Serine and arginine rich splicing factor 1: a potenti al target for neuroprotection and other diseases
Ana M.Sandoval-Castellanos ,Anushka Bhargava ,Min Zhao,Jun Xu,Ke Ning
Abstract Alternative splicing is the process of producing variably spliced mRNAs by choosing distinct combinations of splice sites within a messenger RNA precursor.This splicing enables mRNA from a single gene to synthesize different proteins,which have different cellular properties and functions and yet arise from the same single gene.A family of splicing factors,Serine-arginine rich proteins,are needed to initiate the assembly and activation of the spliceosome.Serine and arginine rich splicing factor 1,part of the arginine/serine-rich splicing factor protein family,can either activate or inhibit the splicing of mRNAs,depending on the phosphorylation status of the protein and its interaction partners.Considering that serine and arginine rich splicing factor 1 is either an activator or an inhibitor,this protein has been studied widely to identify its various roles in different diseases.Research has found that serine and arginine rich splicing factor 1 is a key target for neuroprotection,showing its promising potential use in therapeutics for neurodegenerative disorders.Furthermore,serine and arginine rich splicing factor 1 might be used to regulate cancer development and autoimmune diseases.In this review,we highlight how serine and arginine rich splicing factor 1 has been studied concerning neuroprotection.In addition,we draw attention to how serine and arginine rich splicing factor 1 is being studied in cancer and immunological disorders,as well as how serine and arginine rich splicing factor 1 acts outside the central or peripheral nervous system.
Key Words:alternative splicing;autoimmune disorders;cancer;hypertension;mRNA;neuroprotection;splicing factors;SRSF1
Introduction
In eukaryotic gene expression,splicing describes the removal of introns (noncoding regions) from messenger RNA precursors (pre-mRNAs) and is a crucial step to produce mature mRNA molecules.The alternative splicing process allows messenger RNA (mRNA) from a single gene to synthesize different variants of a protein that have different cellular properti es and/or functions for a particular cellular stage or process (Blanco and Bernabéu,2012).Alternative splicing occurs through using alternative splice sites in the premRNA,which are recognized by the spliceosome.The spliceosome catalyzes the removal of introns (the non-coding regions of a gene) to form different arrangements of exons (the coding regions of a gene) in the final mRNA.This results in the modification of the mRNA coding sequence and different combinations of the mRNA and therefore resulting in distinct translated proteins (Jakubauskienė et al.,2021;Wu et al.,2021).
For splicing to take place,a family of splicing factors named serine-arginine (SR)proteins are needed for the initi al assembly and activation of the spliceosome and play a key role in RNA metabolism (Aubol et al.,2018).A member of this family,serine-arginine splicing factor 1 (SRSF1) is essenti al for either activating or repressing splicing (post-translational modifications) of mRNA (Ghosh and Adams,2011;Paz et al.,2021).This depends on its phosphorylation state and interactive partners.SRSF1,classified under SR proteins,contains RNA recognizing motifs at the N-terminal,which governs the specificity of RNA binding,and the RS domain at the C-terminal which is rich in Arg-Ser repeats,allowing protein-protein interactions.Many serines in the RS domain of the SRSF1 protein are regio-specifically phosphorylated by the SR-specific protein kinase 1 (SRPK1) and this is vital for cytoplasmic-nuclear localization,and alternative splicing.This phosphorylation is a complex reaction and has recently been elucidated.The mechanism of phosphorylation is dependent on the binding cavity of SRPK1 which binds with high affinity to SRSF1,the Arg-Ser repeats in the RS domain,and a directional processing mechanism (Ghosh and Adams,2011;Qian and Liu,2014).
SR proteins have critical roles during development,however,due to their complex structure,and overlapping acti vities and functions,SR proteins also have a role in diseases (Wagner and Frye,2021;Yu et al.,2022).First identified as a splicing regulator,SRSF1’s phosphorylation state and interaction with a variety of proteins allow it to regulate several cellular functions,including mRNA transcription,nuclear export,nonsense-mediated mRNA decay,translation,and protein covalent attachment known as SUMOylation (Das and Krainer,2014).Furthermore,SR proteins can assist with early spliceosome assembly.This assembly event is mediated by the RNA recognition motifs(RRM) domains of SRSF1.Mutations in the RRM of SRSF1 inhibit the formation of spliceosome E complex and splicing.Furthermore,it is shown that hypo-phosphorylation or hyper-phosphorylation of the RS domain of SRSF1 induces a key molecular switch from intramolecular to intermolecular interactions,therefore suggesting a mechanism for the phosphorylation/dephosphorylation cycle during the pre-mRNA splicing (Cho et al.,2011).Moreover,Matsumoto et al.(2020) showed that the AMP-activated protein kinase-dependent phosphorylation of SRSF1 at Ser133 obstructed the binding of SRSF1 to RNA and regulated alternative splicing of pre-mRNA.In a recent study,Haward et al.(2021) identified that the transport and shuttling of SRSF1 between the nucleus and cytoplasm is essenti al for coordinating and affecting nuclear and cytoplasmic post-splicing processes like gene expression.They showed that the physiological significance of SRSF1 shuttling is used to reprogram gene expression networks regarding development and cilia function (Haward et al.,2021).Costa et al.(2021) investigated and described the interaction of transpoting3 (TNPO3),a nuclear carrier,and SRSF1 during myogenesis.They found that SRSF1 was present in the nucleus during cell differentiation into myoblasts.However,TNPO3 appeared to be initially in the nucleus,then TNPO3 was similarly expressed between cytoplasm and nucleus,and finally,TNPO3 was expressed mainly in the nucleus.This behavior encouraged the expression of muscle-specific microRNAs involved in muscular atrophy and myogenesis (Costa et al.,2021).Additionally,Yu et al.(2022) observed,in a mouse incisor model,that SRSF1 determines the alternative splicing mechanism behind the survival and proliferation of dental progenitor cells,finding that SRSF1 assists in tissue renewal.
Gene expression changes during aging are related to splicing changes due to the effects caused by the splicing factors,like SRSF1,in gene expression.Furthermore,this also has been suggested during or before the onset of disease due to its relationship with the transcriptomic and proteomic profiles(Blanco and Bernabéu,2012).Furthermore,SRSF1 can stimulate mRNA decay,a mechanism that corrupts mRNA with premature termination codons(Aznarez et al.,2018).This behavior has moti vated research to further study the effects of SRSF1 and to create therapeutic approaches using SRSF1.Castelli et al.(2017) suggested that reducing the expression levels of SRSF1 was functionally associated with the suppression of chromosome 9 open reading frame 72 (C9ORF72)-amyotrophic lateral sclerosis (ALS) neurotoxicity,encouraging the survival of motor neurons.Saito et al.(2022) showed that retinoic acid increased the phosphorylation of SRSF1,which subsequently played an important role in encouraging cell differentiation and inhibiting cell proliferation.Feng et al.(2021) showed that SRSF1 regulates alternative splicing of cyclin D1 induced by mechanical stress,resulting in abnormal cell proliferation.Additionally,Qian and Liu (2014) highlighted that SRSF1 stimulates the inclusion of exon 10 during the alternative splicing of the protein tau.Aberrant exon 10 splicing of protein tau causes Alzheimer’s disease and other related neurodegenerative disorders (Qian and Liu,2014).
In this review,we are going to highlight how the splicing factor SRSF1 has been studied regarding neuroprotection.Furthermore,we are going to show how SRSF1 is being studied in cancer and immunological disorders,as well as the importance of how SRSF1 acts outside the central or peripheral nervous system.Table 1summarizes the genes that have their expression modulated by SRSF1 and also highlights the mechanisms that may be involved.
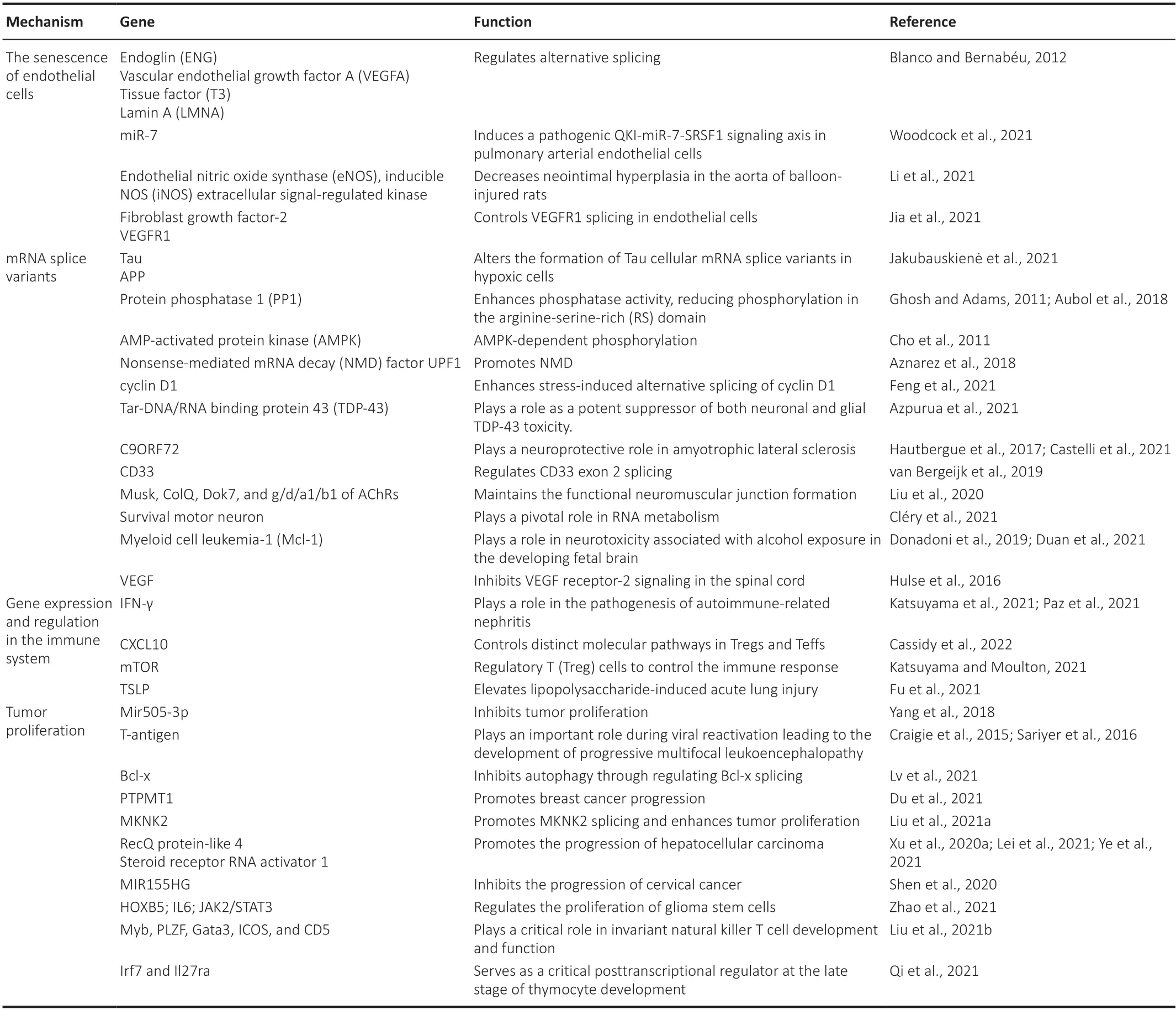
Table 1|SRSF1 modulates the expression of genes through multiple mechanisms
Search Strategy and Selection Criteria
We performed the literature review on the Google Scholar academic research engine.We used the following keywords: “SRSF1”,“SRSF1 AND neuroscience”“SRSF1 AND neuroprotection”,“SRSF1 AND neurodegeneration”,“SRSF1 AND cancer”,“SRSF1 and autoimmune diseases”,“SRSF1 AND lungs”,“SRSF1 AND Alzheimer’s disease”,“SRSF1 AND diseases”.Articles were initi ally screened inti tle and abstract;then,if they fell within the scope of our review,the articles were read in full.We searched for articles published in English,between 2015 and 2022 for research articles,and between 2005 and 2022 for articles to define SRSF1.We searched for these articles from December 2021 to June 2022.
SRSF1 Has a Strong Neuroprotective Effect
Previously,it has been shown that SRSF1 can alter the splicing of diseaserelated genes and exerts neuroprotective effects in neurodegenerative disorders,such as Alzheimer’s disease (AD) and ALS (Castelli et al.,2017,2021;Jakubauskienė et al.,2021).SRSF1 is essenti al for activating,modifying,or repressing the splicing of mRNA precursors.However,this is a complexprocess and highly depends on the splicing and the distinct variants produced,where different outcomes can be obtained.For instance,Jakubauskienė et al.(2021) studied the influence of cellular hypoxia on mRNA splice variation from AD-related Tau genes.They found that SRSF1 modifies the formation of Tau cellular mRNA splice variants in hypoxic cells and can influence disease development in AD.Additionally,Azpurua et al.(2021) screened 2700 Drosophila genes,identifying over 30 genes that suppressed locomotion phenotypes,even in aged animals.They found that SRSF1 acts as a toxicity suppressor of cytoplasmic aggregation of Tar-DNA/RNA binding protein 43,both in neurons and glial cells,in neurodegeneration disorders such as ALS.Interestingly,Castelli et al.(2017,2021) demonstrated that inhibiting SRSF1 conferred neuroprotection in C9ORF72 ALS patient-derived neurons,as the nuclear export of pathological C9ORF72-repeat transcripts was inhibited.Furthermore,they showed that the reduced levels of SRSF1 stimulated the expression of transcripts implicated in synaptic transmission,axonogenesis,and neuron differenti ation.These findings suggest that this approach could be used as a therapeutic target.
To further add to this,Hautbergue et al.(2017) showed that a decrease in SRSF1 levels avoids neurodegeneration and locomotor deficit in a Drosophila model of C9ORF72-related ALS-FTD.This is due to the inhibition of the SRSF1-dependent nuclear export of C9ORF72 repeat transcripts.Depletion of SRSF1 inhibits cell death and,when disabling its interaction with nuclear export factor 1 (NXF1),nuclear export of pathological C9ORF72 transcripts was suppressed,thus,reducing neurotoxicity in Drosophila,ALS patient-derived neurons,and neuronal cell models (Hautbergue et al.,2017).
Van Bergeijk et al.(2019) showed that SRSF1 prevents the formation of CD33 splicing isoform,which is related to the reduced risk of developing late-onset AD.A single nucleotide polymorphism in exon 2 of CD33 causes the production of high levels of mRNA that lack exon 2 and is related to the reduced predisposition to late-onset AD.In comparison to full-length CD33,CD33 lacking exon 2 forms a CD33 protein that inhibits activation responses in microglia (van Bergeijk et al.,2019).The findings by van Bergeijk et al.(2019)showed that PTBP1 and SRSF1 behave as splicing enhancers to elevate CD33 exon 2 inclusion in the mRNA,which may relate to the risk of developing lateonset AD.Furthermore,the SRSF1 binding sequence at the 3’ end of exon 2 allows CD33 exon 2 inclusion into the mRNA,showing that SRSF1 encourages full-length isoform expression,highlighting the signaling regulation in microglia,underlying the AD genetic relationship (van Bergeijk et al.,2019).The findings implied that SRSF1 could be a therapeutic target for AD.Using an anti sense oligonucleoti de or small molecule to block SRSF1 from binding to the CD33 exon 2 pre-mRNA might be a potenti al therapeutic implementation.
Myosatellite cells,also known as satellite cells,are the predominant muscle multipotent stem cells that provide myonuclei for myofiber growth and synaptic-specific gene expression in the course of postnatal development(Liu et al.,2020).Liu et al.(2020) found that myoblasts express SRSF1 at high levels and that its expression is linked to the activation and proliferation of satellite cells.They showed that when SRSF1 is deleted in myogenic progenitors,satellite cell proliferation does not occur.Moreover,SRSF1 is critical for the formation of the functional neuromuscular junction.Not only does it stimulate cell proliferation,but also contributes to muscle innervation.In mice deficient SRSF1 model,mice failed to form mature neuromuscular junction,leading to muscle weakness and premature death (Liu et al.,2020).It was reported that SRSF1 is essenti al for neuromuscular junction formation and function,and its deletion can cause serious consequences such as muscular dystrophy and peripheral neuropathies,highlighting the unique role of SRSF1 in postnatal skeletal muscle growth and function.
Following on from highlighting the unique roles of SRSF1,Cléry et al.(2021) shed fresh light on the mechanism of action of SRSF1 in cells.They showed that the inter-RRM linker of SRSF1 allows for a bimodal mode of interaction with RNA and might be used as a therapeutic approach to treat spinal muscular atrophy.Spinal muscular atrophy is caused by a mutation or deletion in the survival of the motor neuron 1 gene (Cléry et al.,2021).Furthermore,Donadoni et al.(2019) showed that alcohol exposure can cause downregulation of SRSF1,which then subsequently leads to the missplicing of the pre-mRNA of myeloid cell leukemia-1 (Mcl-1) (a pro-survival member of the Bcl-2 family).Their data suggested that immature neurons and neural progenitors are susceptible to the ethanol-associated toxicity and downregulation of SRSF1,and the following alternative splicing of Mcl-1 has an essenti al role in ethanol neurotoxicity.
Yang et al.(2018) studied that SRSF1 self-binding mechanism inhibits Mir505-3p,hindering its ability to impede the proliferation of neuronal tumor cell lines.They showed that the environment in which the tumor is growing (in this study by serum-rich or serum-reduced environment) influences SRSF1 interactions with Mir505-3p,suggesting that these conditions should be taken into account while developing anticancer therapies in the neural system.
Hulse et al.(2016) studied that the controlled alternative splicing of premRNA through SRSF1 could be used for the development of analgesics for neuropathic pain.They observed that high expression of anti-nociceptive VEGF-Axxxb isoform was related to a decrease in neuropathic pain.Furthermore,the VEGF-A165a isoform was associated with hyperalgesia,while the VEGF-A165b isoform was linked to anti -nociception.These results suggested that pre-mRNA splicing could be modulated for the treatment of chronic neuropathic pain.
Sariyer et al.(2016) studied the interaction of Pur-alpha and SRSF1 in the development of progressive multifocal leukoencephalopathy (PML).PML is a rare and critical demyelinating disease of the brain and spinal cord caused by the human polyomavirus,JC virus (JCV).Patients with HIV and those using immunosuppressive monoclonal anti body therapies may develop this disease(Sariyer et al.,2016).Sariyer et al.(2016) observed the overexpression of SRSF1 in glial cells suppressed JCV gene expression and viral replication.Furthermore,they identi fy that Pur-alpha (an important regulatory partner)enhances JCV gene expression and viral replication while suppressing SRSF1 at mRNA and protein levels in glial cells.Their observations suggested that an interactive behavior exists between SRSF1 and Pur-alpha and is suggestive of their role during viral reactivation leading to the development of PML.This mechanism might be considered when designing novel treatments for PML (Sariyer et al.,2016).In addition,Craigie et al.(2015) showed that T-antigen,a protein encoded by JCV,stimulated JCV gene expression while suppressing SRSF1 in glial cells.This further suggests a novel mechanism for the development of PML treatments.
These multiple studies on SRSF1 and its relation to neurodegenerative diseases suggest the potenti al of targeting splicing factors to understand the relationship between gene expression and AD and Parkinson’s disease (La Cognata et al.,2015).In the case of Parkinson’s disease,a high expression of nuclear paraspeckle assembly transcript 1 has been partially identified for Parkinson’s disease,indicating the role of SRSF1 as a downstream product of Parkinson’s-related genes (Zhang et al.,2020).
SRSF1 Has a Regulative Effect on Cancer Development
Srsf1 has recently been discovered to be an oncogene that is ti ghtly regulated for normal physiology.SRSF1 has various effects on the development of cancer.Research has been done to study how SRSF1 affects cancer development and if these discoveries may help develop anticancer therapies.SRSF1 has been found to inhibit autophagy by regulating Bcl-x splicing,suggesting a therapeutic role in tumorigenesis (Lv et al.,2021).However,SRSF1 regulates the aberrant alternative splicing of PTPMT1,stimulating breast cancer progression (Du et al.,2021).Moreover,improved SRSF1 phosphorylation and nucleus translocation resulted in the alternative splicing of the MKNK2 gene,enhancing colon adenocarcinoma proliferation (Liu et al.,2021a).It is interesting to see that depending on the nature of cancer,SRSF1 can act differently,obtaining a plethora of outcomes that can decrease or induce cancer progression.Hence,the importance of studying its role is in all the stages of cancer development.
Ye et al.(2021) studied that SRSF1 binds to RecQ protein-like 4,improving its stability and stimulating the migration,proliferation,and invasion of hepatocellular carcinoma.Furthermore,Zhang et al.(2021) showed that when LINC01296 binds to SRSF1,proliferation,and migration of oral squamous cell carcinoma cells were accelerated.Therefore,LINC01296 binding to SRSF1 promotes tumor formation.
Duan et al.(2021) showed that long noncoding RNAs DiGeorge syndrome critical region gene 5 promotes the formation of esophageal squamous cell carcinoma via SRSF1-alternative splicing of the Mcl-1 gene.In addition,Shen et al.(2020) evaluated that among long noncoding RNAs,which are differentially expressed in healthy tissue and cervical cancer,MIR155HG is upregulated in cervical cancer tissue,enhancing proliferation and invasion of cancerous cells.They demonstrated that MIR155HG knockdown decreased cell invasion by binding to SRSF1 (Shen et al.,2020).Therefore,the progression of cervical cancer could be inhibited by regulating the binding of MIR155HG to SRSF1.Furthermore,Xu et al.(2020a) highlighted the importance of long non-coding RNA (lncRNA) in tumorigenesis and metastasis in hepatocellular carcinoma,as LINC02580 is a metastasis suppressor when bonded to SRSF1.
Furthermore,Lei et al.(2021) found upregulated expression of SRSF1 and steroid receptor RNA activator 1-long isoform (SRA1-L) in metastatic hepatocarcinoma.In vitro,overexpression of SRSF1 and SRA1-L stimulated hepatocarcinoma migration,whereas the short isoform (SRA1-S) reversed SRA1-L and SRSF1 effects (Lei et al.,2021).Furthermore,Zhao et al.(2021)investigated the expression of SRSF1 during glioma proliferation.They found that HOXB5 regulates SRSF1 expression and that SRSF1 could bind to circATP5B and stimulate the proliferation of glioma stem cells.This provides a novel biomarker for glioma diagnosis and can be used as a potenti al tool for prognostic evaluation (Zhao et al.,2021).
Interestingly,Anczuków et al.(2015) identified SRSF1 binding sites and their effects.If SRSF1 bound close to the 3’ splice site,exon inclusion or exon skipping was promoted and if SRSF1 bound close to the 5’ splice site,exon inclusion was promoted.In addition,overexpression of CASC4-FL isoform encouraged cell proliferation and increased acinar size.Furthermore,SRSF1 has different cues for splicing activation or repression,regulating its own oncogenic role (Anczuków et al.,2015).
Dong et al.(2019) showed that the lncRNA MIR205 host gene (MIR205HG)regulated KRT17 expression,which affected migration,proliferation,and apoptotic activity in cervical cancer cells.Moreover,when MIR205HG was downregulated,KRT17 expression was decreased,but this was recovered when SRSF1 was inhibited.Therefore,MIR205HG regulated KRT17 via binding to SRSF1,highlighting a promising therapeutic approach for cervical cancer(Dong et al.,2019).
A miR-1246 probe was used to identify RNA binding proteins responsible for the enrichment of exosome miRNA in pancreatic cancer cells (Xu et al.,2020b).Xu et al.(2020b) identified and verified that SRSF1 is bound to the miR-1246 probe,concluding that SRSF1 regulates exosome miRNA enrichment,which then initi ates signaling in pancreatic cancer cells.
In contrast,Yu and Fang (2022) studied that circRPAP2,a circular noncoding RNA inhibited migration and proliferation of breast cancer cells,bothin vitroandin vivo,when bound to SRSF1 to regulate PTK2 alternative splicing.They explained that circRPAP2 binds to SRSF1,then,as the latter did not bind to PTK2 pre-mRNA,the alternate splicing of PTK2 does not occur,leading to a decrease in protein expression,inhibiting the migration and proliferation of breast cancer cells.Hence,circRPAP2 could have a therapeutic role in tumor suppression.
Additionally,Broggi et al.(2020a) analyzed clinical data and histological characteristics in patients with pancreatic cancer,finding that SRSF1 was correlated with Ki-67 expression and microvasculature presence.Furthermore,as SRSF1 regulates the alternate splicing of vascular endothelial growth factor,it was expected to find new blood vessels in the tissue samples.These results suggested the influence of SRSF1 in pancreatic cancer development.An extensive review from Lo Giudice et al.(2022) covered the clinical role of SRSF1 in cancer.
Martínez-Terroba et al.(2018) found a connection between SRSF1 and DNA ligase 1 (LIG1) in non-small cell lung cancer.When SRSF1 binds to LIG1 mRNA,the expression of LIG1 was stabilized and its translation to mTOR was improved.This means that LIG1 might be regulated with SRSF1 to reduce non-small cell lung cancer proliferation and stimulate non-small cell lung cancer apoptosis.
Chen et al.(2017) reported that SRSF1 regulation of DBF4B pre-RNA splicing is the key to tumor formation of colorectal cancer cells.Alternate splicing of DBF4B pre-mRNA can generate full-length (FL) and short (s) length variants.This alternate splicing of DBF4B exon 6 affects tumorigenesis in colorectal cancer.Isoform DBF4B-FL and SRSF1 promote the proliferation of colon cancer cellsin vitroand in mice.Hence,knockdown of DBF4B-FL inhibits the proliferation of cancerous cells.Furthermore,overexpression of DBF4B-FL protects against DNA damage caused by low levels of SRSF1.They demonstrated that alternate splicing caused by SRSF1 on DBF4B,generating DBF4B exon 6 inclusion (DBF4B-FL) could impact cancer development in colorectal cancer patients.
A glioma is an abnormal mass of glial components of the central nervous system.They are heterogeneous in nature,as they are classified into ependymomas,oligodendrogliomas,and astrocytomas,among others.Therefore,there is a need to find specific markers to identify each glioma subtype (Broggi et al.,2021b).It has been shown that SRSF1 is present in glioma tissue;hence,Broggi et al.(2021b) studied 102 cases to determine if SRSF1 could be used as a potential tool for diagnostic purposes.Using immunohistochemical assays,they evaluated the expression of SRSF1 in glioblastomas,ependymomas,oligodendrogliomas,pilocytic astrocytomas,sub-epidermal giant cell astrocytoma,and pleomorphic xanthoastrocytomas(Broggi et al.,2021b).SRSF1 was strongly expressed in 81% of glioblastoma cases,80% of sub-epidermal giant cell astrocytoma cases,71% of oligodendroglioma cases,and 75% of pleomorphic xanthoastrocytomas cases.In contrast,SRSF1 was not detected in 87% of ependymoma cases and 67% of pilocytic astrocytomas cases (Broggi et al.,2021b).Although different studies have shown that SRSF1 is upregulated in grade IV glioblastoma because SRSF1 is responsible for the alternative splicing of vascular endothelial growth factor A,generating the pro-angiogenic and anti -angiogenic isoforms (Broggi et al.,2021b).Based on these findings and from the literature,it is challenging to use SRSF1 as a histological marker for gliomas as it was not detected in all subtypes.
SRSF1 Plays a Role in the Regulation of Autoimmune Disorders
SRSF1 also plays a role in autoimmune disorders.It was found that SRSF1 acts as a post-transcriptional regulator in invariant natural killer T cell functional differentiation and development,encouraging to study of invariant natural killer T-correlated diseases (Liu et al.,2021b).Furthermore,it has been reported that SRSF1 regulates gene networks that stimulate thymocyte apoptosis,proliferation,and differenti ation (Qi et al.,2021).In addition,it has been studied how SRSF1 affected IFN-γ production and Th1 differentiation as these contribute to the pathogenesis of lupus nephritis.It was found that deletion of SRSF1 decreased Th1 differenti ation and IFN-γ production by the control of RhoH (Katsuyama et al.,2021).
Cassidy et al.(2022) studied that SRSF1 regulates the expression of 189 and 582 genes related to dysfunctional regulatory T cells and hyperactive effector T cells respectively.Furthermore,SRSF1 controls different pathways in hyperactive effector T cells,such as cytokine production,whereas it controls chemokine signaling for dysfunctional regulatory T cells (Cassidy et al.,2022).Furthermore,their findings show that aberrant SRSF1 promotes de immunopathogenesis of autoimmune diseases (Cassidy et al.,2022).Katsuyama and Moulton (2021) showed that SRSF1 is crucial for the homeostasis and proper functioning of regulatory T cells,as the absence of SRSF1 from these cells causes autoimmunity.This autoimmunity is caused by elevating mTORC1 acti vity,production of proinflammatory cytokines,and glycolytic metabolism (Katsuyama et al.,2019;Katsuyama and Moulton,2021).
In addition,low levels of SRSF1 in T cells are related to lymphopenia in systemic lupus erythematosus patients,which also correlates with low levels of anti -apoptotic Bcl-xL.Moreover,overexpression of SRSF1 encouraged T cell survival in systemic lupus erythematosus patients (Katsuyama and Moulton,2021).This finding suggests an important role of SRSF1 in treating systemic lupus erythematosus patients.
RIG-I is upregulated in psoriasis lesions,which stimulates the production of type-I interferons.Xue et al.(2015) studied if RIG-I activity was regulated by SRSF1.They found that the interaction of SRSF1 with RIG-I encouraged the production of type-I interferon triggered by cytosolic DNA.Therefore,the interaction between SRSF1 and RIG-I may be a therapeutic approach to regulating how cytosolic DNA initi ates aberrant type-I interferon production.
SRSF1 Plays Roles in Other Diseases
SRSF1 plays a very interesting role in other diseases,where SRSF1 could be targeted to control or mitigate the effects of such pathological conditions.As an example,Woodcock et al.(2021) demonstrated that increased SRSF1 and RNA-binding protein quaking were observed in patients with pulmonary arterial hypertension.Furthermore,they demonstrated that SRSF1 overexpression encouraged the migration of human pulmonary arterial endothelial cells.The increase in migration is an important cause of pulmonary arterial hypertension.Therefore,SRSF1 can be modulated to inhibit endothelial cell migration.
Sun et al.(2021) showed that a novel pulmonary fibrosis inhibitor (a long noncoding RNA (lncRNA NONMMUT060091)) binds to SRSF1,downregulating its pro-fibrotic activity.This indicates that lncRNA NONMMUT060091 significantly reduces pulmonary fibrosis by decreasing the formation of the EDA+Fn1 splicing isoform,through the negative regulation of the expression and activity of SRSF1.Additionally,Fu et al.(2021) showed that SRSF1 expression increased in lipopolysaccharide-induced acute lung injury.When treatment with SRSF1 anti bodies was performed,inflammation was reduced in the acute lung injury mouse model.SRSF1 blocking might be an alternative in the treatment of acute lung injury.
Li et al.(2021) assessed the role of valsartan in neointi mal hyperplasia and the toll-like receptor 4-nitric oxide synthase pathway in the balloon-injured rat aorta.They found that 20 mg/kg/day of valsartan decreased neointi mal hyperplasia in the aorta of balloon-injured rats by inhibiting the expression of SRSF1.Furthermore,Jia et al.(2021) showed that fibroblast growth factor-2 encouraged endothelial cell proliferation,survival,and sprouting during angiogenesis by activating the SRSF1-dependent axis,which regulates VEGRF1 alternating splicing in endothelial cells.Additionally,Sun and Hu(2020) studied the role of lncRNA HOTAIR in myocardial ischemia-reperfusion(IR).Myocardial IR limits the recovery of impaired cardiac function in acute myocardial infarction patients.This study evaluated that when HOTAIR was upregulated,miRNA-126 was downregulated in IR mice.The relationship between HOTAIR-miRNA-126-SRSF1 affects IR,where HOTAIR worsens myocardial IR by competi tively binding miRNA-126 with SRSF1.
Costa et al.(2021) found that transpoti n3 (TNPO3) shuttles SRSF1 from the cytoplasm to the nucleus.SRSF1 is involved in controlling protein diversity in muscle and satellite cell differenti ation through alternative splicing.They showed that,during myogenesis,SRSF1 remain in the nucleus,whereas TNPO3 stayed in the cytoplasm and its expression was decreased.Therefore,the interactions between TNPO3 and SRSF1 may influence the production of proteins during myogenesis.
Malakar et al.(2016) explained that peripheral insulin resistance in diabetes type 2 is caused by defective insulin signaling through the insulin receptor(INSR).Due to the skipping or inclusion of exon 11,the INSR has two isoforms:INSR-A and INSR-B respectively.Exon 11 inclusion,caused by insulin,occurs through the activation of the RAS-MAPK/ERK signaling pathway and the increased expression of SRSF1.When the exon 11 was skipped by a splice site competi tive anti sense oligonucleoti de,the MAPK/ERK signaling pathway was inhibited,sensiti zing pancreatic β-cells to lipotoxicity and apoptosis (Malakar et al.,2016).
A study revealed how SRSF1 stimulated vascular smooth muscle cell proliferation and neointima formation after injury (Xie et al.,2017).It was found that,after injury,SRSF1 expression in rat arteries improved neointi ma formation (Xie et al.,2017).Moreover,SRSF1 knockdown inhibited the migration and proliferation of human coronary arterial smooth muscle cells.These findings suggest a therapeutic use of SRSF1 for vascular hyperplastic diseases (Xie et al.,2017).
Paz et al.(2015) showed that HIV-1 replication was inhibited by the expression of SRSF1 mutants with the RRM2 domain alone or in combination with the RRM1 domain.Furthermore,this viral inhibition did not affect the cell viability of the leukocyte-derived H9 cell line.These results show promise,as they showed that SRSF1 mutants could be used to inhibit replication of multiple viral strains without affecting cell viability,in comparison to current drugs,which are not designed to target proteins from multiple viral strains (Paz et al.,2015).
Conclusion
Beyond the role of SRSF1 during development and tissue renewal,the increasing interest in SRSF1 has shown that SRSF1 can be targeted to modulate alternative splicing,highlighting the possibility of achieving different cellular functions and responses.SRSF1 has been shown to have neuroprotective effects,which could lead to the development of novel and promising therapeutics for the treatment of neurological disorders,such as ALS,Alzheimer’s disease,and Parkinson’s disease.Furthermore,SRSF1 is not only limited to the central nervous system or the peripheral nervous system,as it is found in many other tissues,such as the lungs,pancreas,and heart.Also,SRSF1 plays a significant role during tumorigenesis and the development of other diseases.Therefore,SRSF1 can also be regulated to aid in cancer,autoimmune disorders,and hypertension treatments,to mention some of the diseases that can benefit from SRSF1 regulation.It will be very interesting and insightful to see research regarding how environmental or epigenetic factors affect SRSF1 regulation as well as new approaches to therapeutics,for example,the use of viral vectors or gene therapy to modify the mechanism behind SRSF1 regulation that could change the development of a disease.
Nevertheless,further studies must be carried out when developing such therapeutics,because inhibiting or suppressing SRSF1 may have different effects depending on the molecular targets.Therefore,it is critical to conti nue further expand our understanding of the role of SRSF1 in different pathological conditions.This review highlights the potential benefits of targeting the SRSF1 splicing factor,and how this knowledge can be applied to elucidate the role of other splicing factors in disease progression.
Author contributions:AMSC and AB drafted and wrote the manuscript,and performed data collection.MZ,JX and KN helped to revise the article critically.All authors approved the final version of the manuscript.
Conflicts of interest:The authors declare no conflicts of interest.
Availability of data and materials:All data generated or analyzed during this study are included in this published article and its supplementary information files.
Open access statement:This is an open access journal,andarticles are distributed under the terms of the Creative Commons AttributionNonCommercial-ShareAlike 4.0 License,which allows others to remix,tweak,and build upon the work non-commercially,as long as appropriate credit is given and the new creations are licensed under the identical terms.
Open peer reviewers:Giuseppe Broggi,University of Catania Department of Surgical and Medical Sciences Advanced Technologies GF Ingrassia: Universita degli Studi di Catania,Italy;Toshio Nakaki,Teikyo University,Japan.
Additional file:Open peer review reports 1 and 2.
- 中国神经再生研究(英文版)的其它文章
- Bystanders or not? Microglia and lymphocytes in aging and stroke
- Alzheimer’s disease risk after COVID-19: a view from the perspective of the infecti ous hypothesis of neurodegeneration
- Can glial cells save neurons in epilepsy?
- Lights for epilepsy: can photobiomodulation reduce seizures and offer neuroprotection?
- The landscape of cognitive impairment in superoxide dismutase 1-amyotrophic lateral sclerosis
- CMT1A current gene therapy approaches and promising biomarkers

