPad Prism 8软件进行作图。按照DNase Ⅰ(TaKaRa,Dalian,China)试剂盒步骤除去基因组DNA污染。根据SYBR? Primescript miRNA RT-PCR Kit(TaKaRa,Dalian,China)试剂盒说明书进行第一链cDNA的合成。定量RT-PCR反应体系为20 μL,含有1 × ROX参比染料,10 μL 2 × SYBR预混物Ex Taq Ⅱ(TaKaRa),每个引物浓度0.4 μmol·L-1,模板cDNA 100 ng。所有分析均采用3个生物学重复,qPCR所用引物见表1。
1.5 叶绿素含量的测定
称取0.3 g左右的桃叶片放入干净无水的研钵内,加入少量的碳酸钙粉末和3 mL 95%乙醇,使用干净的研磨棒进行充分研磨,再向其中加入10 mL 95%乙醇,继续研磨至组织变白,放置在避光条件下,静置5 min后,使用滤纸将研磨液过滤至25 mL的棕色容量瓶中,并用少量的95%乙醇冲洗滤纸定容至25 mL。以95%乙醇作为空白对照,分别在波长665 nm、649 nm下测定提取液的吸光度[16-17]。
根据以下公式计算叶绿素含量:
C叶绿素a/(mg·L-1)=13.95×A665-6.88×A649;
C叶绿素b/(mg·L-1)=24.96×A649-7.32×A665;
w(叶绿素a)/(mg·g-1)=C叶绿素a×V×N/W×0.001;
w(叶绿素b)/(mg·g-1)=C叶绿素b×V×N/W×0.001;
w(总叶绿素)/(mg·g-1)=w(叶绿素a)+w(叶绿素b)。
式中,V为提取液体积,N为稀释倍数,W为鲜质量(g)。
1.6 乙烯释放量的测定
取处理后生长良好的桃幼苗5 g;将上述材料分别放入100 mL 色谱瓶中,使用气相色谱分析仪(岛津GC-2010 Plus)进行乙烯测定,每个处理3次重复。色谱条件:WBI为110 ℃,柱温 50 ℃,FID为200 ℃,氢气流速为50 mL·min-1,进样量:1000 μL,运行时间5 min。
乙烯释放量/(ng·kg-1·s-1)=[乙烯质量浓度(ng·mL-1)×容器体积(mL)]/[样品质量(g)×密封时间(s)]。
1.7 生物信息学分析
利用Phytozome网站(https://phytozome.jgi.doe.gov/pz/portal.html)获得PpSGR和PpSGRL序列信息,同时根据文献下载其他物种中的同源序列。通过DNAMAN获得不同物种中SGR氨基酸序列比对图,并结合MEGA 7.0的Clustal W进行聚类分析。使用ProtParam(https://web.expasy.org/protparam/)在线分析,对PpSGR蛋白的分子质量、等电点、氨基酸数目、氨基酸组分、脂肪指数、亲水性等进行分析。用SOPMA(https://npsa-prabi.ibcp.fr/)在线软件对PpSGR蛋白的二级结构进行预测;使用Phyre2软件对PpSGR蛋白的三级结构进行预测。
2 结果与分析
2.1 PpSGR基因的序列特征
PpSGR基因组序列全长1115 bp,包含4个外显子和3个内含子。编码区全长831 bp,编码277个氨基酸。通过Protparam进行预测,等电点pI为8.05,属于碱性蛋白,相对分子质量约为31.18 ku。由20种氨基酸组成,带负电荷氨基酸残基数量为30,带正电荷氨基酸残基数量为32;不稳定系数为47.54,属于不稳定蛋白;脂肪系数为77.04,总平均亲水性为-0.386,属亲水蛋白(+值为疏水蛋白,-值为亲水蛋白)。
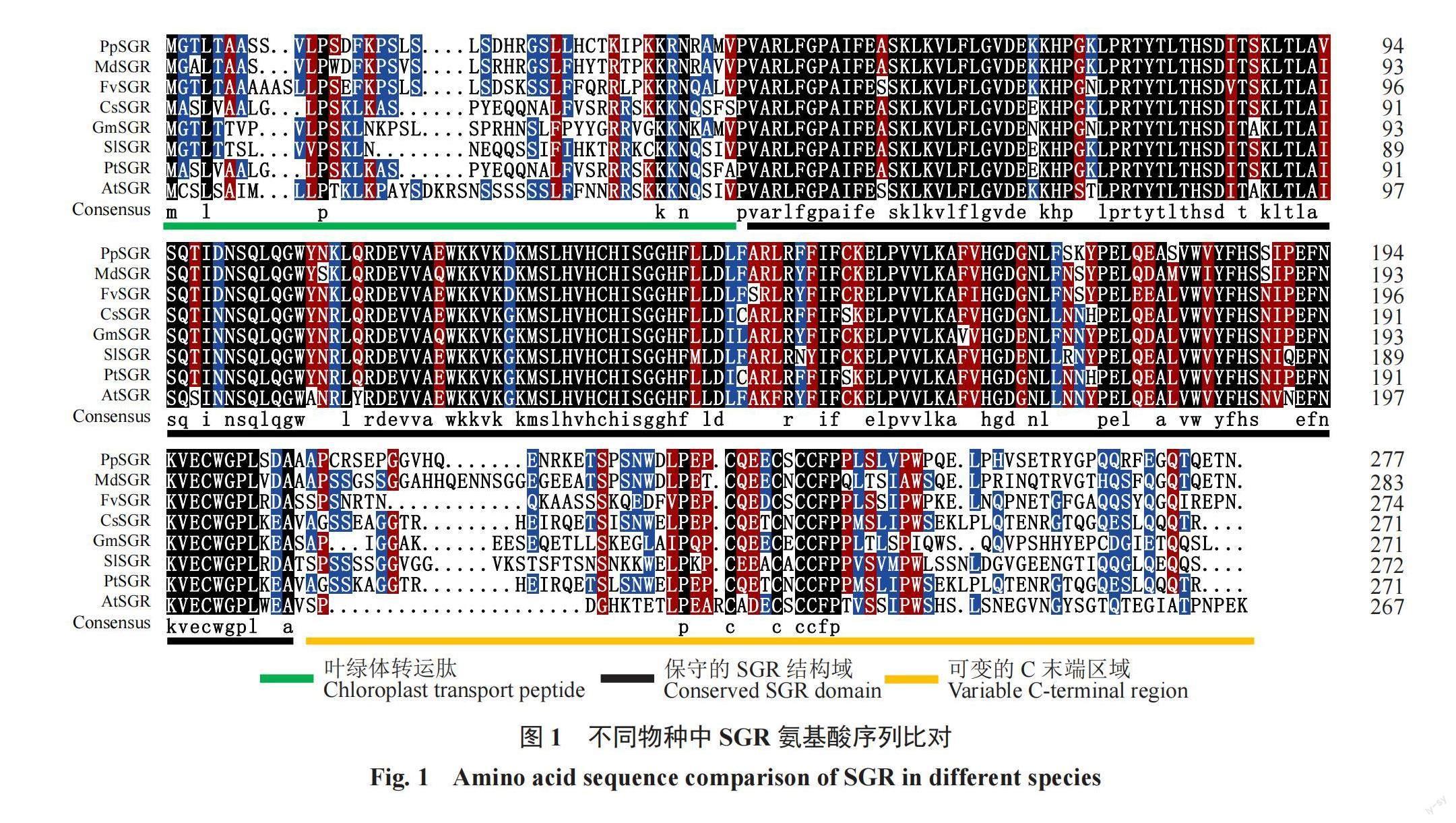
不同物种SGR氨基酸序列比对的结果显示,PpSGR蛋白与其他物种中SGR蛋白具有较高的同源率,且都含有保守的SGR保守域(SGR domain),N端的叶绿体信号肽(chloroplast transit peptide)和C端多变区域(variable C-terminal region)(图1)。其中PpSGR与苹果MdSGR蛋白的同源率最高,为77.89%,与草莓FvSGR蛋白的同源率为70.41%,与柑橘CsSGR蛋白和杨树PtSGR蛋白的同源率为64.29%,系统进化树的结果显示,桃PpSGR蛋白与同属蔷薇科的苹果、草莓的亲缘关系最近(图2)。
对PpSGR蛋白的二级结构和三级结构进行预测,蛋白质的多肽链通过折叠、螺旋和卷曲等二级结构形成比较稳定的空间结构,利用Sopma网站分析桃PpSGR的二级结构(图3-A),发现多个氨基酸参与α-螺旋、无规则卷曲和延伸链等二级结构的形成,其中α-螺旋占比27.44%、延伸链占比12.64%、无规则卷曲占比57.04%(表2)。采用在线软件Phyre2对PpSGR蛋白三级结构进行预测的结果(图3-B)显示,PpSGR蛋白的三级结构主要由α-螺旋和无规则图卷曲构成,与二级结构预测结果一致。
2.2 PpSGR在不同发育时期桃果实中的表达分析
叶绿素降解主要发生在果实成熟及叶片衰老的过程中。因此,笔者选用花后20 d、50 d、80 d、120 d、160 d、180 d,共6个时间点的桃果实,进行PpSGR转录水平分析。qRT-PCR分析的结果(图4)显示,在果实发育的前期果肉呈现明显绿色时,PpSGR转录水平较低。随着果实逐步成熟,果肉褪绿,表达量不断升高。在花后120 d,PpSGR的表达量快速上升,达到花后80 d的16倍,并在180 d达到最高。基于以上结果,推测PpSGR基因可能是调控桃果实褪绿的重要基因。
2.3 瞬时转化桃幼苗验证PpSGR的功能
为了验证PpSGR基因的功能,利用桃实生幼苗瞬时转化法对PpSGR基因的功能进行初步的验证。半定量结果显示,瞬时过表达的材料中PpSGR的转录水平明显上升(图5-A)。为进一步确认PpSGR瞬时转化的效果,随机选取7个单株,分别进行定量检测。结果显示,在每一个单株中都实现了PpSGR的过表达(图5-B)。对转基因植株表型分析,发现瞬转PpSGR植株叶片颜色呈现淡绿色,而对照植株呈现深绿色(图6-A),PpSGR植株葉片叶绿素含量较低而对照植株叶片叶绿素含量较高(图6-B)。上述结果表明,PpSGR具有促进叶绿素降解,进而导致桃叶片褪绿的功能。
2.4 盐胁迫下PpSGR促进叶片黄化
有研究表明,过表达SGR基因的植物植株叶片在非生物胁迫条件下会表现出早期衰老叶片褪绿的现象[9-10]。笔者在本研究中的结果显示,在300 mmol·L-1 NaCl的盐胁迫条件下,与空载相比,转PpSGR基因的桃叶片在盐处理5 d后出现明显的黄化表型(图7-A),且转PpSGR的桃叶片随着胁迫处理时间的延长,PSⅡ原初光能转化效率(Fv/Fm值)显著降低(图7-B)。
2.5 PpSGR基因对乙烯释放量的影响
乙烯是促进果实成熟、叶片衰老的重要内源激素。因此,推测瞬转PpSGR促进桃叶片褪绿的过程可能会对乙烯合成产生影响。乙烯合成酶(ACS)是乙烯合成途径中的关键酶,ACS基因家族是乙烯合成途径中的重要基因家族,它们编码乙烯合成酶,参与乙烯的生物合成。前人研究显示桃中共有6个ACS基因[18-19]。笔者在本研究中对转PpSGR桃幼苗中乙烯合成相关基因进行了定量分析。半定量和定量结果(图8)显示,除了ACS2和ACS3未检出转录水平外,乙烯合成基因PpACS1、PpACS4和PpACS6在转PpSGR桃幼苗中被显著诱导上调表达,其中转PpSGR桃幼苗中PpACS1表达量是对照的32倍以上;PpACS5表达量在转PpSGR桃幼苗中显著下调。
为确认PpSGR对桃苗乙烯合成的调控,设计了一个乙烯含量测量的容器,进行桃幼苗乙烯释放量的测定(图8-B)。结果显示,瞬转PpSGR的桃苗,其乙烯释放量呈现逐渐上升的趋势。在侵染4 d和6 d时,试验组与对照组乙烯释放量达到极显著差异水平。该结果表明,过表达PpSGR具有促进桃幼苗乙烯合成的功能。
3 讨 论
叶绿素是存在于类囊体中的一种色素蛋白,也是绿色植物吸收光能的重要物质,大量分布于植物的叶片和茎中[20-21]。植物绿色组织褪绿是植物在成熟和衰老过程中的叶绿素发生降解而引起的,例如果实成熟时果皮颜色变化及叶片老化黄化等[22-23]。在柑橘成熟过程中,果实外表皮的叶绿素发生降解,表皮变黄[24]。
乙烯对叶绿素的降解有一定的影响[25]。乙烯释放量增加会促进果实的成熟,导致叶绿素降解,如:成熟的葡萄果实会释放大量乙烯,引起葡萄穗梗中的叶绿素含量明显降低,并导致穗梗发生褐化[26],其中使得调控叶绿素降解速率的关键基因,如SGR1等的表达量明显升高[27],暗示着乙烯与SGR1基因之间可能存在着调控关系。此外,有研究表明,乙烯对SGR基因的表达也具有促进作用。在拟南芥中乙烯可以迅速诱导AtSGR1的表达[28];在豆科植物中间锦鸡儿中乙烯能通过诱导CiNAC1基因上调促进叶片衰老,进而促进叶绿素降解相关基因SGR1、SGR2的表达量升高[29]。Shimoda等[30]报道了STAY-GREEN(SGR),也称为NON-YELLOWING(NYE),Wei等[31]从高羊茅中分离得到FaNYE1,与大麦中HvSGR具有较高的同源率,在自然老化和黑暗条件下FaNYE1诱导表达,FaNYE1蛋白含量与植株中的叶绿素含量成反比,过表达FaNYE1会加速叶绿素的降解。SGR在植物叶绿素降解过程具有非常重要的作用。有研究者在拟南芥中发现SGRs编码脱镁螯合酶(MCS),能嵌入到Chl-蛋白酶复合体中提取Mg2+,具有酶的活性,从而对植物叶绿素降解具有重要调控作用[8,32]。目前,已有大量研究对SGR基因的褪绿功能进行验证。例如,当莱茵衣藻CrSGR在拟南芥细胞中过表达时,拟南芥叶片中叶绿素含量下降,证实了SGR基因在叶绿体中具有解镁活性,从而使得拟南芥叶片褪绿表现黄化[32];Sato等[33]在水稻和豌豆中找到了孟德尔的绿子叶突变体,即SGR,它能减缓在暗条件下对叶绿素的分解,结果表明,SGR参与叶绿素降解主要是通过叶绿素降解酶基因CNYC1/NOL、PAO實现的。Sakuraba等[15]发现在盐胁迫下拟南芥SGR1和SGRL可在衰老前快速降解叶绿素。笔者的研究结果表明,随着果实成熟PpSGR基因表达量逐渐升高,过表达PpSGR基因后桃叶片出现褪绿表型,并且在盐胁迫条件下,PpSGR促进了叶片黄化。上述结果证实了PpSGR具有促进桃叶片褪绿的功能。
在桃果实成熟过程中会产生大量乙烯,同时果皮和果肉褪绿也相伴发生。前人的大量研究显示,乙烯可以上调SGR1表达促进褪绿发生[27-28]。但也有研究指出,在番茄中沉默SlSGR1能通过改变乙烯相关基因的表达进而延长果实货架期。这意味着SGR1可能具有反馈调节乙烯合成或乙烯信号通路的功能[34-35]。乙烯合成酶基因家族成员在乙烯的生物合成和信号传递过程中发挥重要功能,如Zeng等[19]的结果表明,PpACS1在调控油桃成熟阶段的乙烯产量方面发挥着关键作用。本研究中,过表达PpSGR后,乙烯合成关键基因ACS表达量也随之变化,除了ACS2和ACS3未检出转录水平外,乙烯合成基因PpACS1、PpACS4和PpACS6在转PpSGR桃幼苗中均显著诱导上调表达,PpACS5表达量在转PpSGR桃幼苗中却显著下调。这与桃果实成熟后乙烯合成相关基因的表达量变化类似,笔者的结果表明,转PpSGR的桃苗中PpACS1的表达量超出对照32倍。乙烯含量测定结果显示,在过表达PpSGR后,桃幼苗乙烯的释放量显著增加。上述结果表明SGR可能具有促进乙烯合成的功能。
SGR除了具有酶的活性,另外也有研究指出,SGRs蛋白还可以通过与LHCII互作或招募主要的Chl降解酶来实现调控过程[36]。也正是由于SGR在叶绿素降解中充当的角色尚不够清晰,因此,进一步解析SGR是如何调控ACS基因的表达从而影响乙烯的合成具有重要意义,这将加深对SGR在叶绿素降解以外的生理过程中潜在作用的理解。
4 结 论
本研究表明,PpSGR基因随着果实成熟转录水平逐渐上升;瞬时过表达结果表明,PpSGR具有促进叶片褪绿的功能。此外,发现过表达PpSGR后,桃苗乙烯合成限速基因PpACS1的表达显著上调32倍,且内源乙烯释放量显著增多。基于本研究的结果,初步推测PpSGR具有促进桃幼苗乙烯合成的功能,这对桃果实成熟以及果实采后保鲜等方面的研究具有一定的启发意义,也为SGR基因功能研究提供了一定的理论参考。
參考文献 References:
[1] SAKURABA Y,PARK S Y,KIM Y S,WANG S H,YOO S C,H?RTENSTEINER S,PAEK N C. Arabidopsis STAY-GREEN2 is a negative regulator of chlorophyll degradation during leaf senescence[J]. Molecular Plant,2014,7(8):1288-1302.
[2] SAKURABA Y,PARK S Y,PAEK N C. The divergent roles of STAYGREEN (SGR) homologs in chlorophyll degradation[J]. Molecules and Cells,2015,38(5):390-395.
[3] WANG C,GAO L E,LI R Z,WANG Y E,LIU Y Y,ZHANG X,XIE H. High-throughput sequencing reveals the molecular mechanisms determining the stay-green characteristic in soybeans[J]. Journal of Biosciences,2020,45:103.
[4] PENG Y Y,LIAO L L,LIU S,NIE M M,LI J A,ZHANG L D,MA J F,CHEN Z C. Magnesium deficiency triggers SGR-mediated chlorophyll degradation for magnesium remobilization[J]. Plant Physiology,2019,181(1):262-275.
[5] ZHU K J,ZHENG X J,YE J L,HUANG Y,CHEN H Y,MEI X H,XIE Z Z,CAO L X,ZENG Y L,LARKIN R M,XU Q,PEREZ-ROMAN E,TAL?N M,ZUMAJO-CARDONA C,WURTZEL E T,DENG X X. Regulation of carotenoid and chlorophyll pools in hesperidia,anatomically unique fruits found only in Citrus[J]. Plant Physiology,2021,187(2):829-845.
[6] ONO K,KIMURA M,MATSUURA H,TANAKA A,ITO H. Jasmonate production through chlorophyll a degradation by Stay-Green in Arabidopsis thaliana[J]. Journal of Plant Physiology,2019,238:53-62.
[7] 唐玉凤,姚敏,何昕,官梅,刘忠松,官春云,钱论文. 甘蓝型油菜SGR基因家族的全基因组鉴定与功能分析[J]. 作物学报,2023,49(7):1829-1842.
TANG Yufeng,YAO Min,HE Xin,GUAN Mei,LIU Zhongsong,GUAN Chunyun,QIAN Lunwen. Genome-wide identification and functional analysis of SGR gene family in Brassica napus L.[J]. Acta Agronomica Sinica,2023,49(7):1829-1842.
[8] SHIMODA Y,ITO H,TANAKA A. Arabidopsis STAY-GREEN,Mendels green cotyledon gene,encodes magnesium-dechelatase[J]. The Plant Cell,2016,28(9):2147-2160.
[9] REN G D,AN K,LIAO Y,ZHOU X,CAO Y J,ZHAO H F,GE X C,KUAI B K. Identification of a novel chloroplast protein AtNYE1 regulating chlorophyll degradation during leaf senescence in Arabidopsis[J]. Plant Physiology,2007,144(3):1429-1441.
[10] PARK S Y,YU J W,PARK J S,LI J J,YOO S C,LEE N Y,LEE S K,JEONG S W,SEO H S,KOH H J,JEON J S,PARK Y I,PAEK N C. The senescence-induced staygreen protein regulates chlorophyll degradation[J]. The Plant Cell,2007,19(5):1649-1664.
[11] BARRY C S,MCQUINN R P,CHUNG M Y,BESUDEN A,GIOVANNONI J J. Amino acid substitutions in homologs of the STAY-GREEN protein are responsible for the green-flesh and chlorophyll retainer mutations of tomato and pepper[J]. Plant Physiology,2008,147(1):179-187.
[12] YANG X T,ZHANG Z Q,JOYCE D,HUANG X M,XU L Y,PANG X Q. Characterization of chlorophyll degradation in banana and plantain during ripening at high temperature[J]. Food Chemistry,2009,114(2):383-390.
[13] SAKURABA Y,LEE S H,KIM Y S,PARK O K,H?RTENSTEINER S,PAEK N C. Delayed degradation of chlorophylls and photosynthetic proteins in Arabidopsis autophagy mutants during stress-induced leaf yellowing[J]. Journal of Experimental Botany,2014,65(14):3915-3925.
[14] 蔣亚君. 桃PpIDD4、PpIDD12和PpIDD13参与DELLA依赖的赤霉素合成反馈调控及PpIDD11调控雌蕊柱头伸长的功能[D]. 郑州:河南农业大学,2022.
JIANG Yajun. PpIDD4,PpIDD12 and PpIDD13 are involved in DELLA-dependent feedback regulation of gibberellin biosynthesis and PpIDD11 regulates pistil stigma elongation in peach[D]. Zhengzhou:Henan Agricultural University,2022.
[15] SAKURABA Y,KIM D,KIM Y S,H?RTENSTEINER S,PAEK N C. Arabidopsis STAYGREEN-LIKE (SGRL) promotes abiotic stress-induced leaf yellowing during vegetative growth[J]. FEBS Letters,2014,588(21):3830-3837.
[16] 项倩,吴磊,徐若涵,杨再强. 不同温度下染病番茄叶片SPAD和叶绿素含量的相关性[J]. 北方园艺,2022(18):8-15.
XIANG Qian,WU Lei,XU Ruohan,YANG Zaiqiang. Correlation between SPAD and chlorophyll content in infected tomato leaves at different temperatures[J]. Northern Horticulture,2022(18):8-15.
[17] 张宪政. 植物叶绿素含量测定:丙酮乙醇混合液法[J]. 辽宁农业科学,1986(3):26-28.
ZHANG Xianzheng. Determination of plant chlorophyll content by acetone ethanol mixed solution[J]. Liaoning Agricultural Science,1986(3):26-28.
[18] 陈星星,张斌斌,郭绍雷,马瑞娟,姜卫兵. 不同肉质型桃果实成熟过程中乙烯生物合成相关基因的表达差异[J]. 南京农业大学学报,2020,43(4):637-644.
CHEN Xingxing,ZHANG Binbin,GUO Shaolei,MA Ruijuan,JIANG Weibing. The expression differences of genes related to ethylene biosynthesis in different fleshy peach fruits during ripening[J]. Journal of Nanjing Agricultural University,2020,43(4):637-644.
[19] ZENG W F,PAN L,LIU H,NIU L,LU Z H,CUI G C,WANG Z Q. Characterization of 1-aminocyclopropane-1-carboxylic acid synthase (ACS) genes during nectarine fruit development and ripening[J]. Tree Genetics & Genomes,2015,11(2):18.
[20] ZHU X Y,CHEN J Y,QIU K,KUAI B K. Phytohormone and light regulation of chlorophyll degradation[J]. Frontiers in Plant Science,2017,8:1911.
[21] KUAI B K,CHEN J Y,H?RTENSTEINER S. The biochemistry and molecular biology of chlorophyll breakdown[J]. Journal of Experimental Botany,2018,69(4):751-767.
[22] LAI B,HU B,QIN Y H,ZHAO J T,WANG H C,HU G B. Transcriptomic analysis of Litchi chinensis pericarp during maturation with a focus on chlorophyll degradation and flavonoid biosynthesis[J]. BMC Genomics,2015,16(1):225.
[23] LIRA B S,DE SETTA N,ROSADO D,ALMEIDA J,FRESCHI L,ROSSI M. Plant degreening:Evolution and expression of tomato (Solanum lycopersicum) dephytylation enzymes[J]. Gene,2014,546(2):359-366.
[24] YIN X R,XIE X L,XIA X J,YU J Q,FERGUSON I B,GIOVANNONI J J,CHEN K S. Involvement of an ethylene response factor in chlorophyll degradation during citrus fruit degreening[J]. The Plant Journal,2016,86(5):403-412.
[25] 李志谦,李靖雯,邹东方,黄莹莹,叶霞,冯建灿. 葡萄采后贮藏过程中穗梗褐变的研究进展[J]. 河南农业大学学报,2021,55(5):815-820.
LI Zhiqian,LI Jingwen,ZOU Dongfang,HUANG Yingying,YE Xia,FENG Jiancan. Research progress of rachis browning in postharvest storage of grape[J]. Journal of Henan Agricultural University,2021,55(5):815-820.
[26] LI L,KAPLUNOV T,ZUTAHY Y,DAUS A,PORAT R,LICHTER A. The effects of 1-methylcyclopropane and ethylene on postharvest rachis browning in table grapes[J]. Postharvest Biology and Technology,2015,107:16-22.
[27] 李具鹏,傅茂润,杨晓颖. 1-MCP处理对采后葡萄果梗褐变及叶绿素降解相关基因的影响[J]. 食品工业科技,2018,39(20):268-273.
LI Jupeng,FU Maorun,YANG Xiaoying. Effect of 1-MCP treatment on postharvest browning and chlorophyll breakdown pathway related genes in grape rachis[J]. Science and Technology of Food Industry,2018,39(20):268-273.
[28] QIU K,LI Z P,YANG Z,CHEN J Y,WU S X,ZHU X Y,GAO S,GAO J,REN G D,KUAI B K,ZHOU X. EIN3 and ORE1 accelerate degreening during ethylene-mediated leaf senescence by directly activating chlorophyll catabolic genes in Arabidopsis[J]. PLoS Genetics,2015,11(7):e1005399.
[29] 岳文冉,岳俊燕,張秀娟,杨杞,韩晓东,王瑞刚,李国婧. 中间锦鸡儿CiNAC1基因促进转基因拟南芥叶片的衰老[J]. 中国生物工程杂志,2018,38(4):24-29.
YUE Wenran,YUE Junyan,ZHANG Xiujuan,YANG Qi,HAN Xiaodong,WANG Ruigang,LI Guojing. The CiNAC1 from Caragana intermedia promotes transgenic Arabidopsis leaf senescence[J]. China Biotechnology,2018,38(4):24-29.
[30] SAKURABA Y,SCHELBERT S,PARK S Y,HAN S H,LEE B D,ANDR?S C B,KESSLER F,H?RTENSTEINER S,PAEK N C. STAY-GREEN and chlorophyll catabolic enzymes interact at light-harvesting complex II for chlorophyll detoxification during leaf senescence in Arabidopsis[J]. The Plant Cell,2012,24(2):507-518.
[31] WEI Q,GUO Y J,KUAI B K. Isolation and characterization of a chlorophyll degradation regulatory gene from tall fescue[J]. Plant Cell Reports,2011,30(7):1201-1207.
[32] MATSUDA K,SHIMODA Y,TANAKA A,ITO H. Chlorophyll a is a favorable substrate for chlamydomonas Mg-dechelatase encoded by STAY-GREEN[J]. Plant Physiology and Biochemistry,2016,109:365-373.
[33] SATO Y,MORITA R,NISHIMURA M,YAMAGUCHI H,KUSABA M. Mendels green cotyledon gene encodes a positive regulator of the chlorophyll-degrading pathway[J]. Proceedings of the National Academy of Sciences of the United States of America,2007,104(35):14169-14174.
[34] LUO Z D,ZHANG J H,LI J H,YANG C X,WANG T T,OUYANG B,LI H X,GIOVANNONI J,YE Z B. A STAY-GREEN protein SlSGR1 regulates lycopene and β-carotene accumulation by interacting directly with SlPSY1 during ripening processes in tomato[J]. New Phytologist,2013,198(2):442-452.
[35] BARRY C S. The stay-green revolution:Recent progress in deciphering the mechanisms of chlorophyll degradation in higher plants[J]. Plant Science,2009,176(3):325-333.
[36] SAKURABA Y,SCHELBERT S,PARK S Y,HAN S H,LEE B D,ANDR?S C B,KESSLER F,H?RTENSTEINER S,PAEK N C. STAY-GREEN and chlorophyll catabolic enzymes interact at light-harvesting complex II for chlorophyll detoxification during leaf senescence in Arabidopsis[J]. The Plant Cell,2012,24(2):507-518.

