Spatiotemporal pharmacometabolomics based on ambient mass spectrometry imaging to evaluate the metabolism and hepatotoxicity of amiodarone in HepG2 spheroids
Limei Li ,Qingce Zng ,Xinzhu Li ,Ying Zhu ,Shnjing Wen ,Jiuming He ,Ruiping Zhng ,*,Zeper Abliz ,b,c,**
a State Key Laboratory of Bioactive Substance and Function of Natural Medicines,Institute of Materia Medica,Chinese Academy of Medical Sciences and Peking Union Medical College,Beijing,100050,China
b Key Laboratory of Mass Spectrometry Imaging and Metabolomics(Minzu University of China),National Ethnic Affairs Commission,Beijing,100081,China
c Center for Imaging and Systems Biology,College of Life and Environmental Sciences,Minzu University of China,Beijing,100081,China
Keywords:
Mass spectrometry imaging
HepG2 spheroids
Hepatotoxicity
Drug metabolism
Amiodarone
ABSTRACT
Three-dimensional (3D) cell spheroid models combined with mass spectrometry imaging (MSI) enables innovative investigation of in vivo-like biological processes under different physiological and pathological conditions.Herein,airflow-assisted desorption electrospray ionization-MSI (AFADESI-MSI) was coupled with 3D HepG2 spheroids to assess the metabolism and hepatotoxicity of amiodarone(AMI).High-coverage imaging of >1100 endogenous metabolites in hepatocyte spheroids was achieved using AFADESI-MSI.Following AMI treatment at different times,15 metabolites of AMI involved in Ndesethylation,hydroxylation,deiodination,and desaturation metabolic reactions were identified,and according to their spatiotemporal dynamics features,the metabolic pathways of AMI were proposed.Subsequently,the temporal and spatial changes in metabolic disturbance within spheroids caused by drug exposure were obtained via metabolomic analysis.The main dysregulated metabolic pathways included arachidonic acid and glycerophospholipid metabolism,providing considerable evidence for the mechanism of AMI hepatotoxicity.In addition,a biomarker group of eight fatty acids was selected that provided improved indication of cell viability and could characterize the hepatotoxicity of AMI.The combination of AFADESI-MSI and HepG2 spheroids can simultaneously obtain spatiotemporal information for drugs,drug metabolites,and endogenous metabolites after AMI treatment,providing an effective tool for in vitro drug hepatotoxicity evaluation.
1.Introduction
Evaluation of potential hepatotoxicity is a critical step during initial drug development.In vitro experimental systems are vital components for selecting the most promising drug candidates that show high efficacy and have minimal safety concerns for use in clinical trials [1-3].Currently,several forms of hepatocyte monolayers are commonly used to screen for drug-induced hepatotoxicity or investigate the underlying molecular mechanisms as these systems are amenable to the rapid screening of large numbers of compounds[4].However,mounting evidence indicates that the biochemical cues and cell-to-cell communication necessary to maintain the physiological phenotypes and liver-specific functions of cells are lost in two-dimensional monolayer cultures and these may thus be poor predictors for drug-induced liver injury-implicated compounds[2,5].In contrast,three-dimensional(3D) cell-culture systems have recently attracted considerable attention in drug development.These 3D systems offer unprecedented insights into hepatotoxicity as they more closely reproduce liver biology.Thus,drug effects can be studied in more controllable microenvironments,which is expected to generate more robust data on the potential risks of pharmaceuticals [6,7].
The most commonly used toxicity assessment method in 3D spheroid models is the cell viability assay based on the reactions catalyzed by cellular metabolic enzymes,which indirectly reflect cellular metabolic activity.However,this has been limited by the inability to measure multiple mechanistic parameters that capture a wide spectrum of potential cytopathological changes [8].Currently developed methods based on high-content imaging[9,10] and flow cytometry [11] can simultaneously perform highthroughput determination of multiple toxicological endpoints,such as cell death,intracellular lipid accumulation,mitochondrial membrane changes,and reactive oxygen species.However,these methods require fluorescent labeling,affording limited detection coverage.Metabolomics can generate large amounts of data that represent the complex regulations involved in endogenous metabolism and thereby reveal information on the precise metabolic pathways and processes when hepatotoxicity occurs [12-16].
Mass spectrometry imaging (MSI)-based spatially resolved metabolomics can preserve the spatial location information of a various known and unknown molecules in heterogeneous biological samples without labeling [17-21] and has been used to spatially resolve the distributions of endogenous and exogenous species in micro-3D cell spheroids [22-24].For instance,the penetration and distribution of perifosine[25]and epidermal growth factor receptor targeting antibody cetuximab [26] were successfully mapped to increase understanding of the heterogeneous distribution of these drugs in multicellular tumor spheroids (MCTS).Furthermore,the spatial characteristics of endogenous metabolites in the glycolysis and tricarboxylic acid(TCA)cycle pathways and of lipids in different proliferating microregions in MCTSs have been mapped [23,27],offering unprecedented opportunities for elucidating metabolic mechanismsduringcancerprogression.Caiandcoworkers [28,29] applied matrix-assisted laser desorption/ionization mass spectrometry (MS) imaging to successfully elucidate the distribution and metabolism of bisphenol S [28] and hydroxychloroquine[29]as well as the effects of these on metabolite and lipid alterations in MCTSs,which helped reveal the underlying molecular mechanism of environmental pollutants and drug action.This work provides a paradigm to simultaneously study exogenous compound metabolism and the subsequent effects within in vitro models.
Although advancements in MSI technology,particularly achievable spatial resolution,have enabled clearer visualization of endogenous metabolites within a single heterogeneous cell spheroid,the exact number and types of metabolites that can be covered remains unclear.This limits the ability to assess the applicability of these analytical approaches.In particularly,several strongly polar endogenous small-molecule metabolites,such as glutathione,glutathione disulfide,and nucleosides,can reflect the cellular oxidative/redox state,which plays a crucial role in various biological activities [30,31].Currently,there remains an urgent need for a wide-coverage MSI method to stratify the metabolic detection of various cell states.
Amiodarone(AMI)is a widely used antiarrhythmic drug that can cause the liver injury including steatohepatitis,liver fibrosis and cirrhosis[32].Several studies showed that AMI metabolism plays an important role in drug induced hepatic toxicity [33,34],but the molecular mechanisms remain largely unknown.In this study,we propose a strategy to evaluate drug metabolism and hepatotoxicity by coupling a 3D HepG2 model with spatiotemporal pharmacometabolomics.A high-spatial-resolution and wide-coverage airflowassisted desorption electrospray ionization MSI (AFADESI-MSI)method was developed to map metabolites within HepG2 spheroids.Taking AMI as a model drug,the temporal and spatial features of exogenous drug metabolism during AMI treatment were characterized to reveal potential drug metabolic pathways.Furthermore,metabolic alterations after AMI treatment were analyzed to identify progressive changes related to drug action.Finally,we screened metabolic biomarkers related to cell viability and constructed biomarker groups to characterize hepatotoxic effects.The spatiotemporal pharmacometabolomics approach combined with HepG2 spheroids highlights a potential methodology for in vitro applications of toxicology research and pharmaceutical development.
2.Materials and methods
2.1.Chemicals and reagents
Amiodarone (≥99%) and dimethyl sulfoxide (DMSO) were purchased from Sigma-Aldrich (St.Louis,MO,USA).Dulbecco's modified Eagle's medium (DMEM),fetal bovine serum (FBS),penicillinstreptomycin,0.25% trypsin-ethylenediaminetetra-acetic acid,and phosphate-buffered saline were purchased from Gibco (Carlsbad,CA,USA).Ultra-low attachment (ULA) 96-well plates were purchased from Thermo Fisher Scientific Inc.(Waltham,MA,USA).Acetonitrile(ACN,high performance liquid chromatography grade)was purchased from Thermo Fisher Scientific Inc..Pure water was purchased from Wahaha Co.,Ltd.(Hangzhou,China).0.9%NaCl was purchased from Shijiazhuang Pharmaceutical Co.,Ltd.(Shijiazhuang,China).Other materials used for cell culturing were purchased from Corning (North Carolina,NY,USA),unless otherwise noted.
2.2.HepG2 spheroid cultivation and drug treatment
Human HepG2 liver cell line was purchased from American Type Culture Collection (Manassas,VA,USA).Cells were cultured in DMEM containing 15% FBS and 1% penicillin-streptomycin (V/V),maintained in 5% CO2at 37°C,and passaged every two days as adherent monolayers.For 3D spheroid generation,cells were seeded into ULA 96-well plates at 5000 cells/well.Uniform and solid 3D liver spheroids were used for cellular toxicity assessment after an initiation interval of four days and analyzed in sextuplicate.Established HepG2 spheroids were exposed to 50 μM AMI dissolved in DMEM (0.1% DMSO and 2% FBS,V/V) for different time points.Untreated spheroids were cultured with blank DMEM(0.1%DMSO and 2% FBS,V/V) as a reference.Viability was assessed,and the spheroids were harvested for AFADESI-MSI analysis.
2.3.Sample preparation
After drug treatment,spheroids were immediately washed three times with 0.9% (V/V) NaCl to remove the drug from the surface.After being transferred and embedded into Surgipath Cryo-Gel(Leica)at-20°C,micro spheroids were sectioned into 10-μmthick slices using a freezing microtome(CM1860,Leica,Germany).To obtain a complete,well-formed,representative slide,the 4th to 8th slices (middle part of spheroids) were chosen for further analysis.Slices were thaw-mounted on Superfrost Plus microscope slides (Thermo Fisher Scientific Inc.) and stored at -80°C until AFADESI-MSI,hematoxylin and eosin staining,and immunohistochemistry (IHC) analysis.
2.4.AFADESI-MSI analysis
MSI experiments were performed using an AFADESI platform coupled with a quadrupole Orbitrap mass spectrometer(Q Exactive,Thermo Scientific,Bremen,Germany)as previously reported[35].To obtain high-spatial-resolution images,a 20-μm inner diameter spray needle was used to analyze the cell spheroid sections.ACN:water(8:2,V/V)was used as the spraying solvent at a flow rate of 3 μL/min with the aid of 0.6 MPa nitrogen as the spraying gas.The spray voltage was set at±7.0 kV.The extracting gas flow rate was 45 L/min.A 3D electrical moving stage (Beijing Optical Instrument Factory,Beijing,China) was set at a constant rate of 0.04 mm/s in thex-direction and separated by a 0.04-mm vertical step in they-direction.AFADESI-MSI analysis was performed using a full MS scan in the positive and negative ion mode.Parameters of the mass spectrometer:capillary temperature,350°C;S lens voltage,55 V;maximum injection time,200 ms;automatic gain control target,3×106;resolution,70,000;m/zrange,70-1000.
2.5.Data processing
The collected MS data files (.raw format) were formatted (.cdf)and imported into MassImager for image reconstruction and the average mass spectrum data (.txt) obtainment of the region of interest (ROI).The spheroid area was specifically circled as a ROI according to the characteristic images for ion choline ([M+H]+,m/z104.1070) and glutamate ([M-H]-,m/z146.0459).Data were then imported into the Markerview™software 1.2.1 (AB SCIEX,Framingham,MA,USA) for peak picking,peak alignment to obtain a multidimensional data matrix ofm/z,and intensity assessment for statistical analysis.Tentative metabolite identification was performed using the Human Metabolome Database (HMDB,https://hmdb.ca/) and LIPID MAPS®(https://www.lipidmaps.org/) by extracting ions of [M+H]+,[M+Na]+,[M+H-H2O]+,[M+NH4]+,[M+K]+,[M+Cl]-,and [M-H]-withm/ztolerance <10 ppm.The identification process of AMI metabolites is described in the Supplementary data.
3.Result and discussion
3.1.Establishment of a 3D HepG2 spheroid model to evaluate hepatotoxic effect of AMI
To generate a well-formed 3D HepG2 spheroid model,the optimal seeding number and culturing time were investigated by seeding HepG2 cells at 2000,3000,5000,8000,and 10,000 cells/well.Representative images in Fig.S1A show that the spheroids were self-assembled and round-shaped,especially in the lower density model.The diameter of the spheroids gradually increased to >1000 μm when cultured for 21 days (Fig.1A).The relative standard deviation(RSD)of the diameters was <10%after four days of culturing(Fig.S1B),indicating acceptable reproducibility for the spheroid models.We also examined the viability of spheroids on different culturing days.This increased gradually for up to 21 days(Fig.1B),showing stable proliferation and improved longevity of our established model,which is suitable for long-term toxicity testing.In addition,we used IHC to determine the expression of the representative metabolic enzyme CYP3A4 to estimate the liver-like metabolic function.As expected,there was a clear increase in CYP3A4 expression over time,especially from the 4th to 7th day(Fig.1C).
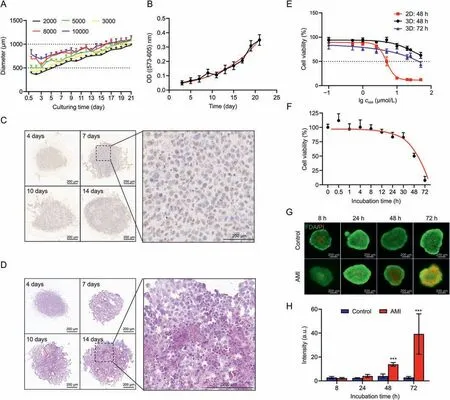
Fig.1.Establishment of HepG2 spheroid models and evaluation of amiodarone (AMI) hepatotoxicity.(A) Growth kinetics represented by the spheroid diameter over 21 days at initial seeding densities of 2000-10000 cells/well.Error bars represent standard deviation (n = 6).(B) The optical density (OD) value was measured to evaluate the cell viability under different culture days(n=6).The black line represents the fitting line of the data.The red line represents nonlinear curve fitting line of the data.(C)Immunohistochemistry(IHC)analysis of CYP3A4 expression in HepG2 spheroids at initial seeding densities of 2000 cells/well cultured for 4,7,10,and 14 days.The darkened brown part of the cytoplasm shows the expression of CYP3A4.(D) Hematoxylin and eosin images of HepG2 spheroids at initial seeding densities of 2000 cells/well cultured for 4,7,10,and 14 days.Darkened cytoplasm staining and karyopyknosis are observed in the inner region of spheroids cultured for 10 and 14 days.(E)AMI dose-response curves of 2D and 3D models after treatment for 48 and 72 h.(F) AMI incubation time-response curves of HepG2 spheroids under 50 μM AMI condition.The black line represents the fitting line of the data.The red line represents nonlinear curve fitting line of the data.(G)Corresponding live/dead fluorescent micrographs in both control group and AMI-treated group after treatment for 8,24,48,and 72 h.Red represents propidium iodide (PI) and green repersents fluorescein diacetate (FDA).(H) Fluorescence semi-quantitative analysis using ImageJ.PI intensity = PI (integrated density)/FDA (area).Error bars represent ± standard deviation (n = 3).***P <0.001.
The HepG2 spheroid developed heterogeneity,similar to other cell spheroids,because of the presence of physiochemical gradients as the spheroid diameter was up to 500 μm wide [36,37].We observed the formation of necrotic cores when the spheroids were cultured for 10 days (Fig.1D),with shrinkage of nuclei and concentration of chromatin for cells in the core areas of the spheroids.However,the necrotic core is undesirable for recreating an in vivolike liver microenvironment for toxicology studies.Therefore,to obtain a homogeneous model that was analogous to the liver both in function and structure,a density of 5000 cells/well was finally chosen for hepatotoxicity assay after four days of culturing when the size of the spheroid was 550-700 μm.
The hepatotoxic effect of AMI on HepG2 spheroids was evaluated to obtain the proper exposure concentration.The median lethal dose (LD50) of the 3D model was ~50 μM after AMI treatment for 72 h (Fig.1E).Less sensitivity was observed in 3D spheroids compared with that in the two-dimensional model,which had an LD50of 4.91 μM.To explore the hepatotoxic state under different viability conditions,50 μM AMI was selected for the timedependent experiment,which showed that cell viability decreased significantly in a time-dependent manner(Fig.1F).These results were confirmed using live/dead staining.Fluorescent imaging and semi-quantitative analyses(Figs.1G and H)showed that signals for propidium iodide-positive cells increased significantly at 48 and 72 h,indicating that AMI induced cell death in the 3D model.
3.2.High-coverage spatially resolved metabolome profiling in HepG2 spheroids
AFADESI-MSI was applied to detect and localize the metabolites within the spheroids.The performance of this method for the detection of endogenous metabolites was evaluated via parallel analyses of six spheroids under both positive and negative modes.The maximum ion intensities exceeded 1×106counts,and typical metabolite ion signals could be easily found in the mass spectra after excluding background ions in both positive and negative detection modes(Fig.S2).Reproduciblem/zfeatures were screened based on whether they were observed in all six samples and the lowest ion intensity was >1,000.Finally,>1100 reproduciblem/zfeatures (530 in positive and 610 in negative ion conditions) were detected (Fig.2A).
Them/zdistribution of these features is shown in Fig.2B.Ions withm/z100-400 accounted for 80% and many ions withm/z700-900 can also be detected.The reproduciblem/zfeatures were then annotated using HMDB and LIPID MAPS®,and 195 and 243 metabolites (positive and negative modes,respectively) were annotated(Fig.2A;Tables S1 and S2).The identified metabolites were grouped according to their HMDB classification.More than 10 classes of metabolites were detected,including amino acids(AAs),organic acids,nucleosides and nucleotides,fatty acids (FAs),and glycerophospholipids (GPs) (Fig.2C).Among these detected metabolites,AAs and FAs were the most abundant,accounting for 40%-50%.GPs were also detected,particularly in the positive mode,and accounted for a large proportion (17.95%).These results demonstrate the high coverage of AFADESI-MSI in the detection of the spheroid metabolites.
The metabolic pathways involved were further analyzed using the Kyoto Encyclopedia of Genes and Genomes database.There were 73 metabolic pathways mapped,and 29 pathways had a coverage of >50% (Table S3).Notably,these pathways included glutamine and glutamate metabolism,linoleic acid metabolism,arachidonic acid metabolism,glycerolipid metabolism,and the TCA cycle,which were all key pathways directly related to the survival and proliferation of cells.
AFADESI-MSI images of representative metabolites showed clear spatial distribution within the spheroids (Fig.2D).There was uniform distribution of many metabolites in the spheroid,such as FAs and carnitine; however,there was spatial heterogeneity of several metabolites: e.g.,AAs and choline were distributed in the center of the spheroid,whereas lysophosphatidylethanolamine 16:0 and lysophosphatidylinositol 16:0 were distributed in the outer layers.These results illustrate the direct and precise visualization of complex spatial distribution of metabolites in spheroids obtained using AFADESI-MSI.In addition,the stability and repeatability of the method were evaluated via the intensity variation of reproduciblem/zfeatures.The RSD values of these features showed that there were 72.3% and 81.2% features with RSD <40% in the stability experiment,and 75.5% and 63.5% features with RSD <40% in the repeatability experiment,demonstrating the acceptable reproducibility and stability of the metabolomic analysis (Fig.S3).These results demonstrate the powerful application potential of AFADESI-MSI in small-scale sample analysis.
3.3.Penetration and metabolism of AMI in HepG2 spheroids
To visualize drug penetration and metabolic processes,HepG2 spheroids were harvested and imaged using AFADESI-MSI after treatment with AMI for 0.5,4,8,24,48,and 72 h.AMI was mainly observed in the spheroid periphery at 0.5 h,and enhanced followed by drug penetration into the spheroid core within 4 h(Fig.3A).The average ion intensity of AMI increased from 0.5 to 72 h,showing that AMI gradually penetrated and accumulated in the spheroids(Fig.S4).The potential AMI metabolite ions were then screened.The tandem MS spectra of these ions were collected using a liquid chromatography-high-resolution tandem mass spectrometer in the parallel reaction monitoring mode(the Supplementary data).Based on the high-resolution MS1 and MS2 spectra of AMI,we deduced the cleavage pathway of AMI(Fig.S5).Based on the similarity of the product ions (Fig.S6) and fragmentation patterns to the drug metabolite parent,a total of 15 phase I metabolites of AMI were identified.The retention times,mass errors,formulas,and reaction types of all identified metabolites are presented in Table S4.These metabolites are involved inN-desethylation,hydroxylation,deiodination,dealkylation,and desaturation metabolic reactions.In previous studies,mono-N-desethylamiodarone was identified as the major metabolite of AMI in human plasma [38].33 AMI metabolites in human bile were identified [39],including 22 phase I and 11 phase II metabolites,and the major metabolites were mono-N-desethylamiodarone and ω-carboxylate AMI.In addition,49 AMI metabolites in rats were identified(all 49 in urine and 17 in serum)[40],and revealed that these metabolites were formed via four primary metabolic pathways:hydroxylation or carboxylation(oxo/hydroxylation) of the butyl-benzoxazole group;N-desethylation;deionization; and glucuronidation.Although the number of AMI metabolites identified in the HepG2 spheroids is less than that in previous reports,nine potential AMI metabolites involved in multistep metabolic reactions,such as deiodination,desaturation,and dealkylation reactions,were found for the first time(Table S4,labeled with#).
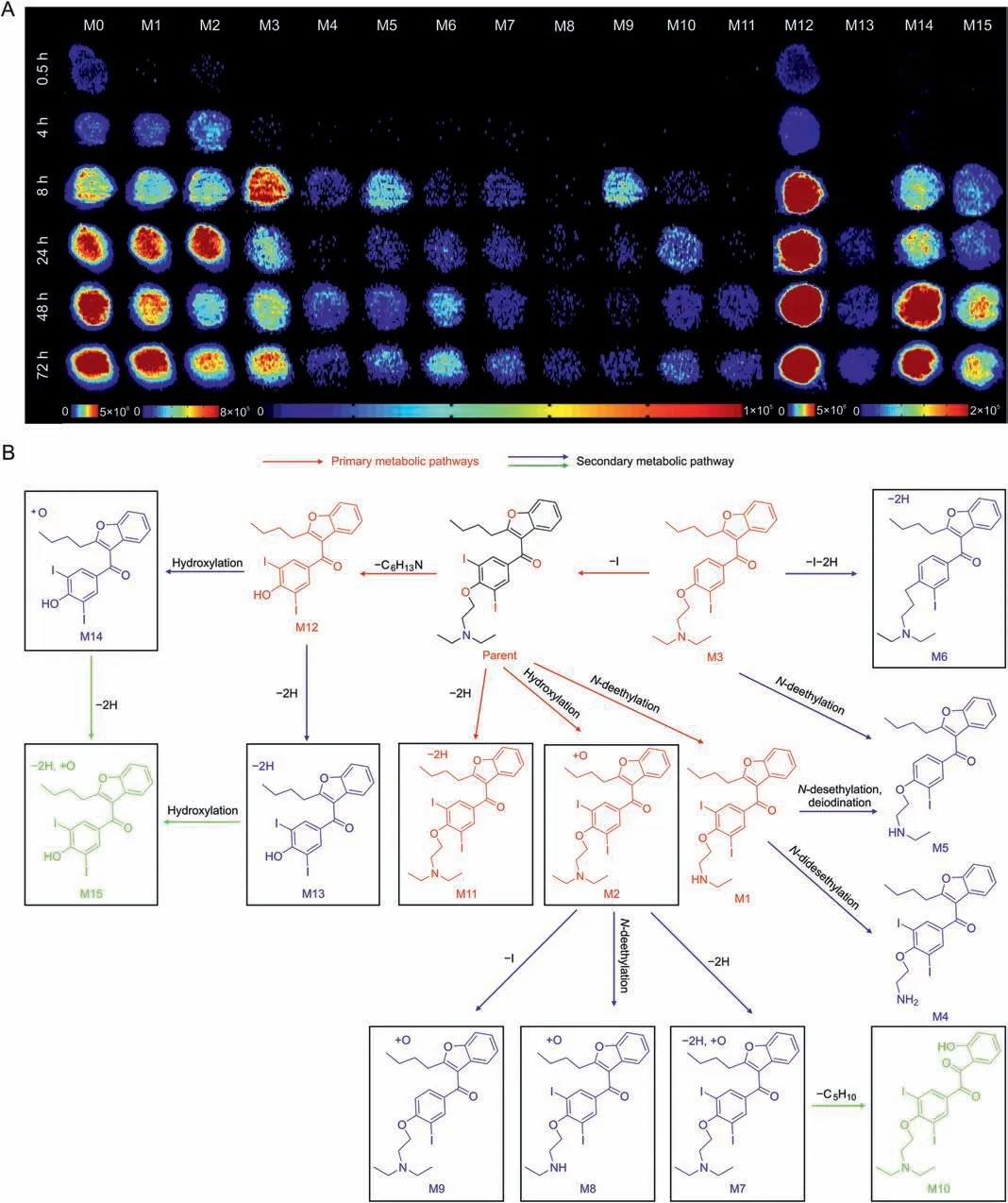
Fig.3.Time-dependent penetration and metabolism of amiodarone (AMI) in HepG2 spheroids analyzed using airflow-assisted desorption electrospray ionization-mass spectrometry imaging(AFADESI-MSI).(A)AFADESI-MS images of AMI and metabolite distribution at different treatment times.HepG2 spheroids were treated with 50 μM AMI for 0.5,4,8,24,48,and 72 h.(B) Postulated metabolic pathways of AMI in the HepG2 spheroids.
The spatiotemporal changes of these identified metabolites were mapped to monitor their location and dynamics for accurate estimation of drug metabolism.From the occurrence time and intensity of these ions in each group,an interesting characteristic was found:M1,M2,M3,and M12 metabolites showed higher intensity and appeared even earlier in the 0.5 and 4 h AMI-treated groups of spheroids compared with other drug metabolites (Fig.3A).Based on the reaction types,these metabolites are the main metabolites directly transformed from AMI through one-step reactions.For instance,M1 was identified as one of the main metabolites in the liver with aN-desethylation transformation[39,41].M11 was found to form from a one-step dehydrogenation reaction,although this metabolite had low intensity,indicating that dehydrogenation was not the main metabolic pathway of AMI.We also found that M4 and M5 underwent anN-desethylation reaction,but the intensities were low and time of appearance was late.Therefore,these metabolites are regarded as M1 derivatives viaN-desethylation or deiodination reactions.Similarly,M2 was produced after AMI had undergone a monohydroxylation reaction,and M7,M8,and M9 were inferred as being derived from M2.Therefore,we used the intensity of the metabolites to reflect the priority of the metabolic reactions and used the time of metabolite occurrence to reflect the metabolic rate.The proposed metabolic networks of AMI are shown in Fig.3B.The pathway activity and relationships between each metabolite are clearly displayed.This spatiotemporal imaging pattern of drug metabolism improves the understanding of the biotransformation process of drugs and potential substance basis of hepatotoxicity.
3.4.Spatiotemporal metabolic response of HepG2 spheroids under AMI treatment
To investigate the toxicity effects induced by AMI exposure on HepG2 spheroids,metabolomic profiles at different treatment time points were analyzed after excluding drug and drug metabolites.The volcano plot in Fig.S7 shows considerable metabolic change after 24 h AMI treatment,and these changes were more pronounced at 48 and 72 h.Notably,most of the ions were significantly downregulated,indicating a decrease in overall cellular metabolism with considerable reduction in cell viability.
The fold change (FC) of reproduciblem/zfeatures in each AMItreated group was assessed using non-supervised principal component analysis(PCA) to search for discriminant metabolomic patterns caused by hepatotoxicity.Each AMI-treated group was well clustered and there was distinct separation between the longand short-term-treated groups(48,72 h and 0.5,4 h,respectively)(Fig.4A).The metabolites that contributed to classification were screened according to the threshold of the Student'st-testPvalue <0.05,FC <0.8 or >1.2.Finally,81 and 138 metabolites were identified with significant FCs in the 24 and 72 h AMI-treated groups,respectively (Table S5).The heatmap (Figs.4B and S8)presents a marked variation in the levels of metabolites in the 24,48,and 72 h AMI-treated groups compared with those in the control groups.AMI treatment induced significant downregulation of lipids,and specifically of FAs,such as monounsaturated FAs,polyunsaturated FAs,hydroxy FAs,FA esters of hydroxyl FAs,monophosphate glycerol,diphosphate glycerol,and phosphocholine(PC)(Fig.4C).However,there was also a marked upregulation of phosphatidylinositol(PI)and phosphatidylglycerol(PG)of 1.2-to 2.2-fold,respectively,in the AMI-treated group in a timedependent manner.Levels of several small-molecule organic acids,AAs,and nucleosides were also significantly downregulated(Figs.4B and S8 and Table S5).
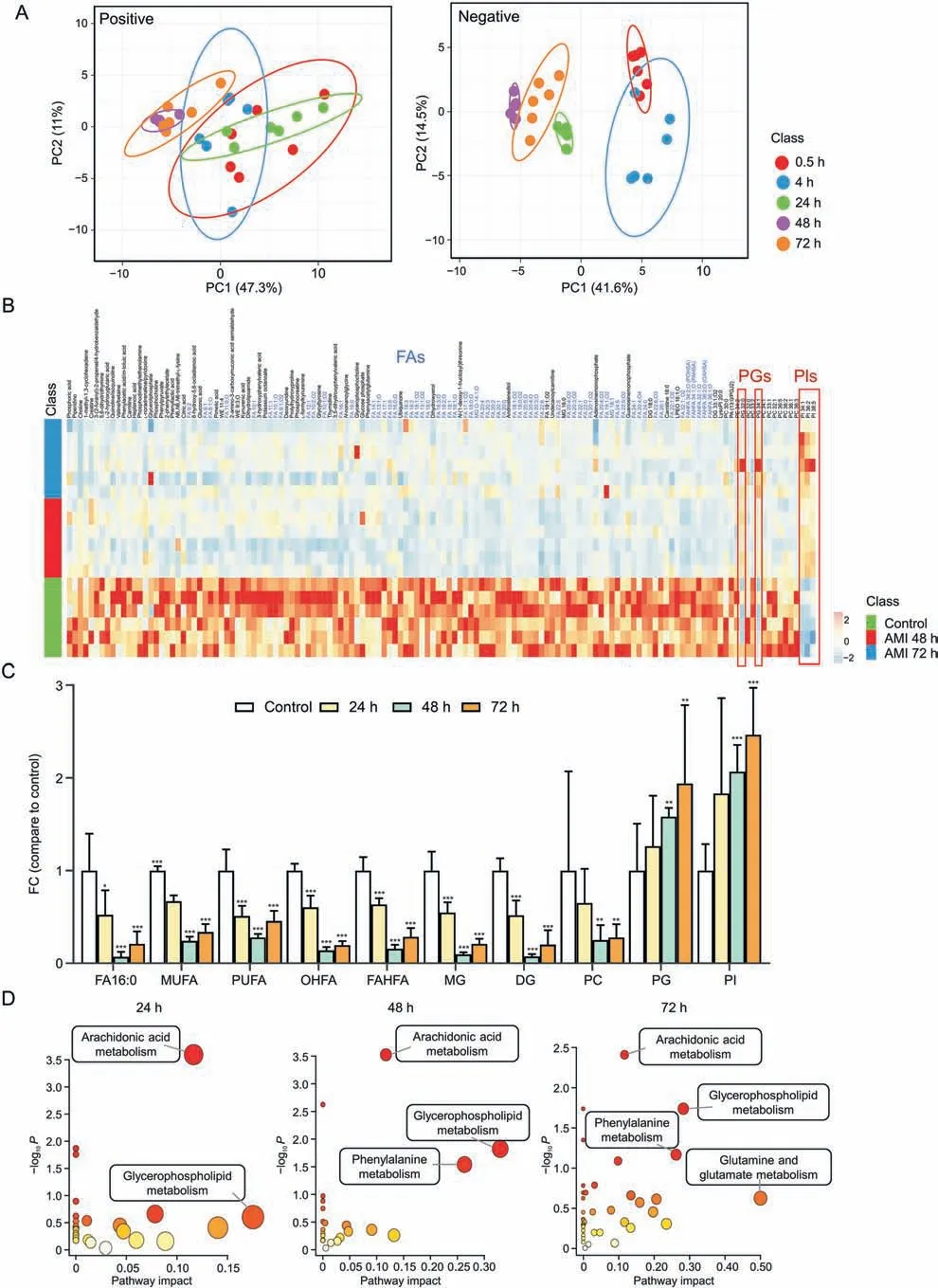
Fig.4.Amiodarone (AMI)-induced metabolic variation in HepG2 spheroids.(A) Principal component analysis (PCA) score plots of spheroids in the 0.5,4,24,48,and 72 h AMItreated groups in positive and negative ionization modes (n = 6).(B) Heatmap analysis of identified metabolites after AMI treatment for 48 and 72 h.(C) Fold change of 24,48,and 72 h AMI-treated groups vs.control group of fatty acids (FAs) and other lipid metabolites.*P <0.05,**P <0.01,and ***P <0.001.(D) Altered metabolic pathways during AMI treatment.WE: wax ester; MG: monophosphate glycerol; DG: diphosphate glycerol; LPEO: lysophosphatidylethanolamine preoxide; FAHFA: fatty acid esters of hydroxyl FAs;POHSA: palmitoleic acid esters of hydroxystearic acid; PAHSA: palmitic acid esters of hydroxy stearic acid; OAHSA: oleic acid esters of hydroxy stearic acid; LysoPI: lysophosphatidylinositol; PC: phosphocholine; PA: palmitic acid; PGJ: prostaglandin J; PE: phosphatidylethanolamine; PG: phosphatidylglycerol; PI: phosphatidylinositol; FC: fold change;MUFA: monounsaturated FAs; PUFA: polyunsaturated FAs; OHFA: hydroxy FAs.
The differential metabolites in the 24,48,and 72 h AMI-treated groups were enriched in various metabolic pathways.Four metabolic pathways,namely arachidonic acid,glycerophospholipid,phenylalanine,and glutamine and glutamate metabolism showed aggressive progression in both number and impact value with increasing exposure time (Fig.4D),reflecting the enhanced exacerbation of metabolic dysregulation.Arachidonic acid metabolism was the earliest and most significant pathway altered during AMI treatment.Therefore,we further mapped MSI images of arachidonic acid and six associated derivatives in each group(Fig.S9).The intensity of these metabolites gradually decreased both spatially and temporally,which was most significant in peripheral areas in the 24 h AMI-treated group and further decreased at 48 and 72 h.AMI is a cationic amphiphilic drug that can directly inhibit the lysosomal lipases or bind to the hydrophobic and hydrophilic moieties of negatively charged cellular phospholipid-producing drug-phospholipid complexes.These complexes are indigestible by lysosomal phospholipases A1,A2,and C and therefore directly inhibit the hydrolysis of arachidonic acid from phospholipids [42].In addition,these changes are related to inflammatory response and lipid peroxidation,where arachidonic acid and its associated derivatives play an important role [43,44].
As the glycerophospholipid metabolic pathway was found to be significantly altered in the 48 and 72 h AMI-treated groups(Fig.4D),we also mapped the glycerophospholipid metabolic pathway network and observed that alternate trends were evident among different phospholipid metabolites(Fig.5).The intensity of PIs and PGs biosynthesized via the cytidine diphosphatediacylglycerol pathway was upregulated but that of PCs,phosphatidylethanolamines (PEs),and phosphatidylserines was downregulated.PCs and PEs are major components of the cell membrane,involved in energy storage,and play an important role in the growth and proliferation of cells [45,46].After AMI treatment,the viability of the spheroids rapidly decreased within a short time;thus,these glycerophospholipids were quickly depleted for energy supply.However,PGs and PIs are important signal transduction molecules,and their increase may be related to the inhibitory effect of AMI on phospholipase because of the cationic amphiphilic nature[42].
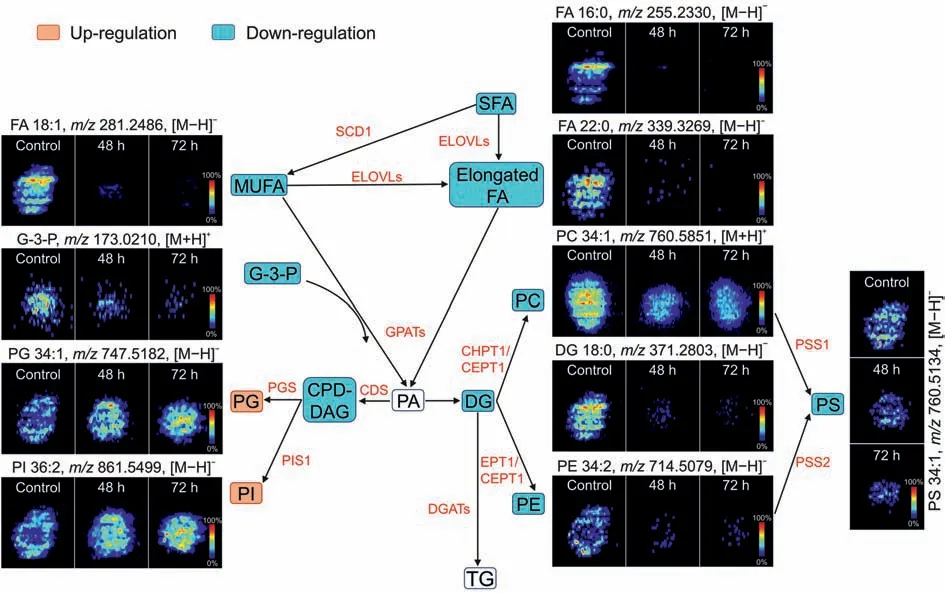
Fig.5.Spatial distribution and changes of metabolites related to phospholipid metabolism in HepG2 spheroids after amiodarone(AMI)treatment(48 and 72 h).SFA:saturated fatty acid; SCD1: stearoyl-CoA desaturase 1; MUFA: monounsaturated fatty acid; FA: fatty acid; ELOVLs: elongase of very-long-chain fatty acids; G-3-P: glycerol-3-phosphate; GPATs:glycerol-3-phosphate acyltransferases;PA:phosphatidic acid;PG: phosphatidylglycerol;PGS:phosphatidylglycerol synthase; CDP-DAG:cytidine diphosphate-diacylglycerol;CDS:CDP-DAG synthase;DG:diphosphate glycerol;CHPT1:choline phosphotransferase 1;CEPT1:choline/ethanolamine phosphotransferase 1;PC:phosphocholine;EPT1:ethanolamine phosphotransferase;PE:phosphatidyl ethanolamine;PS:phosphatidylserine;PSS1:phosphatidylserine synthase 1;PSS2:phosphatidylserine synthase 2;PI:phosphatidylinositol;PIS1: phosphatidylinositol synthase 1; DGATs: diacylglycerol acyltransferases; TG: triglyceride.
3.5.Hepatotoxicity characterization of AMI with endogenous metabolite biomarkers
To explore the relationship between cellular metabolite alteration and cell viability,Pearson correlation analysis was performed to search for metabolic biomarkers closely related to cell viability.This identified 42 biomarkers withP<0.05 and absolute value of Pearsonr>0.5 (Table S6).Among these biomarkers,40 exhibited significant positive correlation,and two phospholipids(PI 36:2 and PG 34:1) exhibited significant negative correlation.Notably,24 biomarkers are FAs,including arachidonic acid and five associated derivatives (Table S6).We speculated that the changes in these metabolites probably reflect the spheroid viability,and thus a combination of these metabolites could effectively provide a more accurate reflection of viability during drug treatment.For this purpose,multiple linear regression (MLR) was introduced to construct a biomarker panel model between metabolite change and cell viability.After optimization,a sparse set of metabolites including eight FAs that were most relevant to cell viability were selected.The correlation scatter plot is shown in Fig.S10,and these metabolites were significantly positively correlated with cell viability.The MLR equation is
E(Y) = 0.473X1+0.438X2+ 0.287X3- 0.108X4- 0.215X5+0.123X6- 0.151X7-0.034X8+ 0.288
WhereYis the predicted value of cell viability andXiis the estimated FC of each metabolite.Table S7 shows the detail of the MLR model between the FC of metabolites and cell viability.The correlation scatter plot between the predictive cell viability model and cell viability is shown in Fig.S11.As expected,the inclusion of multiple metabolites improves on the correlation found with a single metabolite and cell viability.
Similarly,the representative AFADESI-MS images of these metabolites present time-dependent decrease in AMI-treated groups(Fig.6).The FCs of these metabolites in the 48 and 72 h AMI-treated groups decreased <0.5 (except for FA 20:2 and FA 20:1),which correlates with the cell viability results (Fig.1F).Several specific metabolites,such as prostaglandin C1,were heterogeneously distributed,and could only be detected in the core region of the spheroid in the 48 h AMI-treated group,indicating that the gradual processes of hepatotoxicity can be captured by integrating both temporal and spatial dimensions of metabolite biomarkers.These results suggest that the biomarkers are closely related to cell activity and can directly characterize the hepatotoxicity of AMI,providing important molecular clues for evaluating the hepatotoxicity of AMI.
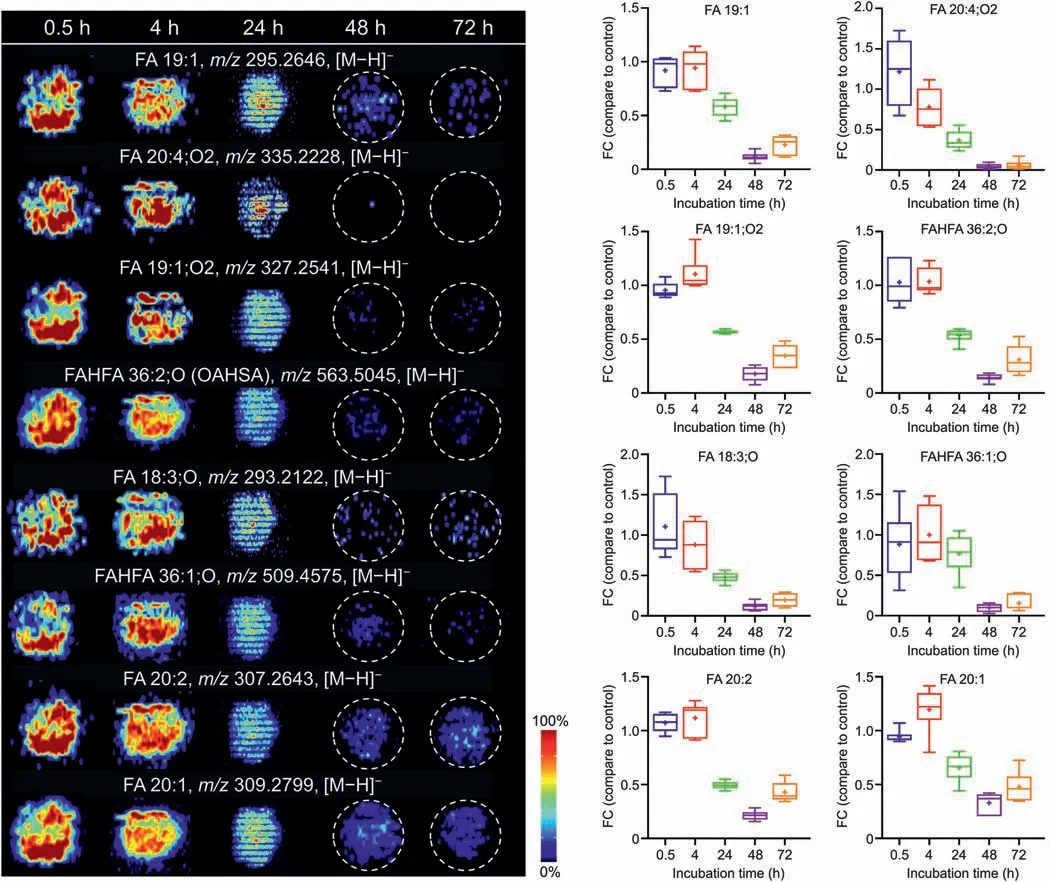
Fig.6.Representative airflow-assisted desorption electrospray ionization-mass spectrometry imaging (AFADESI-MSI) images and changes of eight fatty acids (FAs) after amiodarone (AMI) treatment for 0.5,4,24,48,and 72 h.FAHFA: fatty acid esters of hydroxyl fatty acid; OAHSA: oleic acid esters of hydroxy stearic acid; FC: fold change.
4.Conclusion
In this study,we present a spatiotemporal pharmacometabolomic method to investigate the metabolism and hepatotoxicity of AMI by combining AFADESI-MSI and HepG2 spheroids.The highcoverage AFADESI-MSI method enabled acquisition of spatially resolved information of >1100 endogenous metabolites in HepG2 spheroids,covering >10 metabolite classes and 29 metabolic pathways,providing a powerful tool for the study of hepatotoxicity within an in vitro model.The dynamic features of AMI and 15 associated metabolites could be simultaneously visualized within micro-scale HepG2 spheroids and helped to deduce the drug metabolic network.Using metabolomic analysis,the gradual progression caused by AMI toxicity was clearly demonstrated both in the number and degree of variation of the significantly altered metabolites and metabolic pathways.Using MLR,a group of eight FA biomarkers was constructed that exhibited improved correlation to spheroid viability.Using these methods,specific endogenous metabolic changes related to drug penetration,metabolism,and drug hepatotoxicity can be obtained,provided a new strategy for revealing the potential toxic mechanism and evaluating the hepatotoxic effects of drugs in vitro.
The advantage of this present study is to elucidate the molecular cues of AMI toxicity using MSI of drug-exposed hepatocellular spheroids.However,this method has some limitations.The structural annotations of MSI data are rather challenging,and the accurate information of chemical components is the basis for the subsequent biochemical interpretation.The biomarkers found in this study still need a series of biological validation in vivo and in vitro.In addition,only one drug of AMI was carried out studies,and the potential biomarkers screened by different drug exposures may be different.Therefore,this method can be extended to the study of hepatotoxic drugs with different mechanisms to obtain universal or specific hepatotoxic biomarker models and quantitative indicators,which will help to implement an in vitro modelbased metabolomics method for hepatotoxicity assessment.
CRediT author statement
Limei Li:Investigation,Formal analysis,Writing-Original draft preparation;Qingce Zang:Methodology,Writing- Reviewing and Editing;Xinzhu Li:Validation,Data curation;Ying ZhuandShanjing Wen:Formal analysis;Jiuming He:Resources;Ruiping Zhang:Conceptualization,Supervision;Zeper Abliz:Supervision,Funding acquisition.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This research was funded by the National Natural Science Foundation of China (Grant No.: 21874156),and the Chinese Academy of Medical Science (CAMS) Innovation Fund for Medical Sciences (Grant No.: 2021-1-I2M-028).
Appendix A.Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2023.04.007.
 Journal of Pharmaceutical Analysis2023年5期
Journal of Pharmaceutical Analysis2023年5期
- Journal of Pharmaceutical Analysis的其它文章
- DNA barcoding in herbal medicine: Retrospective and prospective
- Benzodiazepines in complex biological matrices: Recent updates on pretreatment and detection methods
- Ginsenoside Rk3 is a novel PI3K/AKT-targeting therapeutics agent that regulates autophagy and apoptosis in hepatocellular carcinoma
- Host cell protein quantification workflow using optimized standards combined with data-independent acquisition mass spectrometry
- Comprehensive and deep profiling of the plasma proteome with protein corona on zeolite NaY
- A robust luminescent assay for screening alkyladenine DNA glycosylase inhibitors to overcome DNA repair and temozolomide drug resistance
