Purification and Characterization of Hyaluronate Lyases Produced by Two Types of Bacteria
Shuai LI, Xinhui WANG, Yota TATARA, Shigeki HAMADA, Takuya KOZEKI, Kaoru KOJIMA, Takashi YOSHIDA
AbstractHyaluronate lyases were obtained from two types of naturally isolated bacterial strains Paenibacillus yunnanensis and Paennarthrobacter nicotinovorans. PyHL (form P. yunnanensis) in the culture supernatant of the bacteria was purified by two steps of column chromatography. The enzyme showed the molecular mass of 74 kDa by SDS-PAGE and the maximal activity at pH 5.0, 35℃. PyHL maximally degraded hyaluronate by an endo-type manner, and showed low degradation activity toward chondroitin sulfates. Dermatan sulfate was not the substrate. PnHL (from P. nicotinovorans) in the culture supernatant of the bacteria was purified by two steps of column chromatography. The enzyme showed the molecular mass of 70 kDa by SDS-PAGE and the maximal activity at pH 6.0, 30 ℃. Genomic analysis of P. nicotinovorans on the bases of the internal amino acid sequences of PnHL.
Key wordsGlycosaminoglycan; Hyaluronan; Hyaluronate lyase; Paenibacillus yunnanensis; Paennarthrobacter nicotinovorans
DOI:10.19759/j.cnki.2164-4993.2023.04.022
Hyaluronate (HA) is an anionic linear polysaccharide composed of repeating disaccharide units of [(1→3) β-D-N-acetylglucosamine-(GlcNAc)-(1→4) β-D-glucuronic acid(GlcUA)], and classified as a glycosaminoglycan without sulfate residues. HA is abundantly distributed in nearly all vertebrate tissues, especially extracellular matrix (ECM). HA is also produced by some bacteria of the genus Streptococcus[1-2].
HA is degraded by hyaluronidases, which were classified into three main families on the basis of their catalytic mechanisms of action[3]. Hyaluronidases (EC 3.2.1.35) degrade HA by hydrolysis of the B-1,4-glycosidic bond furnishing tetrasaccharide molecule as the main product. These enzymes were extracted from vertebrate tissues and venom. Hyaluronidases (EC 3.2.1.36) are mainly extracted from hookworms and the salivary glands of leeches. These enzymes degrade HA by hydrolysis of the B-1,3-glycosidic bond, thus, yielding sugar fragments having glucuronic acid at the reducing end and generate tetra- and hexasaccharide end products. Hyaluronidases (EC 4.2.2.1), also known as hyaluronate lyases (HL), were isolated from various microorgansims, including e.g. strains of Clostridium, Streptococcus, Streptomyces, Arthrobacter, and Bacillus[4-9]. They degrade HA by a B-elimination reaction to yield unsaturated hyalurono-oligosaccharides as the exhaustive degradation products.
Hyaluronidases are widely used in clinical treatment, medical science, and biochemical research. Hyaluronidases are considered as a "spreading factor", which increases membrane permeability for rendering tissues more permeable to injected fluids by degrading HA in the ECM. Therefore, the enzyme is approved as an adjuvant for increasing analgesic efficacy[10]. In the clinic, hyaluronidases could be applied to vitreous opacity, glaucoma and control vascular complications after HA filler injection in aesthetic dermatology[11-12]. And in biochemical researches, hyaluronidase can be used as tools for the identification of HA[13] and producing HA oligosaccharides[14].
The current commercial hyaluronidases are mainly prepared by extracting bovine testicular tissue. However, the purity and high price of these enzymes limit their usage in pharmaceutical and biochemical engineering[15]. Therefore, more attentions are paid to the hyaluronidases (HL) from bacteria.
In this study, Paenibacillus yunnanensis strain16-6 and Paennarthrobacter nicotinovorans strain 19-1 were isolated from the Lake in Aomori, Japan, and were chosen for those degradation activity toward HA. The hyaluronate lyases produced by the strain (PyHL,PnHL) were purified, characterized. The results showed that PyHL and PnHL could be effective tools for analyses of HA and preparation of HA oligosaccharides, which have the potential to be used in drugs or biochemical research.
Material and Methods
Materials
Bacterial strains(i) P. yunnanensis strain 16-6 was obtained from water of Ketobanoike, a pond in Lakes Juniko (Aomori pref., Japan) by an enrichment culture with HA as the substrate. Subculture was repeated until a single colony was obtained. Bacterial identification on the basis of 16S rRNA analysis was performed by Macrogen Corp. (Japan).
(ii) P. nicotinovorans strain 19-1 was isolated from water of Lake Towada, (Aomori Pref., Japan) by an enrichment culture with PG as the substrate. The rest of the experimental methods were the same as those of P. yunnanensis strain 16-6.
ReagentsLow-molecular weight HA was purchased from Healthy Co. (Japan), and used for bacterial culture. HA from rooster comb (Fujifilm Wako Pure Chemical Co., Japan) was used. Toyopearl HW55F and DEAE Toyopearl 650M were purchased from Tosoh Bioscience Co. (Japan). Proteoglycan (PG) was generously provided by Glycosmo Co. (Japan). Other reagents were commercially supplied by Fujifilm Wako Pure Chemical Co. (Japan).
Microbial cultivation and purification of PyHL and PnHL
(i) P. yunnanensis strain 16-6 was grown in the liquid medium (0.1% HA, 0.67% Yeast Nitrogen Base (w/o) Amino Acid, 0.02% Yeast Extract) with an initial pH 7.0. After 48 h of culture at 27℃ with shaking in a 1 L flask, bacterial cells were removed by a centrifugation of the broth for 20 min at 8 000 rpm at 4 ℃. Following steps were carried at 4 ℃. Proteins in the supernatant were precipitated by ammonium sulfate at the concentration of 70% saturation, then centrifuged for 20 min at 8 000 rpm. The pellets were dissolved in 10 mM acetate buffer (pH 6.0) containing 0.1 M NaCl, then applied to a column of Toyopearl HW55F column that was wan in the same buffer. The enzyme was pooled and concentrated using an ultrafiltration tube (cut by 30kDa, Macrosep Advance Centrifugal Devices 30 K, Pall Co. USA), and then loaded onto a DEAE Toyopearl 650 M column that was pre-equilibrated in 10 mM acetate buffer (pH 6.0). The elution was performed with a liner gradient of NaCl (0.0-0.3 M) in the same buffer. PyHL was concentrated using an ultrafiltration tube (cut by 30 kDa,Nanosep 30 K Omega, Pall Co. USA) and kept frozen until use. Protein concentration was determined by a method developed by Bradford using bovine serum albumins as the standard[16]. SDS-PAGE was performed according to the method described by Laemmli using 10% polyacrylamide gels[17].
(ii) P. nicotinovorans strain 19-1was grown in the liquid medium (0.1% PG, 0.67% Yeast Nitrogen Base (w/o) Amino Acid, 0.02% Yeast Extract) with an initial pH 7.0. After 48 h of culture at 27 ℃ with shaking in a 1 L flask, bacterial cells were removed by a centrifugation of the broth for 20 min at 8 000 rpm at 4 ℃. Following steps were carried out at 4 ℃. Proteins in the supernatant were precipitated by ammonium sulfate at the concentration of 75% saturation then centrifuged for 20 min at 8 000 rpm. And the purification method was basically the same as PyHL above.
Protein concentration and SDS-PAGE, Native-PAGE were performed according to the method as PyHL.
Determination of hyaluronate lyases activity (PyHL, PnHL)
Reaction mixtures containing 0.01% (w/v) HA and the enzyme in total 1.0 ml of 50 mM sodium acetate buffer (pH 5.0), were incubated at 30 ℃ for a period of time. The reactions were stopped by adding 0.5 mL of 1 M NaOH, and then UV absorption of the solutions were measured at 232 nm. One unit (U) of HL activity was defined as the amount of the protein required of the eliminative cleavage of substrate, yielding UV-absorbing materials corresponding to 1 μmol of unsaturated double bond per minute. A millimolar absorption confficient of 5.5 was used in all calculations[18-19].
Effects of pH and temperature on the activity and stability of PyHL and PnHL
(i) The optimum pH of PyHL was determined by measuring the activity of HL with 0.02 M buffer including sodium acetate-HCl (pH 1.0-4.0), sodium acetate-acetate (pH 4.0-6.0), sodium phosphate (pH 6.0-8.0), Tris-HCl (pH 8.0-9.0), disodium carbonate (pH 9.0-10.0), and sodium tetraborate-NaOH (pH 10.0-12.0). Similarly, a range of 10-70 ℃ was used to determine the optimum temperature for PyHL. Thermal stability and pH stability were measured by incubating the enzyme at different temperatures (10-70 ℃) and buffers (pH 1.0-12.0), respectively. The residual activity was measured under standard conditions described above.
(ii) PG was used as the reaction substrate and the experimental method was as same as PyHL.
Substrate specificity
(i) PyHL as 50 μl enzymatic solution was added to a reaction mixture containing 1.95 ml substrate solution (CSA, CSB, CSC, HA, PG, or GAG, 1mg/ml) in the 0.05 M acetate buffer (pH 5.0). After incubation at 30 ℃ for 60 min, the reactions were stopped by adding 0.5 ml of 1 M NaOH. Then the UV absorption of the resultant solutions was measured at 232 nm.
(ii) The experimental method was as same as PyHL.
HPLC of PyHL
A reaction mixture composed of 0.01% HA, 0.05 M acetate buffer (pH 5.0), and the enzyme was incubated at 30 ℃ from 30 min to 12 h. A change in the size of HA was monitored by gel permeation HPLC with TOSOH TSK-gel G5000PWXL column (Tosoh Co., Japan) that was ran in 0.2 M NaCl solution (0.5 ml/min). Effluent was monitored by Refractive Index monitor (Model Chromaster 5450, Shimazu Co., Japan).
Protein analysis of PnHL
The protein analysis of PnHL was recovered from the polyacrylamide gel after performing SDS-PAGE, and subjected to amino acid sequence analysis by LC/MSMS. Internal amino acid sequences of the tryptic peptides were identified by LC-MS/MS (Triple TOF 6600, AB Sciex, Japan) with ProteinPilot Software (AB Sciex) for data processing. The amino acid sequence of PnHL was determined by informatics works with the genomic information of P. nicotinovorans that was used as a database in the MS/MS spectra search. By using the Prosite of ExPASy (http:∥prosite.expasy.org/) the protein function was predicted from the amino acid sequence.
Results and Analysis
Molecular and enzymatic properities of hyaluronate lyases (PyHL, PnHL)
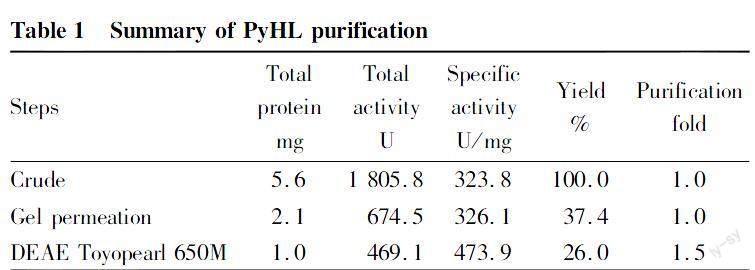
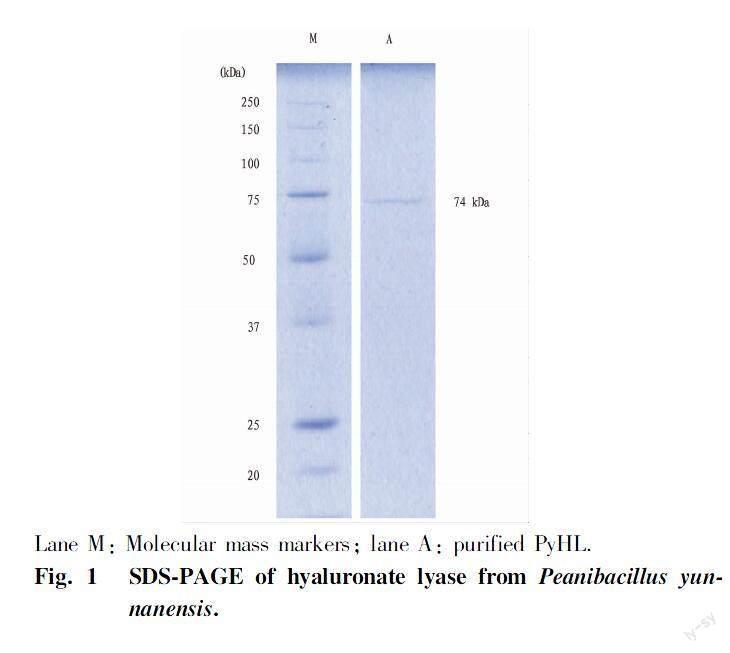
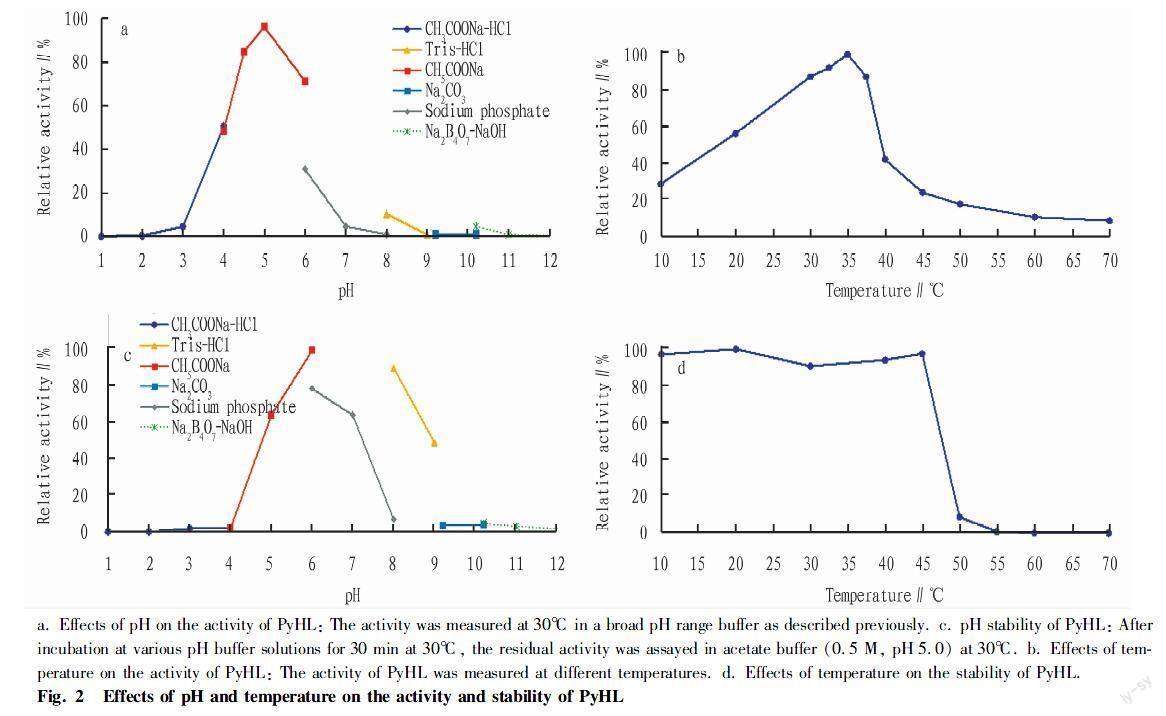
(i) The enzyme PyHL was purified from the culture broth P. yunnanensis by two steps of column chromatography (Table 1). PyHL appeared as a single band on SDS-PAGE and the molecular weight was estimated approximately 74 kDa (Fig. 1). PyHL showed the maximal activity at pH 5.0 in an acetate buffer and almost no activity was shown below pH 3.0 and above pH 9.0 (Fig. 2a, c). We observed an inhibitory effect by a phosphate buffer that reduced the activity by a half of the acetate at pH 6.0. Over 70% of the initial activity was kept after pretreatment at pH between pH 5 and 8 with the maximal at pH 6.0. The activity of PyHL at pH 5.0 was maximal at 35℃ and was stable after a preheating below 45℃ (Fig. 2b, d).
(ii) The enzyme PyHL was purified from the culture broth P. yunnanensis by two steps of column chromatography (Table 2). PyHL appeared as a single band on SDS-PAGE and the molecular weight was estimated approximately 70 kDa (Fig. 3). PnHL showed the maximal activity at pH 5.0 in an acetate buffer and almost no activity was shown below pH 4.0 and above pH 9.0 (Fig. 4a, c). The activity of PyHL at pH 5.0 was maximal at 35 ℃ and was stable after a preheating below 40℃ (Fig. 4b, d).
Substrate specificity of PyHL and PnHL
(i) PyHL exhibited the highest activity toward HA among the GAG substrates (Table 3). The digestion ratios of CSA, CSC were respectively 7% and 9% of HA and the degradation of dermatan sulfate was negligible.
Bacterial HL could be divided into three types of enzymes depending on their substrate specificities. HL from Streptomyces byalurolyticus are specific to HA, while Streptococcus dysgalactiae HL show a little activity toward CSA and CSC[21-22]. Arthrobacter HL degrades CS with nearly one third of the activity toward HA[23]. In this context, PyHL was supposed to have a similar specificity with Peptostreptococcus HA lyase.
(ii) PnHL exhibited the highest activity toward HA among the GAG subatrates (Fig. 5). Then PnHL had degradation activity for CSC and CSA, but had no activity for DS.
Determination of degradation products
The reaction mixtures were analyzed by HPLC, monitored at 232 nm which was specific for unsaturated bands. The time sequence changes in the reaction mixtures when the HA oligosaccharide was used as the substrate are showed in Fig. 6 as an example. Decasaccharides and tetrasaccharides were detected at 232 nm, but no disaccharides were detected by HPLC.
Studies have showed that HA oligosaccharides have important application prospects in the field of medicine[24], such as applications in promotion of angiogenesis[25], immunomodulation, and anti-tumor[26].
Protein analysis of PnHL
PnHL protein was recovered from polyacrylamide gels after SDS-PAGE and amino acid sequence analysis was performed by LC-MS/MS. The identified amino acids were sequenced according to the genomic data of strain 19-1, and the resulting proteins were searched for homology by BLAST (Fig. 7). The results showed that PnHL is highly homologous to P. nicotinovorans[27] polysaccharide lyase 8 family protein (hyaluronate lyase).
Conclusions and Discussion
In this study, two types of extracellular hyaluronate lyase were successfully purified to homogeneity from fermentation broth of the bacteria P. yunnanensis and P. nicotinovorans. These enzymes had the approximate molecular weight of 74 and 70 kDa by SDS-PAGE, which is similar to the known HL, such as 73.7 kDa (Arthrobacter globiformis A152)[8], 77 kDa (Streptomyces hyalurolyticus)[20], but different with known HL, such as 120 kDa (Bacillus niacini strain JAM F8)[9], 50 kDa (group A type 4 Streptococcus)[29], 107 kDa (Streptococcus pneumoniae)[28], and 114 kDa (Clostridium perfringens)[4]. Characteristics of these enzymes were also studied. The hyaluronate lyase also displayed pretty good levels of activity and stability.
Hyaluronate lyases from various species showed different specificities towards polysaccharide substrates[30]. Both PyHL and PnHL showed highest substrate specificity towards HA, which was different from the other conventional hyaluronidases. Generally, the hyaluronate lyase (EC 4.2.2.1) can typically degrades HA and is relatively less active against CSs. Although, the HCLaseM (M, sp. H14) could degrade HA and various types of CSs and showed almost the same enzyme activity towards CS-A, CS-B, CS-C and HA (98, 97, 96, and 100), the PyHL and PnHL (this study) showed lowest activity towards CSs, which showed higher level of substrate specifity on HA. PyHL and PnHL may have the potential to be an effective tool for preparation of functional oligosaccharides and investigation function relationship of HA.
Amino acid sequence analysis was performed on PnHL by LC-MS/MS and the results were checked against the whole genome analysis data of P. nicotinovorans strain 19-1. The results showed that 377 amino acids were identified with 95% confidence, accounting for 82.9% of the protein sequence of PnHL. The protein identified by homology analysis showed high homology to P. nicotinovorans polysaccharide lyase family 8 protein (hyaluronate lyase, WP_189076633.1). Therefore, in this study, we have chosen to refer to it as hyaluronate lyase.
Hyaluronidase as a glycosaminoglycan lyase, has recently attracted attention for its use in the pharmaceutical industry and clinical medicine for the treatment and improvement of joint conditions, and will likely play an important role in biotechnological research[31]. Since aggrecan, which is a PG abundant in joints, is enriched in CSA and CSC, PyHL and PnHL, which showed high degradation activity against them, are suitable for enzymatic therapy to treat and improve herniation in joints. In addition, the biological activity of low molecular weight hyaluronan oligosaccharides is superior to that of natural polymeric hyaluronan, hyaluronidases will be a major tool for the preparation of hyaluronan oligosaccharides. The discovery of new glycosaminoglycan lyases with various properties from nature and a better understanding of their functions and properties would be important not only for basic research but also from an applied perspective.
References
[1] GUNTHER E, OZEGOWSKI JH, KOHLER W. Occurrence of extracellular hyaluronic acid and hyaluronate lyase in Streptococci of Groups A, B, C, and G[J]. Zbl. Bakt, 1996, 285(1): 64-73.
[2] KIM JH, YOO SJ, OH DK, et al. Selection of a Streptococcus equi mutant and optimization of culture conditions for the production of high molecular weight hyaluronic acid[J]. Enzyme and Microbial Technology, 1996, 19(6): 440-445.
[3] EL-SAFORY NS, FAZARY AE, LEE CK. Review Hyaluronidases, a group of glycosidases: current and future perspectives[J]. Carbohydrate Polymers, 2010, 81(2): 165-181.
[4] CANARD B, GARNIER T, JOAINS BS, et al. Molecular genetic analysis of the nagH gene encoding a hyaluronidase of Clostridium perfringens[J]. Mol Gen Genet, 1994, 243(2): 215-224.
[5] BAKER JR, DONG SL, PRITCHARD DG. The hyaluronan lyase of Streptococcus pyogenes bacteriophage H4489A[J]. Biochem. J., 2002, 365(Pt1): 317-322.
[6] OHYA T, KANEKO Y. Novel hyaluronidase from Streptomyces[J]. Biochimica et Biophysica Acta (BBA)-Enzymology, 1970, 198(3): 607-609.
[7] GUO XP, SHI YL, SHENG JZ, et al. A novel hyaluronidase produced by Bacillus sp. A50[J]. PLoS ONE, 2014, 9(4): e94156.
[8] ZHU C, ZHANG JL, LI LY, et al. Purification and characterization of hyaluronate lyase from Arthrobacter globiformis A152[J]. Appl Biochem Biotechnol, 2017, 182(1): 216-228.
[9] KURATA A, MATSUMOTO M, KOBAYASHI T, et al. Hyaluronate lyase of a deep-sea Bacillus niacin[J]. Mar Biotechnol, 2015, 17(3): 277-284.
[10] WOHLRAB J, FINKE R, FRANKE WG, et al. Clinical trial for safety evaluation of hyaluronidase as diffusion enhancing adjuvant for infiltration analgesia for skin with lidocaine[J]. Dermatologic Surgery, 2012, 38(1): 91-96.
[11] GUM GG, SAMUELSON DA, GELATT KN. Effect of hyaluronidase on aqueous outflow resistance in normotensive and glaucomatous eyes of dogs[J]. American Journal of Veterinary Research, 1992, 53(5): 767-770.
[12] KIM DW, YOON ES, JI YH, et al. Vascular complications of hyaluronic acid fillers and the role of hyaluronidase in management[J]. Journal of Plastic, Reconstructive & Aesthetic Surgery, 2011, 64(12): 1590-1595.
[13] YAMADA K. The effect of digestion with streptomyces hyaluronidase upon certain histochemical reactions of hyaluronic acid-containing tissues[J]. The Journal of histochemistry and cytochemistry, 1973, 21(9): 794-803.
[14] KAKIZAKI I, IBORI N, KOJIMA K, et al. Mechanism for the hydrolysis of hyaluronan oligosaccharides by bovine testicular hyluronidase[J]. FEBS Journal, 2010, 277(7): 1776-1786.
[15] SUN JH, HAN X, SONG GR, et al. Cloning, expression, and characterization of a new glycosaminoglycan lyase from Microbacterium sp.H14[J]. Mar Drugs, 2019, 17(12): 681.
[16] LIN B, HOLLINGSHEAD SK, COLIGANS JE, et al. Cloning and expression of the gene for group B Streptococcal hyaluronate lyase[J]. The Journal of Biological Chemistry, 1994, 269(48): 30113-30116.
[17] BRADFORD MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding[J]. Analytical Biochemistry, 1976, 72(1-2): 248-254.
[18] ALLEN AG, LINDSAY H, SEILLY D, et al. Identification and characterization of hyaluronate lyase from Streptococcus suis[J]. Microbial Pathogenesis, 2004, 36(6): 327-335.
[19] LAEMMLI UK. Cleavage of structural proteins during the assembly of the head of Bacteriophage T4[J]. Nature, 1970(227): 680-685.
[20] PARK Y, CHO S, LINHARDT RJ. Exploration of the action pattern of Streptomyces hyaluronate lyase using high-resolution capillary electrophoresis[J]. Biochim Biophys Acta, 1997, 1337(2): 217-226.
[21] OHYA T, KANEKO Y. Novel hyaluronidase from streptomyces[J]. Biochim Biophys Acta., 1970, 198(3): 607-609.
[22] TAM YC, CHAN EC. Purification and characterization of hyaluronidase from oral Peptostreptococcus species[J]. Infect Immun., 1985, 47(2): 508-513.
[23] HIYAMA K, OKADA S. Crystallization and some properties of chondroitinase from Arthrobacter aurescens[J]. Journal of Biological Chemistry, 1975, 250(5): 1824-1828.
[24] KARBOWNIK MS, NOWAK JZ. Hyaluronan: Towards novel anti-cancer therapeutics[J]. Pharmacol Reports, 2013, 65(5): 1056-1074.
[25] WEST DC, KUMAR S. The effect of hyaluronate and its oligosaccharides on endothelial cell proliferation and monolayer integrity[J]. Experimental Cell Research, 1989, 183(1): 179-196.
[26] BENITEZ A, YATES TJ, LOPEZ LE, et al. Targeting hyaluronidase for cancer therapy: Antitumor activity of sulfated hyaluronic acid in prostate cancer cells[J]. Cancer Research, 2011, 71(12): 4085-4095.
[27] JING M, XIAOMENG S, SHUAISHUAI L, et al. Draft genome sequence of paenarthrobacter nicotinovorans Hce-1[J]. Genome Announc, 2017, 5(30): e00727-17.
[28] JEDRZEJAS MJ, MEWBOURNE RB, CHANTALAT L, et al. Expression and purification of Streptococcus pneumoniae hyaluronate lyase from Escherichia coli[J]. Protein expression and purification, 1998, 13(1): 83-89.
[29] HILL J. Purification and properties of Streptococcal Hyaluronate lyase[J]. Infection and Immunity, 1976, 14(3): 726-735.
[30] MISHRA P, KUMAR RP, ETHAYATHULLA AS, et al. Polysaccharide binding sites in hyaluronate lyase-crystal structures of native phage-encoded hyaluronate lyase and its complexes with ascorbic acid and lactose[J]. FEBS Journal, 2009, 276(12): 3392-3402.
[31] ALEXANDRA BB, HOLGER S, NORMAN-PHILIPP H, et al. Hyaluronidse: From clinical applications to molecular and cellular mechanisms[J]. European Journal of Medical Research, 2016, 21(5): 201-205.
Editor: Yingzhi GUANGProofreader: Xinxiu ZHU
- 农业生物技术(英文版)的其它文章
- Optimization of Extraction Conditions for Total Flavonoids from Fructus Aurantii Immaturus and Its Anti-UVB Radiation Activity
- Research on Maize Seed Classification Method Based on Convolutional Neural Network
- Common Species Distribution Models in Biodiversity Analysis and Their Challenges and Prospects in Application
- Vegetable Tunnel House Technology in Tropical Island Countries
- Renewable Energy Seawater Desalination Technology and Application Analysis
- Impact of Water Quality Sampling Process on Environmental Monitoring Results

