Fine-Grained Sleep Apnea Detection Method from Multichannel Ballistocardiogram Using Convolution Neural Network
HUANG Yongfeng(黄永锋) , HUANG Qihong(黄琦洪), SUN Chenxi(孙晨汐), YANG Shuchen(杨树臣), ZHANG Zhiming(张智明)
1 School of Computer Science and Technology, Donghua University, Shanghai 201620, China
2 Shanghai Yueyang Medtech Co., Ltd., Shanghai 201203, China
Abstract:Sleep apnea is a common health condition that can affect numerous aspects of life and may cause a lot of health problems especially in the middle-aged and elderly population. Polysomnography (PSG), as the gold standard, is an expensive and inconvenient way to diagnose sleep apnea. However, ballistocardiogram can be collected by devices embedded in the surrounding environment, enabling inperceptible sleep apnea detection. Moreover, to obtain the fine-grained apnea fragments, a multistage sleep apnea detection model has been proposed. This model firstly uses an improved convolution neural network (CNN) model to coarsely identify apnea events and then a U-Net based model is applied to finely segment apnea fragments. In the experiment, sleep data of 11 patients with apnea for about 70 h have been collected, including BCG data derived from 18 piezoelectric polyvinylidene fluoride (PVDF) sensors embedded in the mattress and PSG data collected synchronously. The results show the accuracy of the classification model as good as 95.7% with 0.818 dice coefficient of the segmentation model, which indicates that the proposed model can almost match the performance of PSG in detecting apnea.
Key words:sleep apnea; ballistocardiogram; convolution neural network (CNN); deep learning
Introduction
Sleep apnea, as an extremely common sleep disorder in modern society, may lead to shortness of breath and affect our sleep quality. Studies have shown that repeated episodes of apnea will lead to nocturnal hypoxia, which in turn can cause many serious complications such as hypertension, diabetes, cardiovascular and cerebrovascular diseases, and even sudden death at night[1]. Moreover, about 10% of population has suffered a lot from sleep apnea, mainly owing to the increasing social pressure and various psychological diseases. Some researchers believe that anxiety and depression are gradually becoming the main or even all inducement of sleep apnea[2]. Due to the universality of above-mentioned problems, sleep apnea has become a nonnegligible disease in modern society.
Since sleep apnea requires monitoring of sleep conditions throughout the night, polysomnography (PSG) has been proposed in clinical medicine for the diagnosis of apnea. PSG, performed in sleep laboratory, is considered the gold standard for sleep apnea diagnosis by recording various physiological signals, including nasal inspiratory airflow, electroencephalogram (EEG) and electrocardiogram (ECG), allowing researchers to achieve accurate results. However, it is uncomfortable and may cause psychological burden of patients. Furthermore, for a severe apnea patient, monitoring at home and inspection in the hospital should be given equal attention. Ballistocardiogram (BCG), as a universal medical technique, records the change of pressure caused by cardiovascular and respiration in a cost-effective and noninvasive way[3-4]. Since the signal can be measured by sensors embedded in the ambient environments without the need for medical staff, it can be a potential method for home sleep apnea monitoring.
However, most of the past studies[5-9]on sleep apnea have focused only on classification tasks using traditional methods such as template matching, heart rate detection paired with machine learning and deep learning methods. Specifically, the classification results can only identify whether or what kind of sleep apnea occurrs in a period. In fact, more detailed information such as the frequency of sleep apnea and the approximate period of sleep apnea is of equally significance. Hence, past studies fail to provide meaningful information for clinical diagnosis in sleep apnea, for example, apnea-hypopnea index (AHI). So, we propose a fine-grained multistage sleep apnea detection model. This model first identifies apnea events using convolutional neural network (CNN), and then uses a U-Net based model to segment the time period during which the apnea occurs.
Furthermore, a high accuracy with single channel BCG signal was achieved in Ref. [10]. However, a single sensor system may cause measurement problems such as low coverage and non-continuity since BCG can only be detected when the human body produces pressure on the mechanical sensor. To tackle this problem, we propose that, due to the noninvasive characteristic, multiple-sensor systems can be used to collect BCG signal at the same time and make full use of BCG signal through channel fusion to improve the accuracy of detection.
The main contributions are the following two points. Firstly, we propose a more fine-grained multistage sleep apnea detection model that can reach the sample point level of resolution, which is at a higher resolution compared with state-of-the-art methods. Secondly, different multichannel data fusion methods are compared, and squeeze-and-excitation net (SENet) shows the best results under the contrast experiment of different parameters.
1 Methods
Figure 1 shows the global architecture of the proposed U-Breath model. The model will settle the multichannel BCG signal fusion problem firstly, and then determine whether sleep apnea events happens or not. If it happens, the fine-grained segmentation is further performed and then the marked apnea segment will be output.
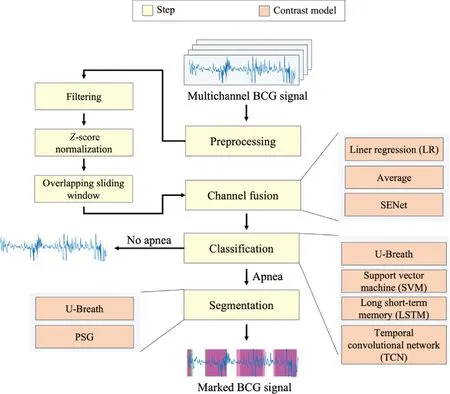
Fig.1 Global model architecture
1.1 Dataset and preprocessing
The BCG data acquisition equipment is composed of 18 PVDF array sensors embedded in the mattress. Eighteen sensors are placed horizontally in two rows, and the position of the sensors in the mattress is roughly equivalent to the head and chest of the human body.
In this study, we collected BCG data from 11 apnea patients. Data acquisition time was approximately 7 h of sleep throughout the night for each subject. The BCG sampling frequency is 50 Hz. To improve the generalization ability of the model, we also added a normal human BCG data collected in the same way.
For comparison, the PSG data of the above 11 patients were also recorded and the segments where apnea occurred were labeled by a professional sleep physician. Professional sleep specialists determined if apnea occurred based on the nasal airflow and thoracoabdominal breathing conditions collected by the PSG. Central apnea is characterized by the absence of nasal airflow and thoracoabdominal breathing, while obstructive apnea is characterized by undetectable nasal airflow and normal thoracoabdominal breathing. Sleep physicians use the above methods to determine whether apnea occurs and how long it has lasted.
Then the Butterworth digital filter is applied to remove high-frequency noise and the cut-off frequency of 3 dB filter is 10 Hz and the order is 7. TheZ-score normalization method is used to normalize the 18 channel signals to accelerate the convergence speed of the model. Moreover, we use the sliding window method to organize the data. Specifically, Fig.2 shows the distribution of the duration of apnea patients in all samples. It can be seen that the duration of most of the samples is concentrated in the time range from 10 to 60, and the duration of the longest apnea sample is around 120. Therefore, the length of each window is 90, and to expand the sample data set, we slide the window in 10-second steps. Within the 90 second window, the data that contain apnea fragments longer than 10 are regarded as positive samples, while the rest are negative samples.
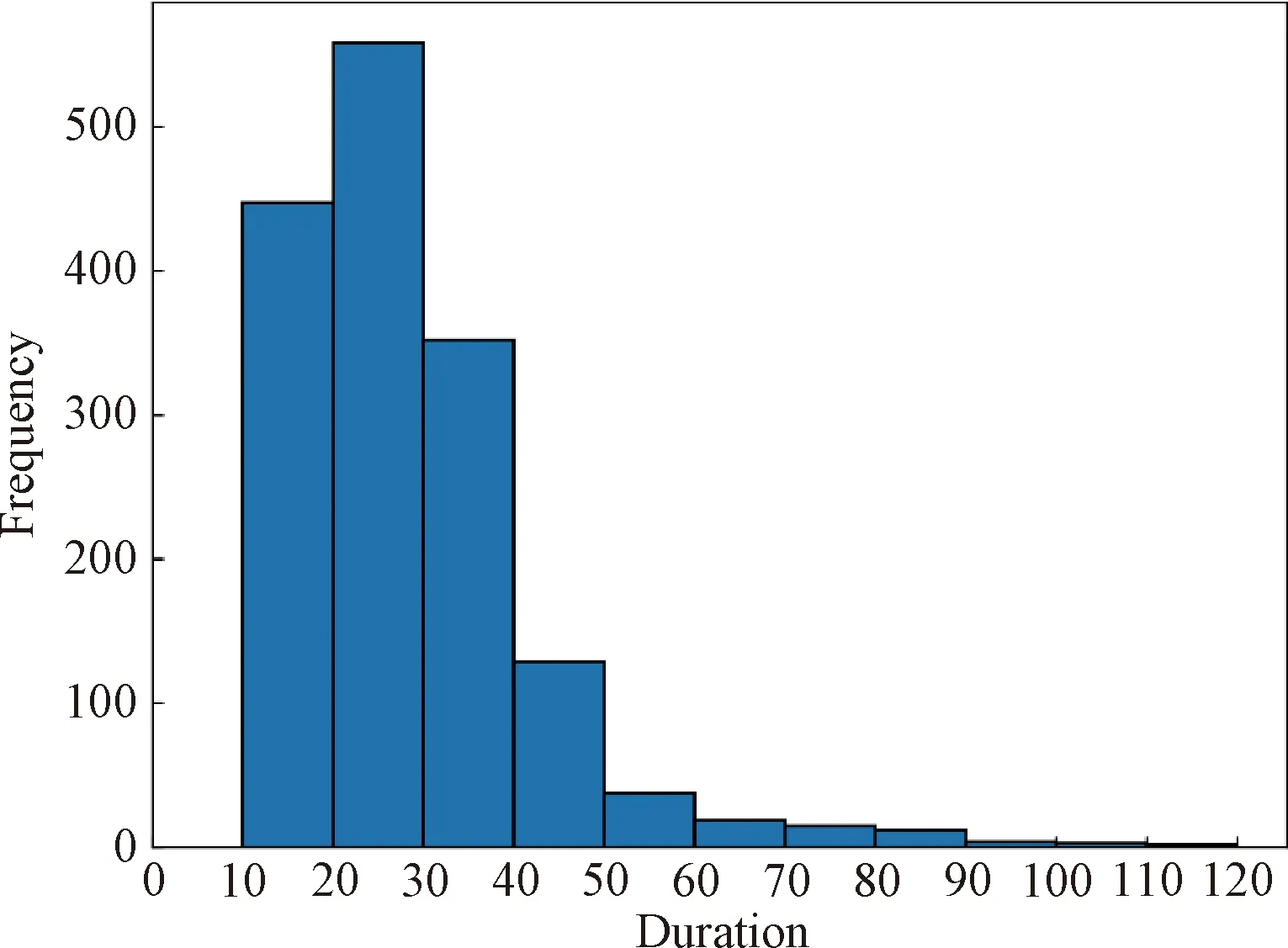
Fig.2 Frequency histogram of all apnea samples
1.2 Channel fusion methods
After data preprocessing, 18 channel BCG signals are sent to the channel fusion layer for gating or fusion. We use LR, average and SENet[10]to process multichannel BCG signals, and the target is to output single channel BCG signal as the input data of subsequent network. The architecture of SENet is illustrated in Fig.3, in which the sign 18×4 500 means that the size of the input signal consists of 18 channels and 4 500 one-dimensional sample points; the signs 18×1,c@ 1×1 and 1×4 500 represent the output signal size after the current layer processing of the model, wherecis a variable parameter. SENet consists of three parts: squeeze, excitation and scale. Its main purpose is to make the model pay more attention to the channel features with the largest amount of information and suppress those unimportant channel features.
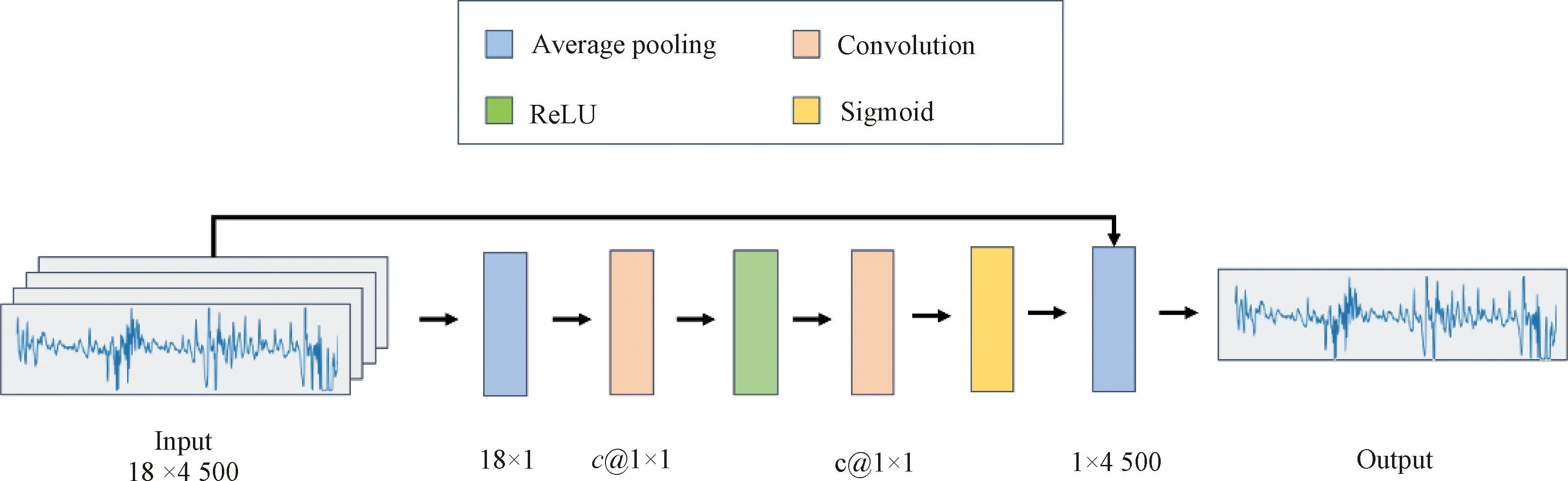
Fig.3 SENet model architecture
1.3 Classification methods
Classification network architecture, inspired by the work of Banluesombatkuletal.[11], is illustrated in Fig.4. In Fig.4, the signs 50@20×1, 30@24×1, 24@10×1 and 12@10×1 represent the channels and size of filter. The sign 192×1 is the full connection block size. The sign 1×4 500 is the input signal size. The goal of classification network is to determine whether BCG data over a window of time contain sleep apnea events. As shown in Fig.4, the classification network is composed of six one-dimensional (1D) convolution layers and one full connection layer. Specifically, each convolution layer contains convolution, activation, batch normalization and pooling operations. In addition to the first convolution layer, the dropout block is added to solve overfitting problem. And the full connection layer is made up of full connection and activation blocks.
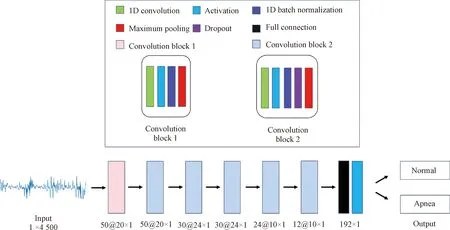
Fig.4 Classification model architecture
1.4 Segmentation methods
As shown in Fig.5, we use a U-Net[12-13]based network to achieve fine-grained segmentation task. U-Net has demonstrated impressive performance on medical image segmentation as well as time series segmentation. Within the U-Net network, it has two main parts, namely an encoder and a decoder, also known as a down-sampling part and a up-sampling part. There are four convolution layers in each part. The meanings of the symbols in Fig.5 are as follows. The numbers 1, 16, 32, 64, 128 and 256 to the right of the down-arrow and to the left of the up-arrow represent the size of the filter. The numbers 2, 4, 6 and 8 to the left of the down-arrow and to the right of the up-arrow represent the size of maximum pooling kernel. And 128+256, 128+64, 64+32 and 32+16 represent the output dimension after concatenation. Besides, 1×4 500 represents the dimension of input data and output data.
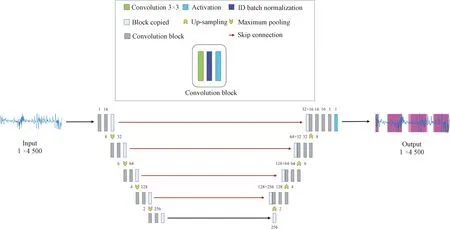
Fig.5 Segmentation model architecture
2 Experiments
2.1 Classification
To verify the effectiveness of the classification network, we manually select one channel signal as the input. Alivaretal.[14]found that the channel with better signal quality should be normal distribution. Therefore, we choose the data that best fit the normal distribution as the input of the classification network in our experiments. Figure 6 shows the data distribution of the 18 sensors for one subject we randomly selected.
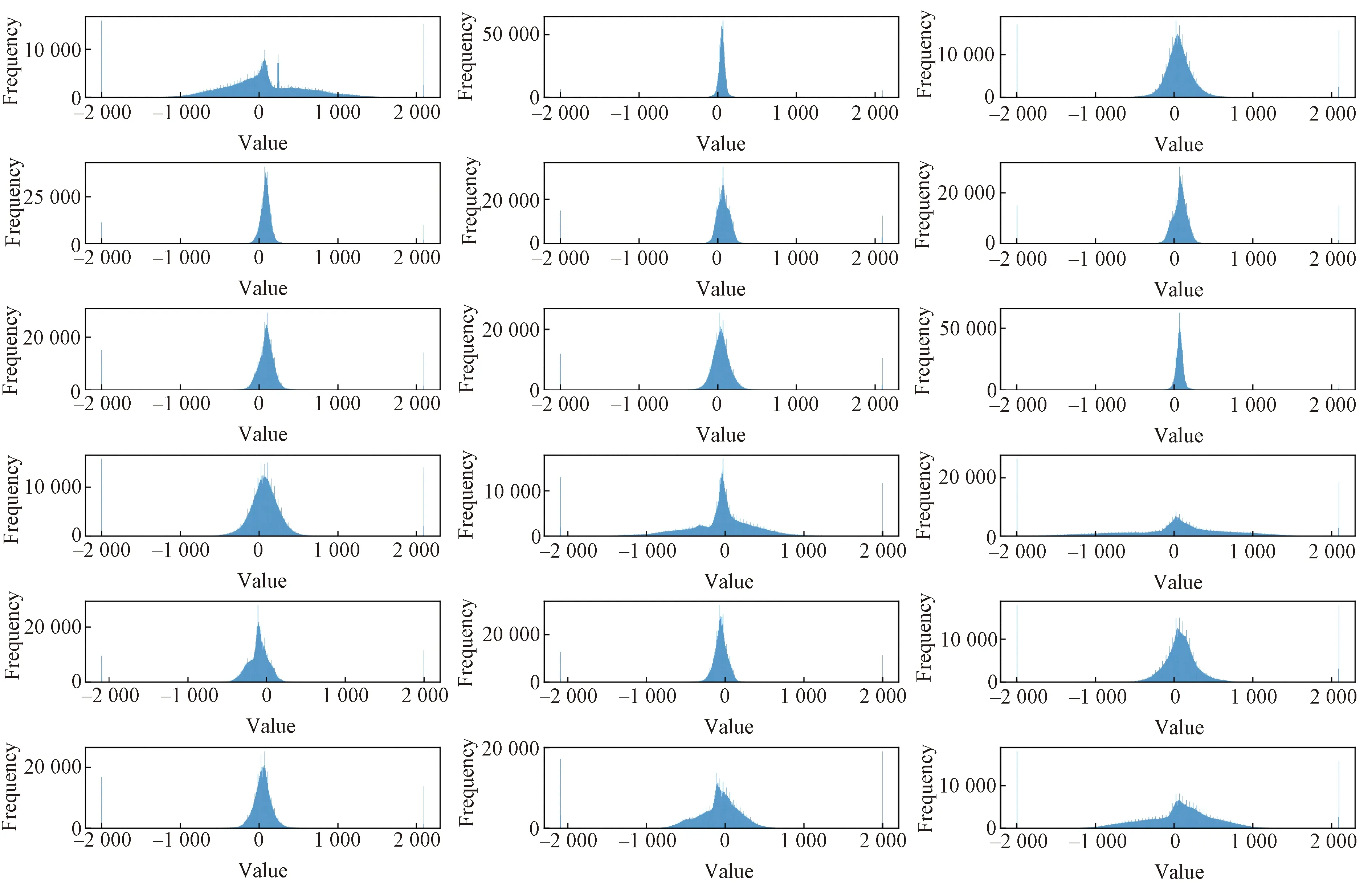
Fig.6 Data distribution of BCG signals of a patient throughout the night
For validating the performance of the proposed model, we trained some different models for comparison. The first one is the U-Breath model proposed in this paper. Next, we reproduced the model in recent study[15], where they proposed a model for apnea event detection based on an SVM implementation. Then, a bidirectional long short-term memory (Bi-LSTM) based[16]model and a TCN based[17]model in the study[18]were also used for comparison. Note that all the above-mentioned models use the same data input format.
In the U-Breath model experiment, we referred to its original study[13]to set our experimental parameters. Specifically, the dropout layer probability is 0.2. That is, 20% of the neuron output value is set to 0 randomly[19]. Then we use a mini-batch method for training[20], and gradient descent method with an Adam optimizer is performed on only 256 samples at a time. The learning rate is 0.001, and the total number of training epochs is 200. And we use cross entropy as the loss function, which can measure the difference between two data distributions. The calculation formula of cross entropy losslcis shown as
(1)
The total number of samples is 23 573, including 10 541 positive samples and 13 032 negative samples. We extracted the training set, the validation set and the test set from the BCG data of 11 subjects in the proportion of 60%, 20% and 20%, respectively, and combined them into the final training set, the final validation set and the final test set. The number of samples of classification network is shown in Table 1.

Table 1 Sample number of each dataset in classification network
After training 200 epochs, the U-Breath model converge. Figure 7(a) shows the train loss and validation loss of the U-Breath model. Meanwhile, the training of the comparison model was also completed according to the respective parameter settings of the comparison model. Table 2 shows the evaluation metrics of all classification models, where the experimental results of the SVM model are averaged after 5k-fold cross validation. Figure 8(a) shows the receiver operating characteristic (ROC) curve of all models and the respective area under curveAROC, both of which indicates that the U-Breath model outperforms other models and achieves elegant results.
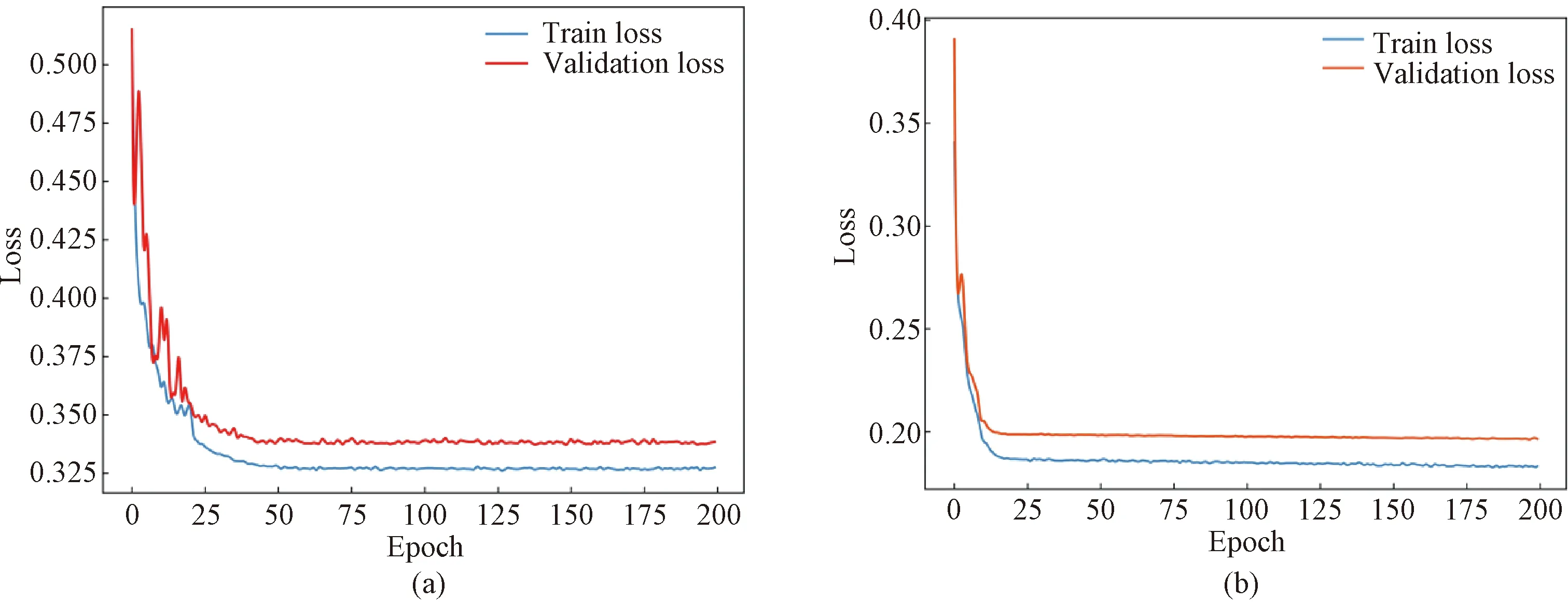
Fig.7 U-Breath model train loss and validation loss: (a) classification result; (b) segmentation result
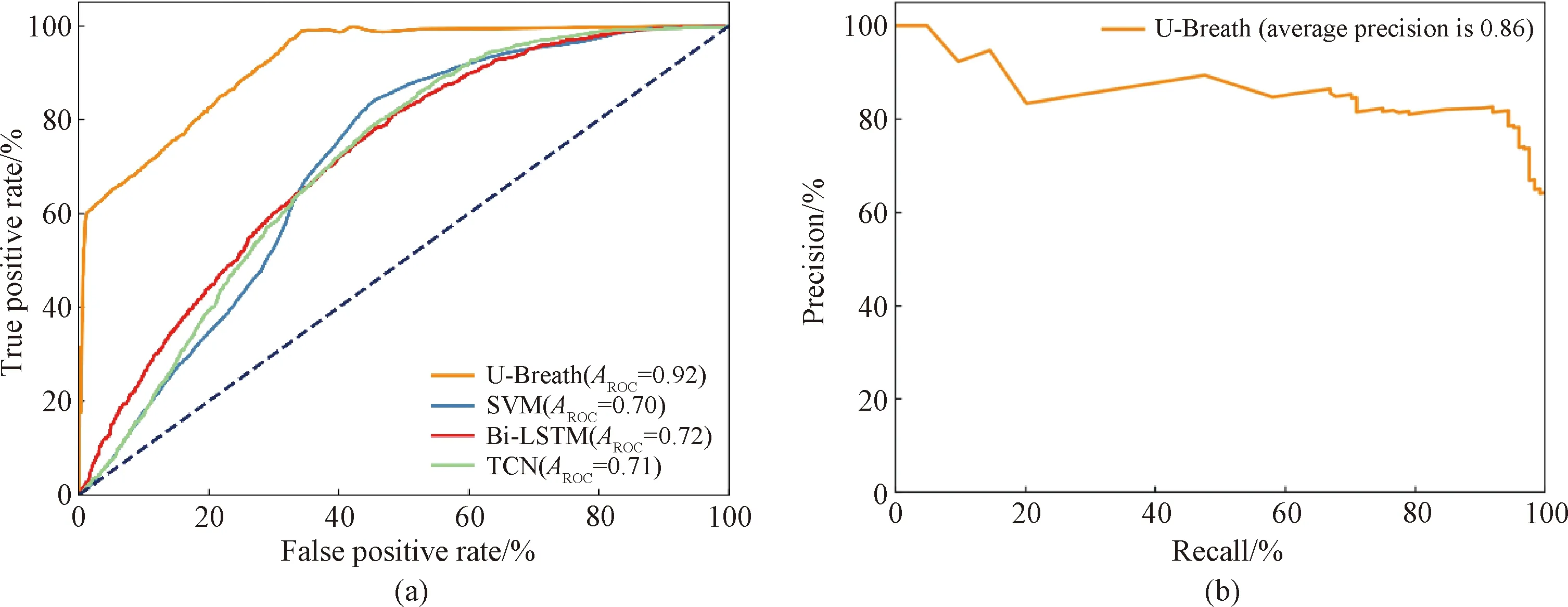
Fig.8 Evaluation metrics for different models: (a) ROC curves for different classification models; (b) precision-recall (PR) curve for U-Breath model
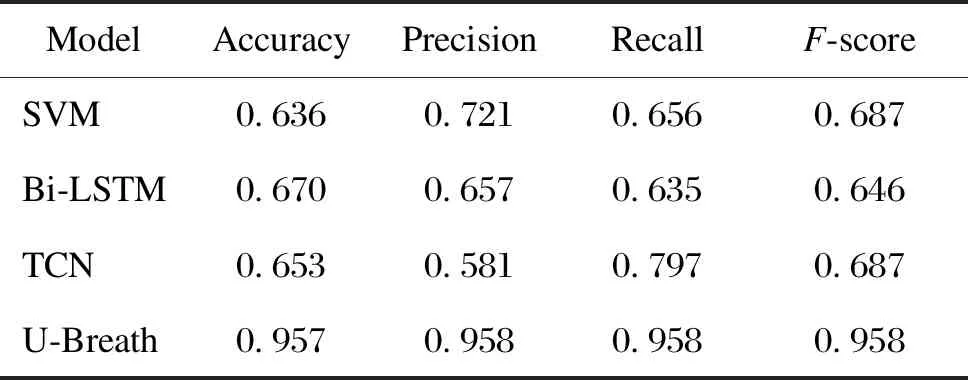
Table 2 Evaluation metrics for different models
2.2 Segmentation
In the segmentation model, we refer to U-Net to set our experimental parameters and the mini-batch method is also used to gradient down 64 samples each time. Then we use the Adam optimizer to update the gradients. The learning rate is 0.001, and the total number of training epochs is 200. In addition, dice coefficient, a widely used loss function in image segmentation, can evaluate the similarity between the prediction set and the real set. The calculation formula of the dice coefficient and the dice doss are given by
(2)
ld=1-cd,
(3)
whereMpis the prediction matrix,Mtis the real matrix,cdrepresents the dice coefficient, andldrepresents the dice loss. If the similarity between two sets is higher, the dice coefficient is closer to 1, while the dice loss is closer to 0.
In the previous classifier, there are 10 541 positive samples, that is, samples with sleep apnea. These samples are sent to the segmentation network, and the sample data are divided into the training set, the validation set and the test set according to the same division method as the classification network is used.
After training 200 epochs, the model converges, and Fig.7(b) shows that there is no over fitting or under fitting happened in the verification set and the test set. The average dice coefficient of the test set is 0.818, which shows that the network achieves good results in the task of BCG signal apnea segmentation. Figure 8(b) shows the PR curve of the segmentation model, which is used for performance evaluation in scenarios with unbalanced samples. Finally, the U-Breath segmentation model proposed in this paper achieves an average precision score of 0.86.
2.3 Channel fusion
In the channel fusion experiments, we evaluate the performance of various channel fusion methods by combining the channel fusion methods with the classification and segmentation networks described above. In this experiment, three channel fusion methods, namely LR, average and SENet, are compared with the manual selection methods.
In the combination experiments with the classification network, we keep the original parameters of the classification network unchanged and only feed the single channel signal into the classification network after the channel selection or fusion process for validation. For SENet, in the experiments, we also verify the performance of the network with different squeeze layersc(SE-c). According to Ref. [10], we verify the network performance whenctakes five parameters of 1, 2, 4, 8 and 16.
Tables 3 and 4 show the experimental results of the U-Breath model after adding different channel fusion layers. Here, for the convenience of comparison, the experimental results of the manual channel selection approach are also added in Tables 3 and 4. Similarly, we also combine the channel fusion layer and segmentation network for the experiment, and the experimental results are shown in Tables 5 and 6.
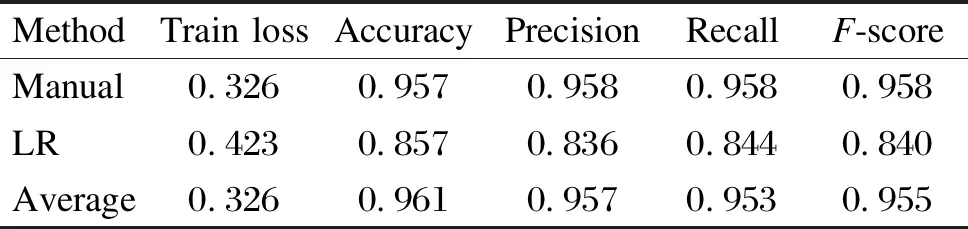
Table 3 Evaluation metrics for the combination of classification networks and different channel fusion methods
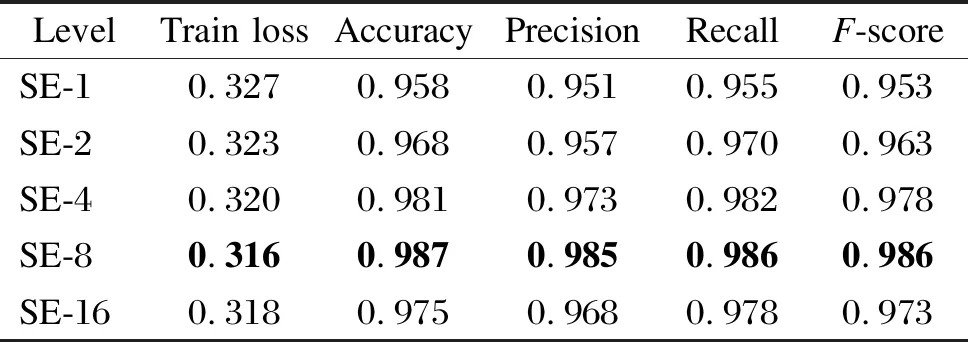
Table 4 Evaluation metrics for the combination of classification networks and SENet under different squeeze level
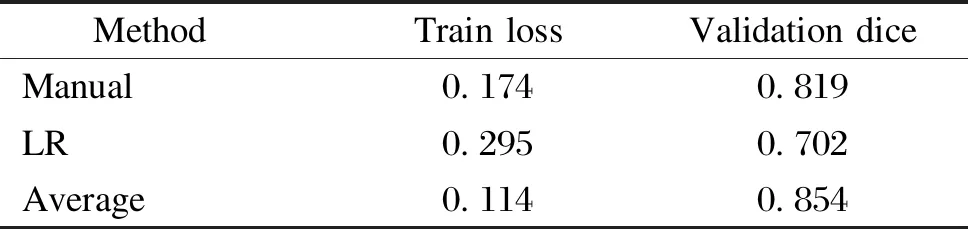
Table 5 Evaluation metrics for the combination of segmentation networks and different channel fusion methods
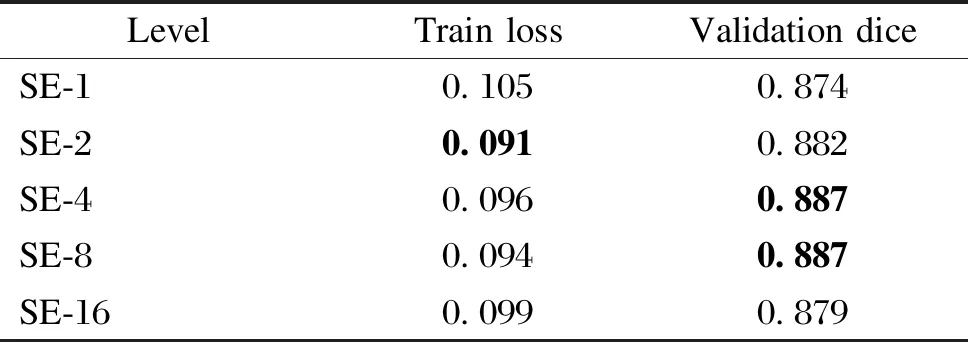
Table 6 Evaluation metrics for the combination of segmentation networks and SENet under different squeeze level
3 Discussion
From the experimental results and evaluation metrics of the classification network, there are obvious differences between the various methods. From a macro point of view, the U-Breath model proposed in this paper is better than other models on the whole, which indicates that the U-Breath model shows excellent classification ability that can accurately judge whether apnea happens or not over a window of time. However, the above approach using manual channel selection requires manual comparison of the data distribution of each channel, which is almost difficult to achieve for home monitoring scenarios. Therefore, although this manual intervention method of channel selection is still accurate, it does not overcome the shortcomings of traditional PSG and ECG methods and lacks practical value.
As shown in Tables 3 and 4, after adding the automatic channel fusion layer, for the U-Breath network proposed in this paper, the channel fusion layer using both the average method and SENet has improved the evaluation metrics compared to the manual channel selection method. This may be due to the fact that the features of multichannel data are more fully utilized compared to those using only one channel data. Somatic motion is an important marker for the occurrence of sleep apnea, while manual channel selection is difficult to utilize somatic motion features because it tends to use data satisfying a normal distribution; whereas for SENet, it can adjust the amount of contribution of each channel, so it can utilize somatic motion features and thus improve the accuracy of sleep apnea detection. SENet which uses eight intermediate layers performs the best, and it has the best value for all evaluation metrics.
Figure 9 shows the comparison between the real sleep apnea segments labeled by PSG and the predicted sleep apnea segments labeled by the proposed U-Breath model. To be specific, the blue line represents the BCG input signal, the green shadow part is the predicted value, the pink shadow part is the real value given by PSG and the overlapping part of the two colors is the accurate prediction part. It can be seen that the difference between the real value and the predicted value is small, which indicates that the segmentation network can roughly locate the time segment of sleep apnea, but there will be some deviation in the beginning and ending parts. Compared with the problem of image segmentation, it is difficult to give accurate edge of segmentation in time series, so it is also difficult to improve the segmentation results. By visualizing test samples, the segmentation network has the ability to locate the general occurrence area of sleep apnea, and the number of sleep apnea occurrence and the general duration of sleep apnea can be obtained from the predicted results. These indicators can provide more fine-grained information for clinical diagnosis of sleep apnea and have a certain reference value for the calculation of AHI and other clinical indicators.
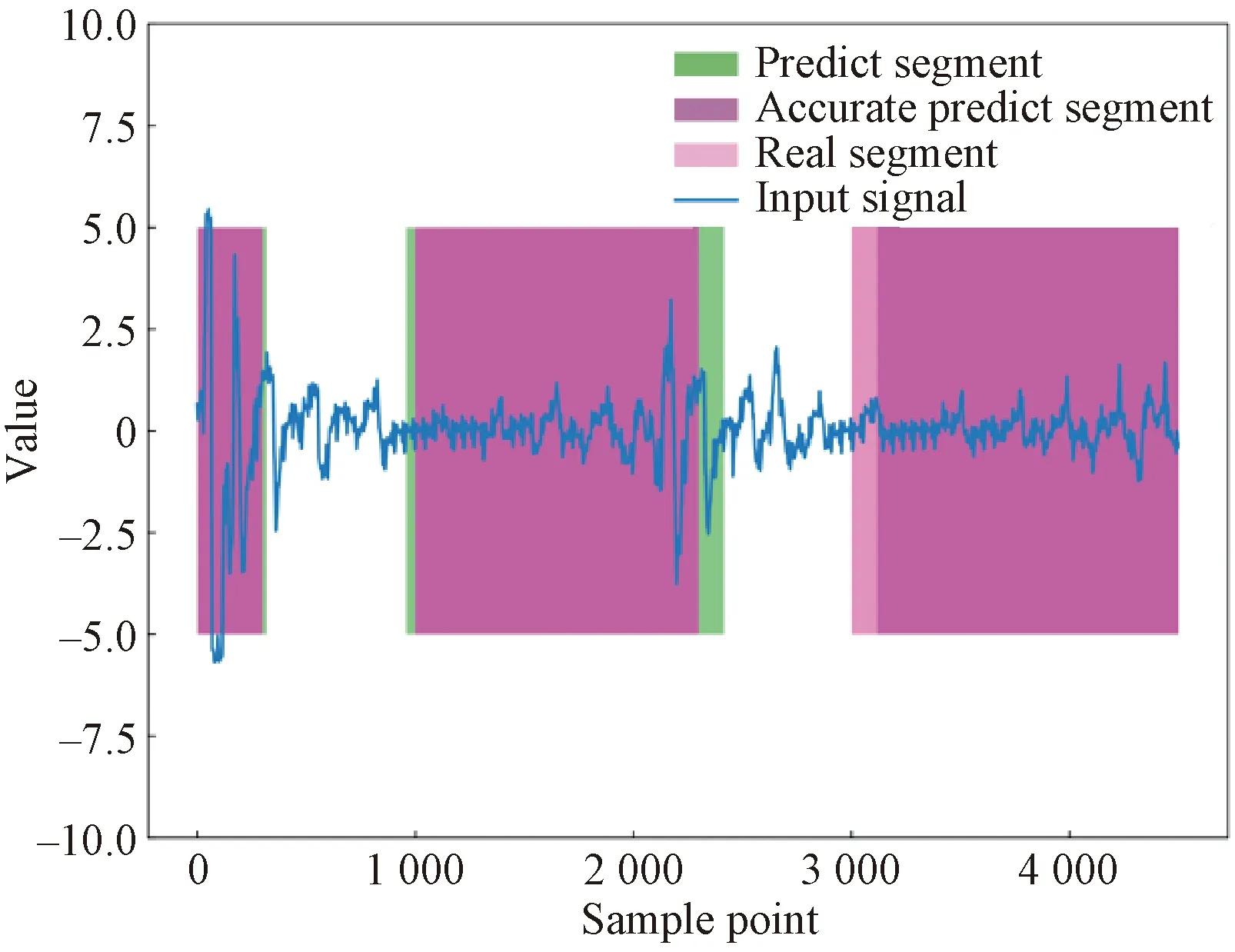
Fig.9 Comparison of real sleep apnea segments labeled by PSG and predicted apnea segments labeled by U-Breath model
4 Conclusions
This paper proposes a new model based on convolution neural network, which can well achieve the detection of sleep apnea events and the extraction of the specific time and duration of the occurrence of sleep apnea. Meanwhile, based on the problem that BCG signals are susceptible to interference, this paper uses multichannel BCG data and compares several channel fusion methods and finds that the SENet-based method can make full use of data from multiple channels and thus improve the performance of the whole model.
However, we also notice that it may take longer time to train the model and detect sleep apnea by using the multistage model, and the reasons may include the redundant calculation caused by the lack of correlation between stages. Therefore, we will focus on how to integrate the multistage model into the one-stage model or how to share the structure and parameters between stages to lighten our model in the future work.
 Journal of Donghua University(English Edition)2023年2期
Journal of Donghua University(English Edition)2023年2期
- Journal of Donghua University(English Edition)的其它文章
- Online Fault Detection Configuration on Equipment Side of a Variable-Air-Volume Air Handling Unit
- Exact Graph Pattern Matching: Applications, Progress and Prospects
- A Class of Simple Modules for Electrical Lie Algebra of Type D5
- Modified Cepstral Feature for Speech Anti-spoofing
- Data-Driven Model for Risk Assessment of Cable Fire in Utility Tunnels Using Evidential Reasoning Approach
- Proportion Integration Differentiation (PID) Control Strategy of Belt Sander Based on Fuzzy Algorithm
