Association between heat shock factor protein 4 methylation and colorectal cancer risk and potential molecular mechanisms: A bioinformatics study
Wen-Jing Zhang,Ke-Lin Yue,Jing-Zhai Wang,Yu Zhang
Abstract BACKGROUND We previously demonstrated that heat shock factor protein 4 (HSF4) facilitates colorectal cancer (CRC) progression.DNA methylation,a major modifier of gene expression and stability,is involved in CRC development and outcome.AIM To investigate the correlation between HSF4 methylation and CRC risk,and to uncover the underlying molecular mechanisms.METHODS Differences in β values of HSF4 methylation loci in multiple malignancies and their correlation with HSF4 mRNA expression were analyzed based on Shiny Methylation Analysis Resource Tool.HSF4 methylation-related genes were identified by LinkedOmics in CRC,and Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analyses were performed.Protein-protein interaction network of HSF4 methylation-related genes was constructed by String database and MCODE algorithm.RESULTS A total of 19 CpG methylation loci were identified in HSF4,and their β values were significantly increased in CRC tissues and exhibited a positive correlation with HSF4 mRNA expression.Unfortunately,the prognostic and diagnostic performance of these CpG loci in CRC patients was mediocre.In CRC,there were 1694 HSF4 methylation-related genes;1468 of which displayed positive and 226 negative associations,and they were involved in regulating phenotypes such as immune,inflammatory,and metabolic reprogramming.EGFR,RELA,STAT3,FCGR3A,POLR2K,and AXIN1 are hub genes among the HSF4 methylation-related genes.CONCLUSION HSF4 is highly methylated in CRC,but there is no significant correlation between it and the prognosis and diagnosis of CRC.HSF4 methylation may serve as one of the ways in which HSF4 mediates the CRC process.
Key Words: Colorectal cancer;DNA methylation;Prognosis;Diagnosis;Bioinformatics;Heat shock factor protein 4
INTRODUCTION
Colorectal cancer (CRC) is a malignant tumor of the digestive tract that occurs in the rectum,cecum,and entire colon,with symptoms such as abdominal pain,difficulty passing stool,constipation,or diarrhea[1].According to the latest statistics from the World Health Organization[2],CRC has become the third most common malignancy worldwide after lung cancer and breast cancer,with about 200000 new cases occurring worldwide each year,of which,916000 die from CRC.According to the American Society of Clinical Oncology[3],the 5-year survival rate for patients with CRC is approximately 65%.Nevertheless,most CRC patients have already developed distal metastases by the time they receive a definitive diagnosis,which leads to a shrinking 5-year survival rate to 14%[3].Consequently,the search for new biomarkers will facilitate the timely diagnosis of CRC and provide new insights into the mechanisms of CRC occurrence and development.
DNA methylation is a process of chemical modification of DNA that affects biological processes such as gene expression,cell differentiation and development[4-6].In epigenetics,DNA methylation is an important marker of cellular genetic information and is widely applied in cancer prediction and diagnosis[7,8].For CRC,the United States Food and Drug Administration currently approves SEPT9 (blood samples) and a combination ofNDRG4andBMP3(stool samples) as commercially available biomarkers related to methylation[9].In addition,APC,SFRP1,SFRP2,SDC2,MGMT,VIMandNDRG4are methylation-related candidate markers of CRC[10].Mechanistically,DNA methylation can inhibit gene transcription or activate gene expression,thereby affecting protein synthesis to mediate the cancer process.For instance,teashirt zinc finger homeobox 3 (TSHZ3) promoter methylation effectively suppresses TSHZ3 expression,which facilitates CRC growth and metastasis[11].Heparanase 2 (HPSE2) is highly methylated in CRC and is associated with poor patient prognosis,and high methylation ofHPSE2reducesHPSE2expression,which inhibits the p53/p21 signaling cascade and facilitates proliferation of CRC cellsin vivoandin vitro[12].Heat shock response (HSR),an ancient cellular self-protective response,helps tumor cells to survive and proliferate smoothly under the stimulation of adverse microenvironment,oxidative stress and other stressors[13].Heat shock factor protein 4 (HSF4),a member of the heat shock transcription factor family,plays an important role in HSR by preventing abnormal protein folding and aggregation to maintain intracellular protein homeostasis[13,14].HSF4 has been identified as a cancer-promoting factor in lymphoma[15],breast cancer[16],and cervical cancer[17].Our previous study demonstrated thatHSF4is significantly upregulated in CRC,which predicts poor patient prognosis,and that it promotes CRC progression by enhancing the activity of c-MET and downstream ERK1/2 and AKT signaling pathways[18].Nevertheless,whether DNA methylation is involved in HSF4-mediated CRC progression remains to be investigated.
This study investigated the correlation betweenHSF4methylation andHSF4expression,and its prognostic and diagnostic value in CRC,and aimed to identify the potential molecular mechanisms associated withHSF4methylation through bioinformatics analysis.The aim was to provide a theoretical basis and a novel perspective forHSF4as a methylation-related biomarker for future CRC diagnosis and treatment.
MATERIALS AND METHODS
Differential analysis of HSF4 methylation and its prognostic and diagnostic value
The Shiny Methylation Analysis Resource Tool (SMART) APP is an interactive and user-friendly web application for comprehensive analysis of DNA methylation in the The Cancer Genome Atlas (TCGA) project,with data from TCGA (https://portal.gdc.cancer.gov/)[19].The level of methylation at each CpG loci ofHSF4was assessed using the β value,which is the ratio of the methylation of the allele to the intensity of unmethylation,ranging from 0 to 1.In this study,we analyzed differences in the β values of 19 methylation probes associated withHSF4in 33 malignancies by SMART,including COAD,READ,BRCA,LAML,LGG,LIHC,BLCA,CESC,CHOL,KIRP,SKCM,LUAD,ACC,DLBC,KIRC,PCPG,OV,ESCA GBM,STAD,UCEC,UCS,HNSC,TGCT,THCA,THYM,KICH,PRAD,SARC,LUSC,MESO,PAAD,and UVM.Wilcoxon rank sum test was performed for difference analysis of β values,and data was adjusted using the Benjamini-Hochberg method.In addition,β values of 19HSF4-related methylation probes were analyzed differentially in COAD stages and their correlation with HSF4 mRNA expression based on SMART.The differential analysis of β values in COAD stages was performed based on ANOVA,and the correlation between β values of each probe andHSF4mRNA expression was performed based on Pearson.The COAD dataset in SMART was extracted andHSF4methylation in prognostic and diagnostic value of COAD was assessed by survival (https://cran.r-project.org/web/packages/survival/index.html) and pROC[20]/timeROC[21] R packages,respectively.Kaplan-Meier survival curves,Receiver operating characteristic (ROC) curves and time-dependent ROC curves were visualized with the ggplot2 R package[22].Patient information is shown in Table 1.

Table 1 Basic information of the dataset in this study
Identification of HSF4 methylation-related genes and their enrichment analysis
LinkedOmics is a publicly available portal that includes three analysis modules,LinkFinder,LinkInterpreter and LinkCompare,to support users in performing multi-omics analysis in cancer,with data from TCGA (https://www.cancer.gov/ccg/ research/genome-sequencing/tcga) and Clinical Proteomic Tumor Analysis Consortium (https://proteomics.cancer.gov/programs/cptac)[23].In this study,genes associated withHSF4methylation were identified at COAD through LinkedOmics.HSF4methylation-associated genes were identified by Spearman and subjected to correction by the Benjamini-Hochberg method.Finally,HSF4methylation-related genes were displayed by volcano plot and heatmap.Enrichment analysis ofHSF4methylation-related genes was performed by hypergeometric distribution algorithm based on Gene Ontology (GO)[24] and Kyoto Encyclopedia of Genes and Genomes (KEGG)[25] databases,and presented by bubble and histogram plots.The above results was visualized with the ggplot2 R package[22].
Protein-protein interaction network construction for HSF4 methylation-associated genes
The protein-protein interaction (PPI) network construction forHSF4methylation-related genes was based on the String database[26],CytoScape software[27] and the MCODE plugin[28].Briefly,HSF4methylation-related genes obtained from LinkedOmics were extracted,and the interactions of these genes were predicted from the String database.The minimum required interaction score of the String database was set to highest confidence.The interactions were imported into CytoScape software (version:3.8.2) for visualization and clustering analysis of the PPI network was performed by the MCODE algorithm.The parameters of MCODE are degree cutoff=2,node density cutoff=0.1,node score cutoff=0.2,Kcore=2,Max depth=100.
RESULTS
Identification of HSF4 methylation levels
HSF4 is located on chromosome 16 with 19 CpG loci,with 14 on CpG island,three on N Shore and two on S Shore (Figure 1).Differential analysis revealed that β values ofHSF4CpG-aggregation methylation were significantly enhanced in most malignancies,including COAD,and READ (Figure 1B).Similarly,the β values of each CpG site were significantly higher in most malignant tumors than in the corresponding paracancerous tissues (Supplementary Figure 1).It is notable that all CpG loci ofHSF4had significantly elevated β values in these malignancies only in COAD (Supplementary Figure 1 and Figure 2).In READ,only two probes,cg07188665 and cg09567485,exhibited no significant difference in β values.We analyzed the methylation levels of HSF4 CpG loci in different tumor stages.The β values of cg06277900,cg03811260,cg04580872,cg06621126,cg03887094 and cg09567485 probes were significantly different at various stages of COAD (Supplementary Figure 2).Therefore,we further explored the correlation betweenHSF4methylation andHSF4expression.In COAD,the β values of the probes displayed a significant positive correlation withHSF4expression,exceptfor cg07188665 (Figure 3).Combined with our previous findings,we believe that HSF4 promotes the CRC process at least through DNA methylation.
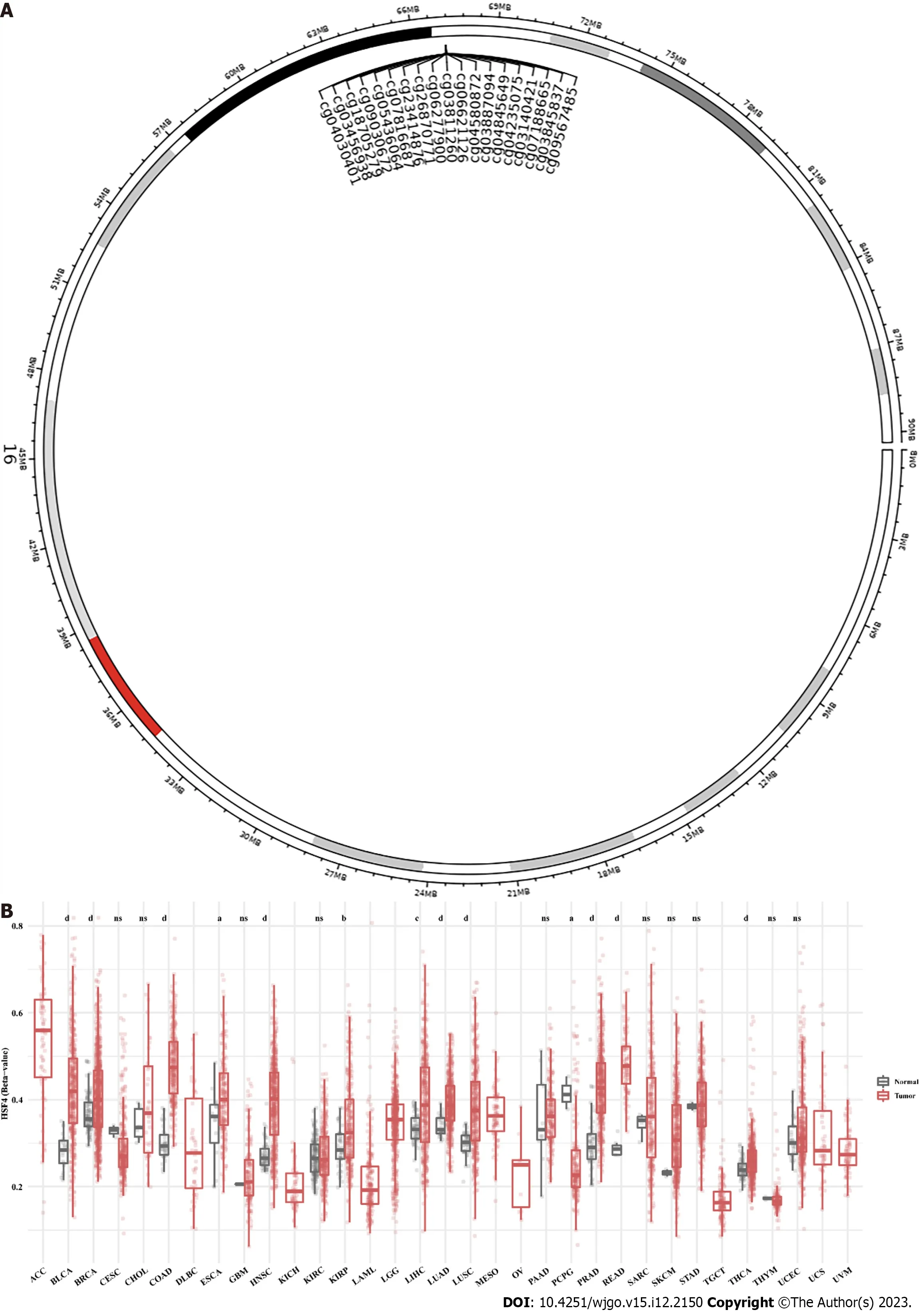
Figure 1 Pan-cancer analysis of heat shock factor protein 4 methylation levels. A: Schematic representation of the distribution of eat shock factor protein 4 (HSF4) methylated CpG loci.B: Differential analysis of the β values of 19 CpG methylation loci of HSF4 in multiple malignancies.aP <0.05,bP <0.01,cP <0.001,and dP <0.0001;ns: No significant difference.
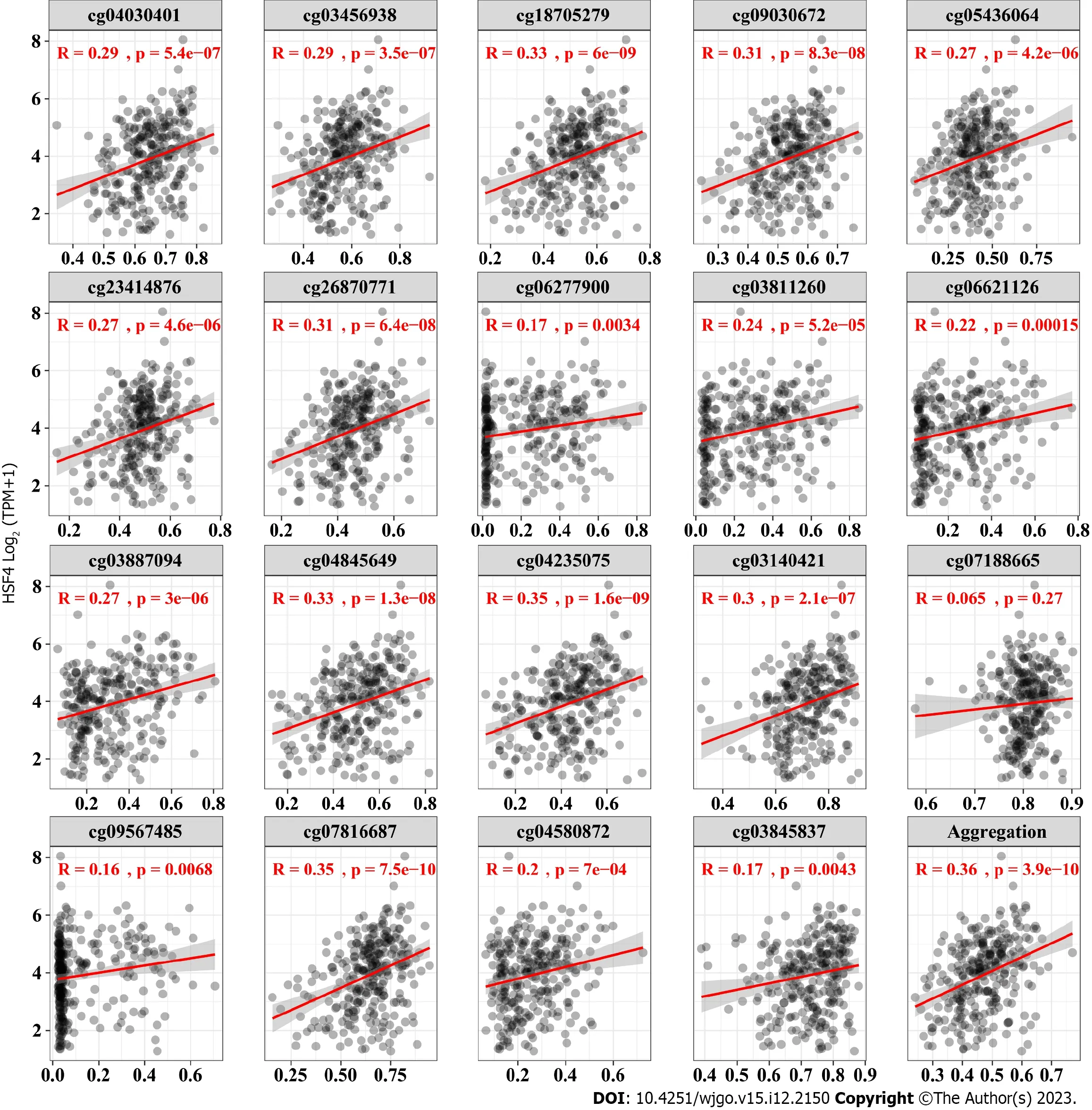
Figure 3 Correlation analysis of heat shock factor protein 4 expression and heat shock factor protein 4 methylation levels. The β values of all 19 probes exhibited a significant positive correlation with the expression of eat shock factor protein 4 (HSF4) mRNA.The x-axis is the β value of 19 probes,and yaxis is log2 (TPM+1) of HSF4 mRNA.
HSF4 methylation correlates poorly with CRC prognosis and diagnosis
In view of the differences inHSF4methylation in CRC,we further analyzed the prognostic and diagnostic value ofHSF4methylation.Kaplan-Meier curves indicated no significant difference in survival among COAD patients with high and low methylation levels for each CpG loci (Figure 4).Nevertheless,most patients with hypermethylated CpG loci had better prognosis.The ROC curve revealed that the area under the curve (AUC) of each CpG loci ranged from 0.498 to 0.574 in COAD patients,suggesting the mediocre diagnostic value ofHSF4methylation in COAD patients (Figure 5A).The time-dependent ROC curves suggested that the AUC of each CpG loci was greater with increasing time (Figure 5B).The above results indicated that the performance ofHSF4methylation as a prognostic and diagnostic biomarker in CRC was ordinary,which may have been caused by relatively low accumulation of single genes.
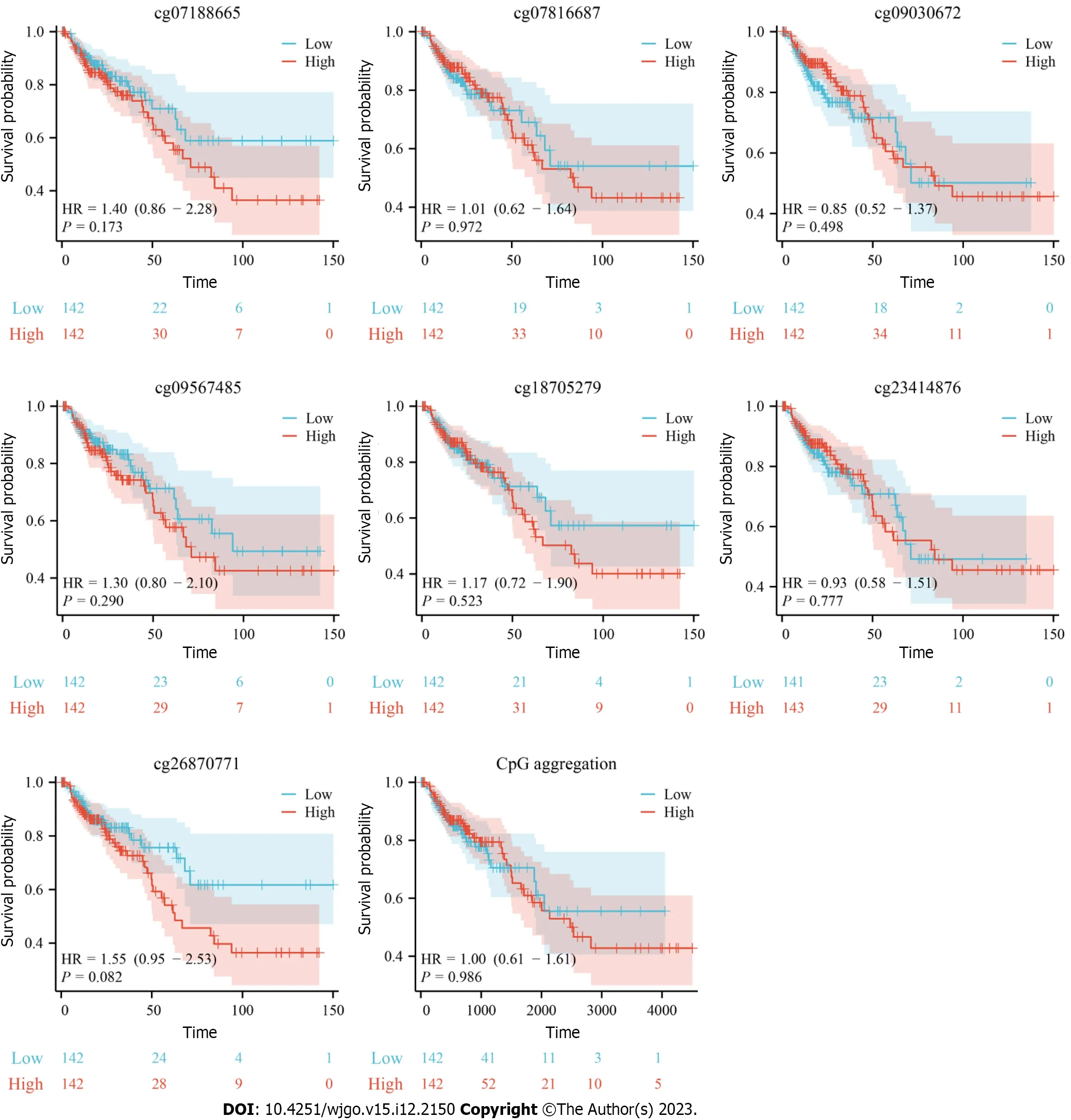
Figure 4 Correlation analysis of heat shock factor protein 4 methylation and prognosis of patients with colon adenocarcinoma. Kaplan Meier survival curves illustrating the survival of colon adenocarcinoma patients with high and low beta values for the 19 probes.The blue curve represents the cohort with low β values,and the red curve stands for the cohort with high β values.
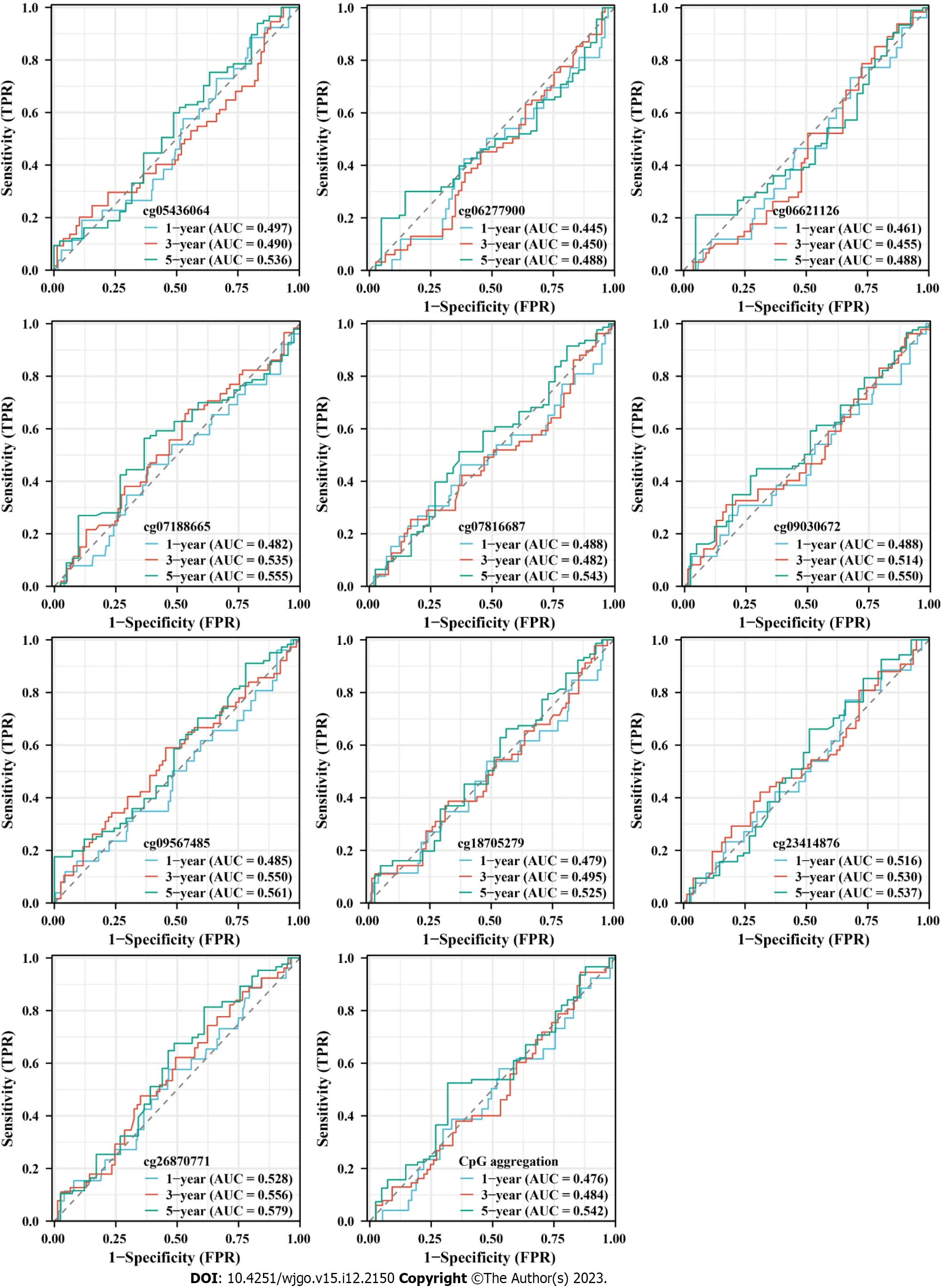
Figure 5 Correlation analysis of heat shock factor protein 4 methylation and diagnosis in patients with colonic adenocarcinoma. A:Receiver operating characteristic (ROC) curves exhibiting the diagnostic value of 19 heat shock factor protein 4 (HSF4) methylation-associated probes in colon adenocarcinoma patients.B: Time-dependent ROC curves displaying the area under the curve of HSF4 methylation at 1,3 and 5 years.AUC: Area under the curve;TPR: True positive rate;FPR: False positive rate.
Identification of HSF4 methylation-related genes in CRC and their functional enrichment analysis
We analyzed the genes associated withHSF4methylation in CRC by LinkedOmics.The expression of 1468 genes was positively correlated withHSF4methylation levels,and expression of 226 genes was negatively correlated withHSF4methylation levels in the COAD cohort (Figure 6A).The heatmap illustrated the top 50 genes with absolute correlation coefficients (Figures 6B and C).To further understand the functions and pathways involved in these genes,we performed GO and KEGG enrichment analysis.GO identified that the proteins encoded by these genes were mainly extracellular matrix,and associated with processes such as positive regulation of mitogen-activated protein kinase cascade,tumor necrosis factor superfamily cytokine production,neutrophil mediated cytotoxicity,and chemokine activity (Figure 6D).KEGG enrichment revealed thatHSF4methylation-related genes were involved in pathways including chemokine signaling pathway,calcium signaling pathway,glycosphingolipid biosynthesis -lacto and neolacto series,inflammatory bowel disease and inflammatory bowel disease (Figure 6E).It is suggested thatHSF4methylation mediates the phenotypic involvement of immune,inflammatory,and metabolic reprogramming in the CRC process.
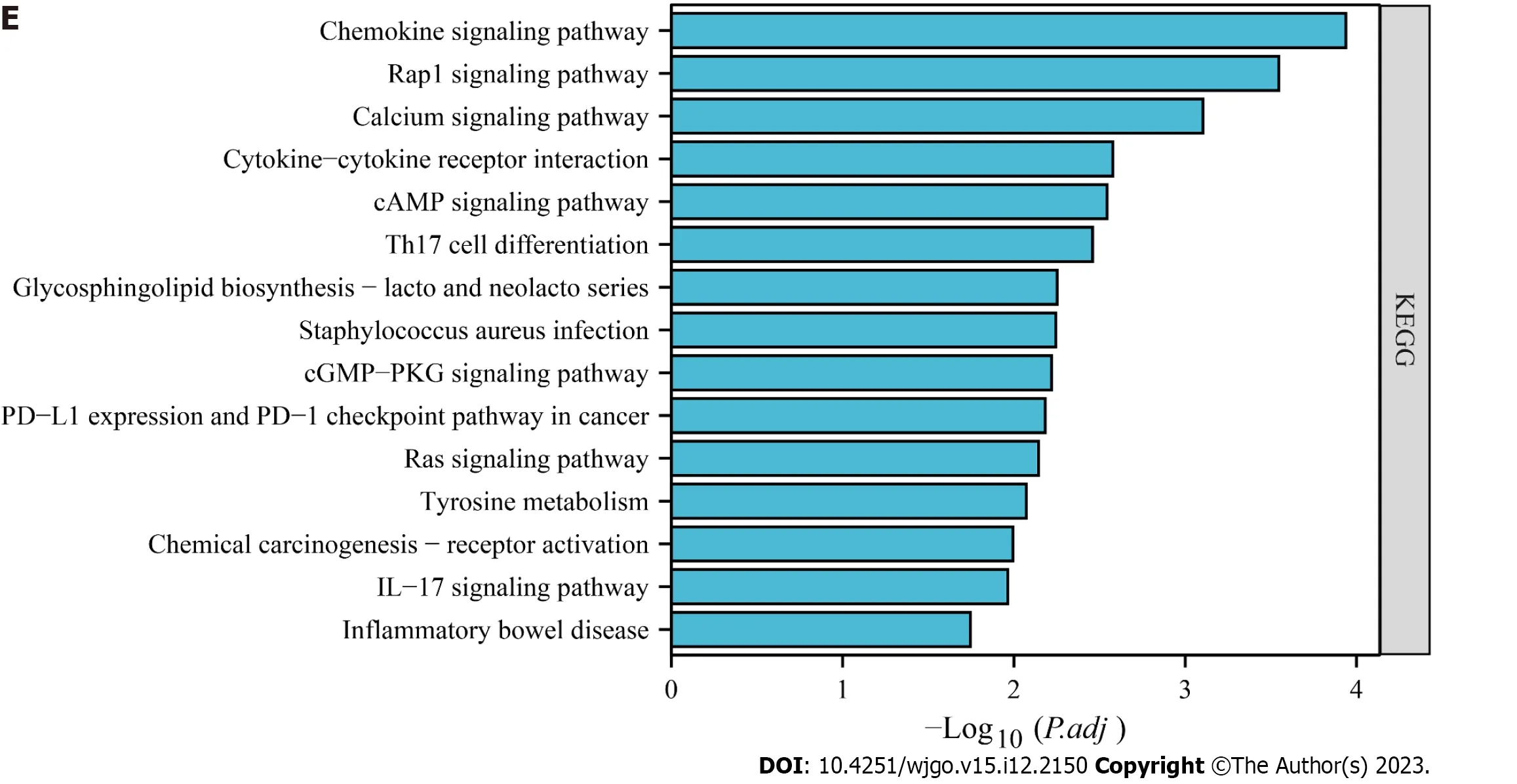
Figure 6 Identification of heat shock factor protein 4 methylation-related genes and their enrichment analysis in colorectal cancer. A:Volcano plot showing genes positively and negatively associated with heat shock factor protein 4 (HSF4) methylation in colorectal cancer.B,C: Expression profiles of the top 50 genes ranked by absolute correlation coefficient of HSF4 methylation-related genes.B is the expression profile of genes positively associated with HSF4 methylation;C is the expression profile of genes negatively associated with HSF4 methylation.D: Bubble plots exhibiting the GO enrichment results of all HSF4 methylation-related genes.E: Possible pathways involved in HSF4 methylation-related genes obtained by Kyoto Encyclopedia of Genes and Genomes enrichment analysis.HSF4: Heat shock factor protein 4;BP: Biological process;CC: Cell component;MF: Molecular function;PD-L1: Programmed cell death-Ligand 1;PD-1:Programmed death 1;IL-17: Interleukin-17.
PPI network of HSF4 methylation-associated genes in CRC
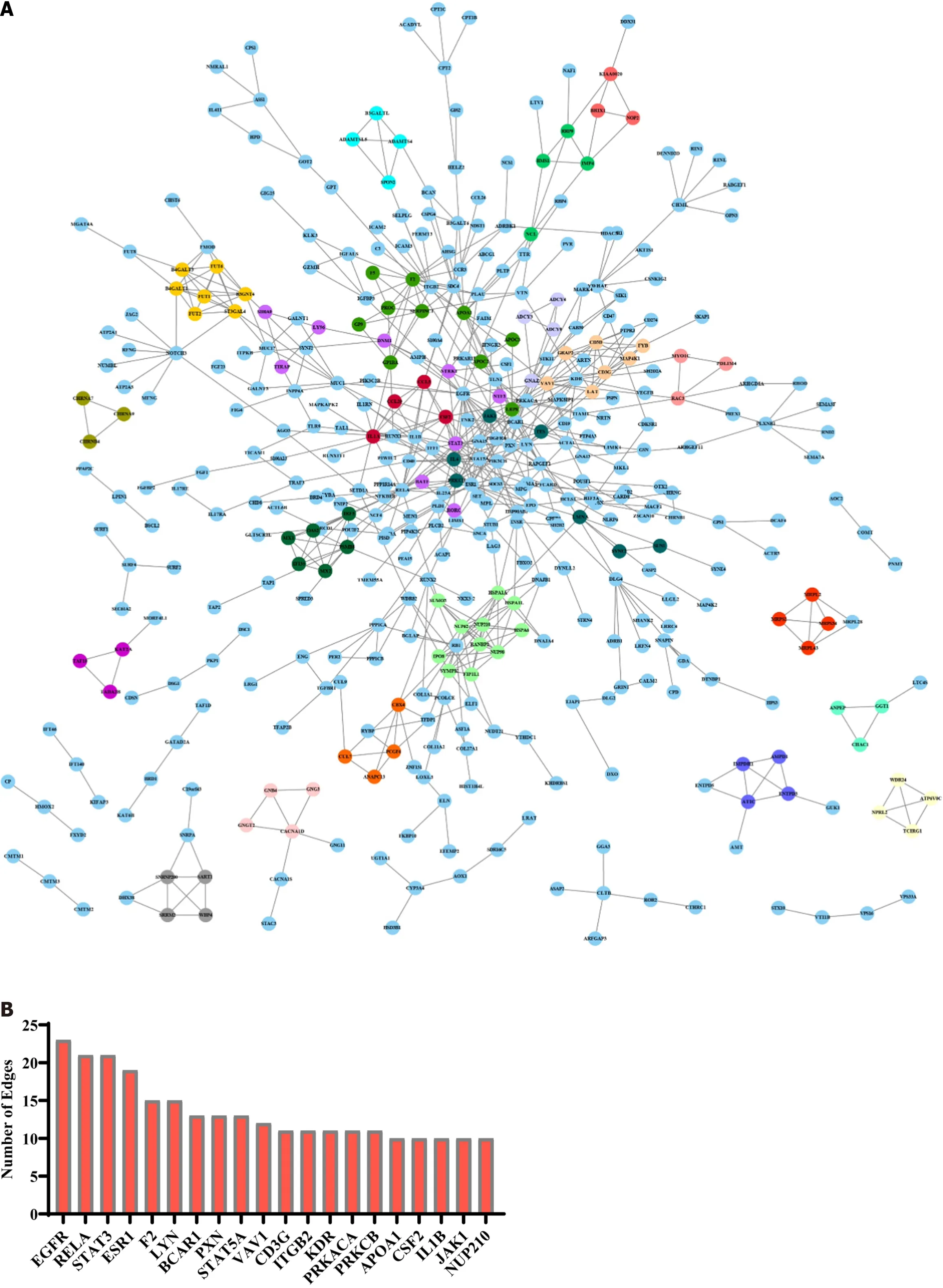
To identify the hub genes inHSF4methylation-related genes,we constructed a relevant PPI network based on the String database and the MCODE algorithm.The PPI network constructed forHSF4methylation positively correlated genes contained 422 nodes and 702 edges,and 22 clustering networks were obtained (Figure 7A).The top 20 genes in this network with the highest number of edges are displayed in Figure 7B,whereEGFR,RELA,STAT3,ESR1,andF2had the highest number of edges.The top 10 interworking networks with clustering scores are illustrated in Figure 7C.The network consisting ofNUP98,SUMO3,IPO8,andHSPA6had the highest clustering score,which contained 11 nodes and 35 edges (Figure 7C).In the same way,the network constructed for negatively associated genes contained 110 genes,122 interactions and five clusters (Figure 8A).The edge numbers TOP5 ofFCGR3A,POLR2K,AXIN1,CCL2and COPS5 had eight,seven,six,five and five edges,respectively (Figure 8B).The five clustering networks composed of genes and interactions are shown in Figure 8C.It is suggested that these genes are involved inHSF4methylation mediation of the CRC process.
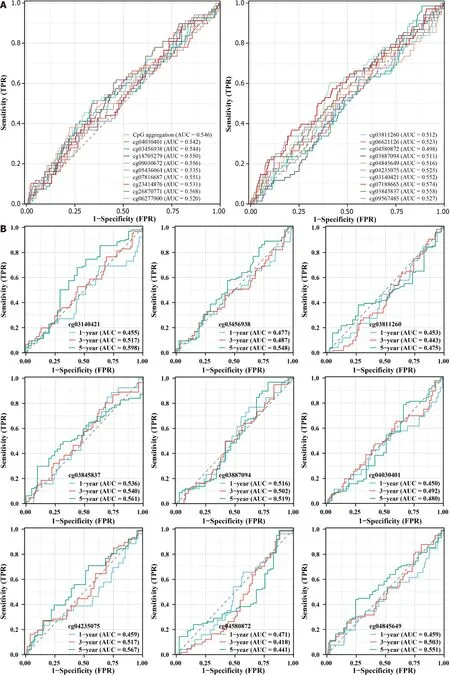
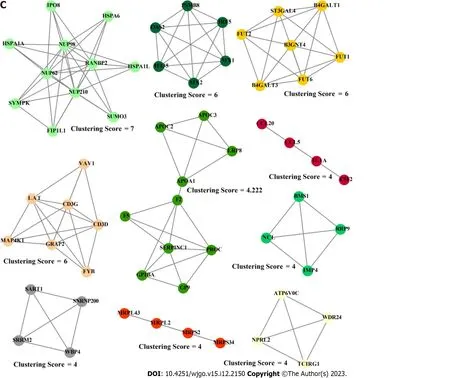
Figure 7 Protein-protein interaction network construction of heat shock factor protein 4 methylation positively associated genes. A:Protein-protein interaction (PPI) network of heat shock factor protein 4 methylation positively related genes constructed based on String database and MCODE algorithm.B: Top 20 genes in PPI network in terms of edge number.C: The top 10 clustering networks in terms of clustering scores obtained by the MCODE algorithm.
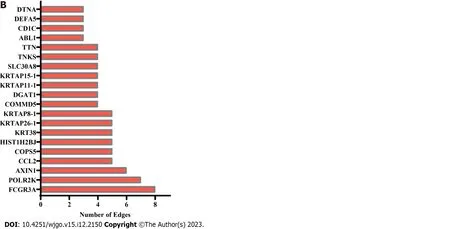
Figure 8 Protein-protein interaction network construction of heat shock factor protein 4 methylation negatively associated genes. A:Representative images of the protein-protein interaction (PPI) network of heat shock factor protein 4 methylation negatively associated genes.B: The bar chart displaying edge number of each gene in the PPI network.C: Gene composition and interactions of clustering networks obtained by the MCODE algorithm.
DISCUSSION
CRC is the second leading cause of cancer-related deaths worldwide.CRC is the outcome of progressive accumulation of a series of mutations and epigenetic changes in the rectum,cecum,and colon,leading to the development of colorectal adenoma and invasive adenocarcinoma.DNA methylation,one of the major epigenetic modifications,has been partially identified as a commercial diagnostic and prognostic biomarker for CRC.We have previously identifiedHSF4as an oncogenic gene in CRC[18].Therefore,we tapped the diagnostic and prognostic value ofHSF4and its possible molecular mechanisms in CRC.Unfortunately,HSF4,like most single-gene markers[29-32],has a mediocre diagnostic and prognostic value for its methylation levels in CRC.As in previous studies[29-32],this may be due to the small sample size analyzed or the insufficient accumulation of single gene methylation.Therefore,we analyzed the role ofHSF4methylation in CRC at the molecular mechanism level.It is noteworthy that we identified 1694 genes associated withHSF4methylation,and their possible involvement in immune,inflammatory,and metabolic reprogramming.In addition,the constructed PPI network demonstrated thatEGFR,RELA,STAT3,ESR1,FCGR3A,AXIN1,CCL2,andCOPS5are hub genes amongHSF4methylation-related genes.
Most of these hub genes have been demonstrated to be involved in the CRC process and have been applied as therapies for CRC.For instance,EGFR is a transmembrane receptor that plays a regulatory role in tumor cell function by binding to EGFs,promoting cell proliferation,differentiation,and survival[33].Currently,monoclonal antibodies against EGFR,such as cetuximab or panitumumab,are utilized in the clinical treatment of patients with metastatic CRC[34,35].RELA,also known as p65 or nuclear factor (NF)-κB p65,is known to be a key transcription factor in tumors,and it mediates immune and inflammatory responses to facilitate cancer cell survival and metastasis,which leads to it being a key target in tumor therapy[36,37].Similarly,FCGR3A belongs to the Fc γ receptor family,which is mainly expressed on the surface of natural killer cells,monocytes,and macrophages and plays an important role in antibody-mediated immune responses[38].Polymorphisms inFCGR3Aare associated with progression-free survival in patients with metastatic CRC treated with cetuximab[39,40].COPS5,also known as CSN5 or JAB1,is one of the constituent proteins of the COP9 signalosome,is a nuclear-plasmid transmembrane protein with multiple functions,and is involved in the regulation of various cellular processes such as cell proliferation,differentiation,apoptosis,and DNA replication and repair[41].It has been demonstrated that COPS5 plays a role as a pro-cancer factor in CRC by regulating Wnt and PI14K/AKT pathways[42-44].Some of these hub genes have also been proven to be related to HSF family proteins.For example,HSR-induced activation of HSP1 is regulated by the NF-κB,which activates transcription of HSPA1A[45].In turn,HSP1 inhibits the activation of NF-κB pathway[46,47].Stephanou and Latchman[48] showed that the activation ofSTAT3alone facilitates the mobilization of the HSP promoter.Nevertheless,whetherHSF4methylation-mediated alterations in HSF4 expression are crosstalk with these hub genes in CRC remains to be further investigated.
Unfortunately,there were some limitations to this study.For instance,the sample size was small,and a larger sample might reveal satisfactory diagnostic and prognostic values[49,50].Exploring the correlation betweenHSF4methylation and CRC subtypes orHSF4-related gene methylation combinations may improve the value ofHSF4in CRC[9,49].It is essential to verifyHSF4methylation in CRC tissues by methylation sequencing or microarrays,which could support the findings of this study[51].Although we demonstrated thatHSF4methylation levels exhibited a positive correlation withHSF4mRNA expression,in vivoandin vitroexperiments are lacking for validation.In the same way,the molecular mechanisms associated withHSF4methylation remain to be exploredin vivoandin vitro.The tumor immune microenvironment (TME) consists of immune cells,blood vessels,and extracellular matrix,and has a dual role in the growth and metastasis of tumor cells[52,53].HSF family proteins,especially HSF1,have been demonstrated to mediate tumor cell associated immune responses in the TME[54,55].This predicts that the function of HSF4 in CRC-associated TME is also worthy of investigation.
CONCLUSION
In conclusion,this study reveals that HSF4 is highly methylated in CRC and is associated withHSF4overexpression.AlthoughHSF4methylation has poor diagnostic and prognostic value,it may be involved in the CRC process by mediating expression ofHSF4or related genes with potential mechanisms.Combined with our previously described findings[18],the present study believes that high expression ofHSF4mRNA and protein and its oncogenic effects are most probably due toHSF4methylation (Figure 9).Specific mechanisms need to be confirmed by morein vivoandin vitroexperiments.
ARTICLE HIGHLIGHTS
Research background
DNA methylation is involved in the regulation of gene expression and has been implicated in development and outcome of colorectal cancer (CRC).
Research motivation
We previously demonstrated that heat shock factor protein 4 (HSF4) expression is abnormally high,and contributes to the malignant biological behavior of CRCin vivoandin vitro.However,the correlation ofHSF4methylation withHSF4expression and prognosis of CRC patients,and other potential molecular mechanisms need to be further investigated.
Research objectives
The present study was proposed to investigate the correlation betweenHSF4methylation and CRC risk,and to uncover the underlying molecular mechanisms.
Research methods
Identification ofHSF4methylation sites,and analysis of the differences in β values ofHSF4methylation sites and their correlation withHSF4mRNA expression were performed using Shiny Methylation Analysis Resource Tool Web.The genes associated withHSF4methylation were identified by LinkedOmics Web for CRC,and Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analyses were performed to reveal the functions and signaling that these associated genes may be involved in.The String database and MCODE algorithm were performed to construct protein-protein interaction (PPI) networks ofHSF4methylation-related genes.
Research results
TheHSF4gene had 19 CpG methylation sites,and their β-values were significantly higher in CRC tissues,positively correlating withHSF4mRNA expression.The β value of theHSF4methylation site was not associated with the prognosis of CRC patients.Notably,there are 1694 genes in CRC tissues whose expression is associated withHSF4methylation and which are involved in immune,inflammatory,and metabolic reprogramming.EGFR,STAT3 and AXIN1 are hub genes in the PPI network constructed by theseHSF4methylation-related genes.
Research conclusions
TheHSF4gene is highly methylated in CRC,and is associated with the overexpression ofHSF4mRNA.HSF4methylation may be involved in the process of CRC by mediating the expression ofHSF4or related genes.
Research perspectives
The finding will provide a theoretical basis and a new perspective onHSF4as a methylation-related biomarker for future CRC diagnosis and treatment.
FOOTNOTES
Author contributions:Zhang WJ and Zhang Y conceived and designed the experiments;Zhang WJ,Yue KL,and Wang JZ analyzed the data;Zhang Y contributed to the data curation;Zhang WJ wrote-original draft preparation;Yue KL,Wang JZ,and Zhang Y participated in the writing-review and editing.
Supported byNational Natural Science Foundation of China,No.82 260601;Joint Foundation of Kunming Medical University and Yunnan Provincial Science and Technology Department,No.202201AY070001-256;Grant for Clinical Medical Center of Yunnan Provincial Health Commission,No.2021LCZXXF-XH03;and Young Academic Talents Cultivation Foundation of Yunnan Province,No.202205AC160070.
Institutional review board statement:This study did not involve any animal and human experimentation.
Conflict-of-interest statement:All the authors report no relevant conflicts of interest for this article.
Data sharing statement:No additional data are available.
Open-Access:This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers.It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license,which permits others to distribute,remix,adapt,build upon this work non-commercially,and license their derivative works on different terms,provided the original work is properly cited and the use is non-commercial.See: https://creativecommons.org/Licenses/by-nc/4.0/
Country/Territory of origin:China
ORCID number:Yu Zhang 0000-0002-1523-3849.
S-Editor:Wang JJ
L-Editor:A
P-Editor:Zhang XD
 World Journal of Gastrointestinal Oncology2023年12期
World Journal of Gastrointestinal Oncology2023年12期
- World Journal of Gastrointestinal Oncology的其它文章
- Dual primary gastric and colorectal cancer: A complex challenge in surgical oncology
- Conversion immunotherapy for deficient mismatch repair locally unresectable colon cancer: A case report
- Association of MBOAT7 rs641738 polymorphism with hepatocellular carcinoma susceptibility: A systematic review and meta-analysis
- Prognostic value of T cell immunoglobulin and mucin-domain containing-3 expression in upper gastrointestinal tract tumors: A meta-analysis
- Intensive follow-up vs conventional follow-up for patients with nonmetastatic colorectal cancer treated with curative intent: A metaanalysis
- Paired-related homeobox 1 induces epithelial-mesenchymal transition in oesophageal squamous cancer
