Neuroprotective effects of long noncoding RNAs involved in ischemic postconditioning after ischemic stroke
Wei Ma, Chun-Yan Li, Si-Jia Zhang, Cheng-Hao Zang, Jin-Wei Yang, Zhen Wu, Guo-Dong Wang Jie Liu Wei Liu Kuang-Pin Liu Yu Liang Xing-Kui Zhang Jun-Jun Li Jian-Hui Guo,, Li-Yan Li
Abstract During acute reperfusion, the expression profiles of long noncoding RNAs in adult rats with focal cerebral ischemia undergo broad changes. However, whether long noncoding RNAs are involved in neuroprotective effects following focal ischemic stroke in rats remains unclear. In this study, RNA isolation and library preparation was performed for long noncoding RNA sequencing, followed by determining the coding potential of identified long noncoding RNAs and target gene prediction. Differential expression analysis, long noncoding RNA functional enrichment analysis, and co-expression network analysis were performed comparing ischemic rats with and without ischemic postconditioning rats. Rats were subjected to ischemic postconditioning via the brief and repeated occlusion of the middle cerebral artery or femoral artery. Quantitative real-time reverse transcription-polymerase chain reaction was used to detect the expression levels of differentially expressed long noncoding RNAs after ischemic postconditioning in a rat model of ischemic stroke. The results showed that ischemic postconditioning greatly affected the expression profile of long noncoding RNAs and mRNAs in the brains of rats that underwent ischemic stroke. The predicted target genes of some of the identified long noncoding RNAs (cis targets) were related to the cellular response to ischemia and stress, cytokine signal transduction, inflammation, and apoptosis signal transduction pathways. In addition, 15 significantly differentially expressed long noncoding RNAs were identified in the brains of rats subjected to ischemic postconditioning. Nine candidate long noncoding RNAs that may be related to ischemic postconditioning were identified by a long noncoding RNA expression profile and long noncoding RNA-mRNA co-expression network analysis. Expression levels were verified by quantitative real-time reverse transcription-polymerase chain reaction. These results suggested that the identified long noncoding RNAs may be involved in the neuroprotective effects associated with ischemic postconditioning following ischemic stroke. The experimental animal procedures were approved by the Animal Experiment Ethics Committee of Kunming Medical University (approval No. KMMU2018018) in January 2018.
Key Words: cerebral infarction; differential expression analysis; expression profiling; GO term; ischemic postconditioning; ischemic stroke; KEGG pathway; lncRNA; mRNA; RNA sequencing
Introduction
Ischemic stroke is a global disease characterized by high morbidity, disability, and mortality. An increasing number of young people are developing this disease (Feigin et al., 2014; Daidone et al., 2021). Currently, thrombolytic therapy has limited efficacy in the treatment of ischemic stroke, and only a small proportion of patients are eligible to receive this therapy due to the limited treatment time window of approximately 4.5 hours (Stapf and Mohr, 2002; Schellinger et al., 2004). Therefore, an urgent need exists to develop new and effective therapies for ischemic stroke.
Endogenous neuroprotection refers to the brain’s ability to respond to external damage, the efficacy of which depends on the intensity and nature of the stimulus. The aim of conditioning is to trigger endogenous protective mechanisms by alternating transient ischemia/reperfusion (IR) before, during, or after ischemia (Bernaudin et al., 1999). Ischemic postconditioning induces endogenous neuroprotection by producing transient ischemia during reperfusion (Halkos et al., 2004), includingin situischemic postconditioning (ISP) or remote ischemic postconditioning (RIP). Cerebral ISP refers to the interruption of blood flow during the early stages of cerebral IR, stimulating the brain tissue to initiate endogenous neuroprotective mechanisms and reduce reperfusion injury (Fan et al., 2017). The neuroprotective mechanisms associated with ischemic postconditioning in rats include improving cerebral blood flow, preventing cytochrome c translocation, and the activation of the protein kinase B (Akt) and phosphoinositide 3-kinase (PI3K) pathways (Li et al., 2017; Tyagi et al., 2019).
Multiple mechanisms underlying the neuroprotective effects induced by ischemic conditioning have been identified, including the regulation of neurotrophic protein expression, the enhancement of neurovascular networks, the alleviation of the inflammatory response and neuronal apoptosis, and the promotion of metabolic responses in the brain (An et al., 2015; Wang et al., 2015). However, the endogenous neuroprotective effects of ischemic postprocessing (RIP and ISP) and the signaling pathways that mediate these effects have not been fully elucidated.
Previous studies have confirmed that long noncoding RNAs (lncRNAs) represent important components at the transcriptional, post-transcriptional, and epigenetic levels, able to regulate gene expression through multiple mechanisms (Knauss and Sun, 2013; Hart and Goff, 2016; Wang et al., 2019; Di et al., 2021). Accumulating studies have shown that lncRNAs are critical genetic regulators of development and disease (Qureshi and Mehler, 2012; Schaukowitch and Kim, 2014; Briggs et al., 2015). Many lncRNAs have been identified in the rat brain, which is important for the development and function of the central nervous system (Qureshi et al., 2010; Knauss and Sun, 2013; Ng et al., 2013; van de et al., 2013; Lipovich et al., 2014). Previous studies have shown that the expression profiles of lncRNAs in adult rats subjected to focal cerebral ischemia changed extensively during acute reperfusion (Dharap et al., 2012; Zhang et al., 2016; Liu et al., 2018b). However, the expression patterns of lncRNAs and their functional roles following ischemic postconditioning in rats subjected to focal ischemic stroke are currently unknown.
The neuroprotective mechanisms of ischemic postconditioning are likely complex and intertwined, with significant crosstalk among signaling pathways. We hypothesized that lncRNA and mRNA expression profiles were significantly altered in rats who underwent ischemic stroke and received an ischemic postconditioning intervention. The aim of the current study was to establish the lncRNA and mRNA expression profiles among rats subjected to ischemic and an ischemic postconditioning intervention and identify novel lncRNAs related to stroke and ischemic postconditioning interventions.
Materials and Methods
Animals
Forty adult male Sprague-Dawley, specific pathogen-free level rats (aged 6-7 weeks, weighing 260 ± 20 g) were purchased from the Animal Department of Kunming Medical University, China [license No. SCXK (Dian) 2015-0002]. To eliminate the potential influence of estrogen and any other sex-related physiological differences, only male rats were used in this study. The study was conducted in strict accordance with the guidelines of Kunming Medical University with regard to the protection and use of experimental animals. The experimental animal procedures were approved by the Animal Experiment Ethics Committee of Kunming Medical University (approval No. KMMU2018018) in January 2018. All experiments were designed and reported according to the Animal Research: Reporting ofIn VivoExperiments (ARRIVE) guidelines. Some possible measures were taken to minimize the rats’ pain. The rats were allowed to eat and drink water freely, and the room temperature was maintained at 23 ± 1°C during the modeling process.
Animal grouping and transient middle cerebral artery occlusion model
The rats were randomly assigned to four groups as follows: sham (n= 10); IR (n= 10); ischemia + ISP (ISP;n= 10); and ischemia + RIP (RIP;n= 10).
Anesthesia was induced with 5% isoflurane (MilliporeSigma, St. Louis, MO, USA) and maintained with 2-3% isoflurane. Throughout the experiment, a rectal probe was used to monitor and maintain the animal body temperature at 37°C. The right middle cerebral artery was occluded for 120 minutes to establish the rat transient middle cerebral artery occlusion (tMCAO) model (Longa et al., 1989; Li et al., 2020). In brief, a surgical incision was made to expose the right common carotid artery, internal carotid artery, and external carotid artery. The proximal common carotid artery was then ligated, and an occlusion filament was inserted into the internal carotid artery through the common carotid artery 19-21 mm distal from the bifurcation to occlude the origin of the middle cerebral artery. After induction of ischemia, the filament was withdrawn, and the rats were placed into a cage to recover from anesthesia at room temperature, with free access to food and water. The rats were subjected to 2 hours of focal cerebral ischemia. In the sham group, rats were subjected to the same procedures without the occlusion of the middle cerebral artery.
Following 120 minutes of tMCAO, the bilateral carotid arteries were occluded using an arterial clamp for 10 seconds, followed by loosening the clamp for 30 seconds. The clamping and releasing procedure was repeated for three cycles in the ISP group. Following 120 minutes of tMCAO, the RIP group was subjected to three cycles in which the bilateral femoral arteries were occluded using an arterial clamp for 10 minutes, followed by loosening the clamp for 10 minutes. The detailed experimental procedure for postconditioning intervention is shown in the schematic diagram found inAdditional Figure 1(Li et al., 2020).
Evaluation of neurological function
The Zea Longa scoring method (Longa et al., 1989) was used to score the neurological function deficits in rats 24 hours after reperfusion in all four groups. Rats with normal neurological function received a score of 0, whereas rats with the most serious neurological deficits received a score of 3 or 4. A score of 5 was assigned to rats that died.
2,3,5-Triphenyltetrazolium chloride staining
At 24 hours after reperfusion, the rats in each group (n= 4) were anesthetized by the intraperitoneal injection of 3% pentobarbital (30 mg/kg; MilliporeSigma) and decapitated, followed by the rapid removal of brain tissues. The rat brains were cut into 2-mm-thick coronal sections and stained with 1% 2,3,5,-triphenyltetrazolium chloride (TTC) solution (MilliporeSigma) for 30 minutes at 37°C, followed by overnight immersion in 4% paraformaldehyde. Finally, the slices were transferred into saline. Six images from each group were captured using an MCID computer imaging analysis system (Image ProPlus software v.6.0, Media Cybernetics, Rockville, MD, USA) (Wang et al., 2014). For TTC staining, the observer was unaware of the treatment the rats received or the grouping.
RNA sequencing and analysis
The cerebral tissues collected from the infarct areas of three rats in each group were used for RNA sequencing. RNA was extracted, quantified, and purified, and the concentration measurement was performed using methods from our previous study (Dai et al., 2020). A 3 μg RNA sample was obtained from each of three rats in each group and subjected to RNA sequencing. The methods used for lncRNA library preparation, lncRNA sequencing, transcriptome assembly, and coding potential analysis were described in our previous study (Dai et al., 2020). In brief, total RNA was extracted from infarcted cerebral tissues using TRIzol (Invitrogen, Carlsbad, CA, USA). Ribosomal RNA was removed using an Epicentre Ribo-zeroTMrRNA Removal Kit (Illumina Inc., San Diego, CA, USA), and linear RNA in the remaining RNA was removed by RNase R treatment (Epicentre, Madison, WI, USA). The libraries were sequenced at the Novogene Bioinformatics Institute (Beijing, China) on an Illumina HiSeq 2500 (PE150) platform, and 150-bp long paired-end reads were obtained. To identify lncRNAs involved in cerebral infarction after ischemic stroke, we used Cufflinks (v2.2.0) software (http://cufflinks.cbcb.umd.edu) to divide all transcripts into different subtypes and established strict screening criteria to filter transcripts that did not have all of the typical characteristics of lncRNA (Trapnell et al., 2010). Transcripts predicted to have coding potential by any of the three following tools were removed, and those without coding potential were retained as our candidate set of lncRNAs. The Coding Potential Calculator (CPC2, 0.9-r2, http://cpc.cbi.pku.edu.cn/) (Kong et al., 2007), the Coding/Noncoding Index (CNCI, v2, http://www.bioinfo.org/software/cnci) (Sun et al., 2013) and Pfam-scan (Pfam, v1.3, http://pfam.sanger.ac.uk/) (Finn et al., 2014) were used to assess the coding potential of transcripts.
Differential expression analysis
TopHat (v2.1.1, http://tophat.cbcb.umd.edu/) and Cufflinks (v2.2.0, http://cufflinks.cbcb.umd.edu) were used to calculate the fragments per kilobase of transcript per million fragments mapped (FPKMs) of both lncRNAs and coding genes in each sample (Trapnell et al., 2012). Gene FPKMs were computed by summing the FPKMs of transcripts in each gene group. Ballgown (v3.4.0, https://bioconductor.org/biocLite.R) provides statistical routines for determining differential expression in digital transcript or gene expression data using a model based on the negative binomial distribution (Pertea et al., 2016). Transcripts with aP-adjust < 0.05 were defined as being differentially expressed. Adjustment ofP-value was performed according to the Benjamini and Hochberg method (Benjamini and Hochberg, 1995). We obtained the differentially expressed transcripts from the comparisons among groups, and then Venn Diagrams (v.1.6.20, https://cran.rstudio.com/web/packages/VennDiagram/index.htm) were used to visualize the common and unique differentially expressed transcripts among the groups.
lncRNA target gene enrichment analysis in cis role
Cisrole refers to lncRNA that acts on neighboring target genes (Ørom et al., 2010). We searched coding genes 100 kb upstream and downstream of each lncRNA and analyzed their functions (Ørom et al., 2010). Gene Ontology (GO) enrichment analysis of the genes associated with the differentially expressed lncRNAs was performed using the GOseq R package (v.release 2.12, http://www.geneontology.org/) (Gene Ontology Consortium et al., 2013). Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of differentially expressed genes was performed using KOBAS software (v.2.0, http://kobas.cbi.pku.edu.cn) (Kanehisa and Goto, 2000). Fisher’s exact test and the Chi-square test were used to select significant GO categories and KEGG pathways. The threshold for significance wasP< 0.05, and the false discovery rate was calculated to correct theP-value.
Construction of lncRNA-mRNA co-expression networks
The LncRNA-mRNA co-expression networks were constructed based on Pearson’s correlation analysis using Cytoscape software (v.3.8.0, https://cytoscape.org/) (Shannon et al., 2003). In the co-expression network, each lncRNA or mRNA corresponded to a node, and the nodes were linked by edges. LncRNAs with greater numbers of connections to mRNAs or genes were considered to have a greater degree of association, indicating their increased importance.
Quantitative real-time reverse transcription-polymerase chain reaction assay
The cerebral tissues from the infarct areas in each of the four groups (n= 3 in each group) were subjected to quantitative real-time reverse transcription-polymerase chain reaction (PCR) to determine the gene levels. In brief, total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and 1 μg RNA from each sample was reverse transcribed into complementary DNA and subjected to quantitative real-time reverse transcription-PCR. The PCR mixture included 5 μM (final concentration) primers in a total volume of 20 μL. The primer information is described inTable 1. The PCR cycles were as follows: enzyme activation at 50°C for 2 minutes, initial denaturation at 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. The relative gene expression levels were determined from three independent samples, with each sample assayed in triplicate. The four independent samples in each group were run in triplicate and analyzed using the 2-∆∆Ctmethod, as previously described (Wang et al., 2020).
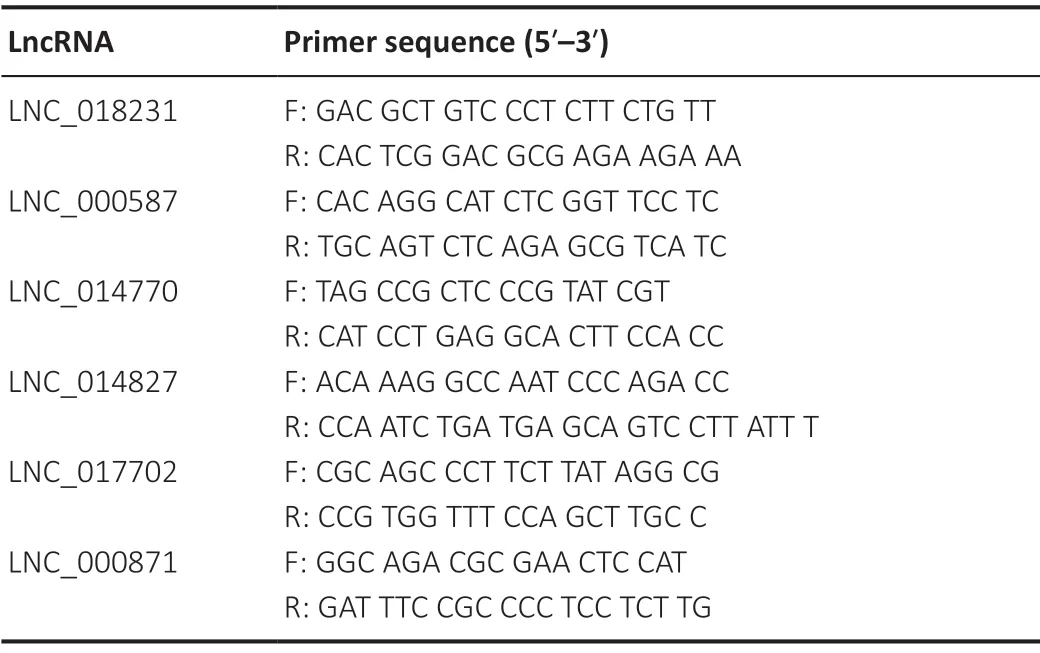
Table 1 | Primer sequences of lncRNAs
Statistical analysis
Data from each group were analyzed by one-way analysis of variance, followed by Tukey’spost hoctest, using SPSS 22.0 statistical software for Windows (IBM, Armonk, NY, USA). The data showed a normal distribution and were homogenous. Data are presented as the mean ± standard error of the mean (SEM). A value ofP< 0.05 was considered significant.
Results
ISP and RIP decrease changes to the cerebral infarction area of IR rats
TTC staining results revealed an obvious infarct area in the left cerebral hemisphere of rats in the IR group. The cerebral infarct area was reduced in the ISP and RIP groups compared with the IR group (Figure 1). No infarct area was observed in the left cerebral hemisphere in the sham group.
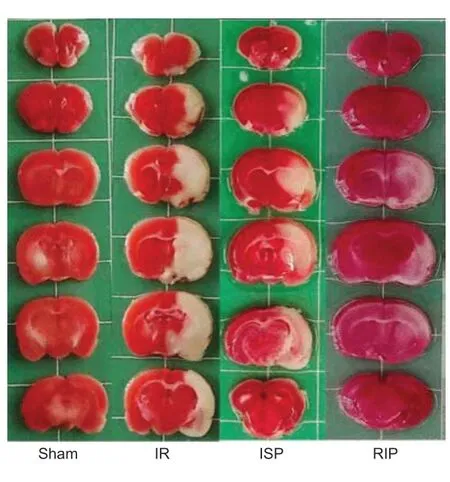
Figure 1|Representative cerebral infarct areas stained by 2,3,5-triphenyltetrazolium chloride (TTC).
ISP and RIP decrease neurological functional deficit scores in IR rats
The neurological functional deficit scores in the IR group were greater than 3, and the neurological functional deficit scores in the ISP and RIP groups were significantly decreased compared with those of the IR group (ISP group:P< 0.05, RIP group:P< 0.01). Rats in the sham group had a score of 0 (Additional Figure 2).
Identification of lncRNAs after ischemic postconditioning in ischemic stroke rats
After screening, 46,101 recognized transcripts were identified as known reference transcripts (Figure 2AandB). The coding potential of novel long transcripts was calculated using coding potential assessment tools (CPC2, CNCI, and Pfam), and potential protein-coding transcripts were removed. Finally, 29,542 novel long transcripts were retained after analyzing the overlap among CPC2, CNCI, and Pfam (Figure 2C). Among these new lncRNAs, 49.0% were long intervening noncoding RNAs, 42.2% were intronic lncRNAs, and 8.6% were antisense lncRNAs (Figure 2D). The lncRNAs identified by the three encoding potential analysis tools (CPC2, CNCI, and Pfam) were used as candidate lncRNAs for subsequent analysis.
Differential expression analysis of lncRNAs and mRNAs after ischemic postconditioning in ischemic stroke rats
Ischemic stroke and ischemic postconditioning significantly altered cerebral lncRNA expression profiles. Compared with the sham group, 2616 lncRNAs were significantly altered in the IR group, including 1421 upregulated lncRNAs and 1195 downregulated lncRNAs. Compared with the sham group, 3447 lncRNAs were significantly altered in the ISP group, including 1429 that were upregulated and 2018 that were downregulated. Compared with the sham group, 873 lncRNAs in the RIP group showed significant changes, including 488 upregulated lncRNAs and 385 downregulated lncRNAs. Compared with the IR group, 68 lncRNAs were significantly altered in the ISP group, including 28 that were upregulated and 40 that were downregulated. Compared with the IR group, 128 lncRNAs in the RIP group showed significant changes, including 46 upregulated lncRNAs and 82 downregulated lncRNAs. Compared with the ISP group, 172 lncRNAs in the RIP group showed significant changes, including 87 upregulated lncRNAs and 85 downregulated lncRNAs (Figure 3).
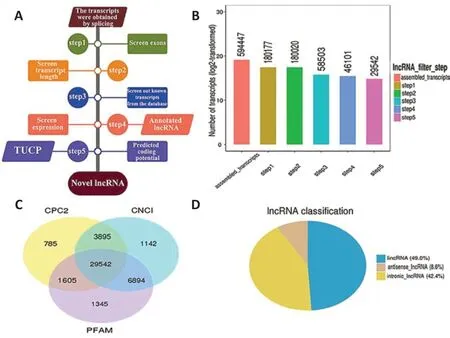
Figure 2|RNA sequencing experiments and bioinformatics analysis of cerebral infarct areas in rats.

Figure 3|Volcano plots of differentially expressed long noncoding RNAs (lncRNAs) in each group after ischemic stroke.
Among the differentially expressed mRNAs, significant changes were found in the expression of 2825 mRNAs in the IR group compared with the sham group, including 2144 upregulated mRNAs and 681 downregulated mRNAs. Compared with the sham group, the expression of 3126 mRNAs was altered in the ISP group, including 2155 upregulated mRNAs and 971 downregulated mRNAs. Compared with the sham group, the expression of 890 mRNAs was altered in the RIP group, including 766 upregulated mRNAs and 124 downregulated mRNAs. Compared with the IR group, the expression of 36 mRNAs was altered in the ISP group, including 14 upregulated mRNAs and 22 downregulated mRNAs. Compared with the IR group, the expression of 89 mRNAs was altered in the RIP group, including 36 upregulated mRNAs and 53 downregulated mRNAs. Compared with the ISP group, the expression of 109 mRNAs was altered in the RIP group, including 53 upregulated mRNAs and 56 downregulated mRNAs (Figure 4).
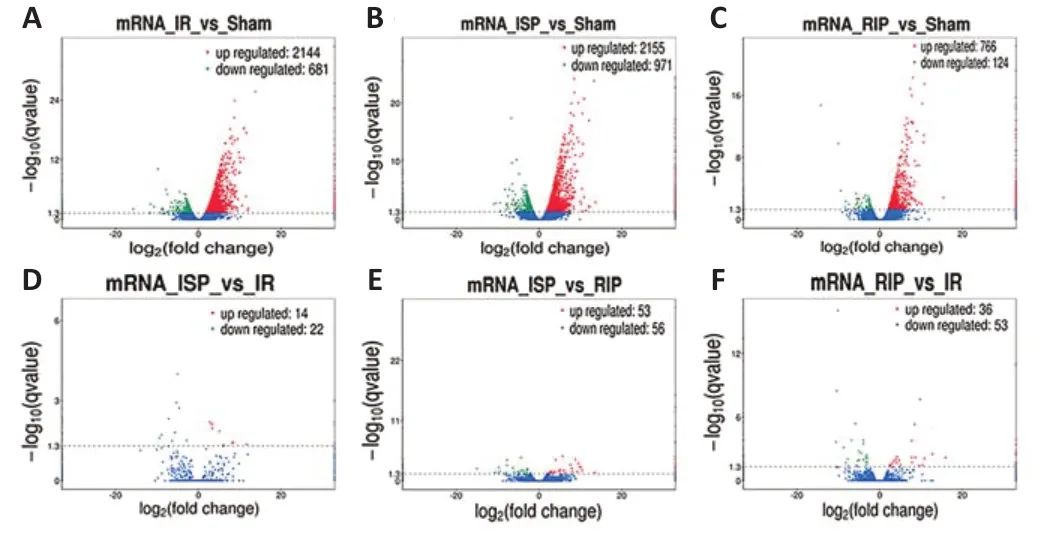
Figure 4|Volcano plots of differentially expressed mRNAs in each group after ischemic stroke.
Hierarchical clustering revealed systematic variations in the expression of lncRNAs and mRNAs in the ischemic cerebral tissue among the different groups. The lncRNA expression profiles of different samples within the same group were similar (Figure 5AandB), indicating that altered lncRNAs within the same group may participate in similar biological processes. We also found that the number of upregulated lncRNAs in the IR group was greater than the number of downregulated lncRNAs compared with the sham group. Compared with the IR group, the numbers of downregulated lncRNAs in the RIP and ISP groups were greater than the numbers of upregulated lncRNAs. The five most upregulated and downregulated lncRNAs in each group are shown inTable 2.
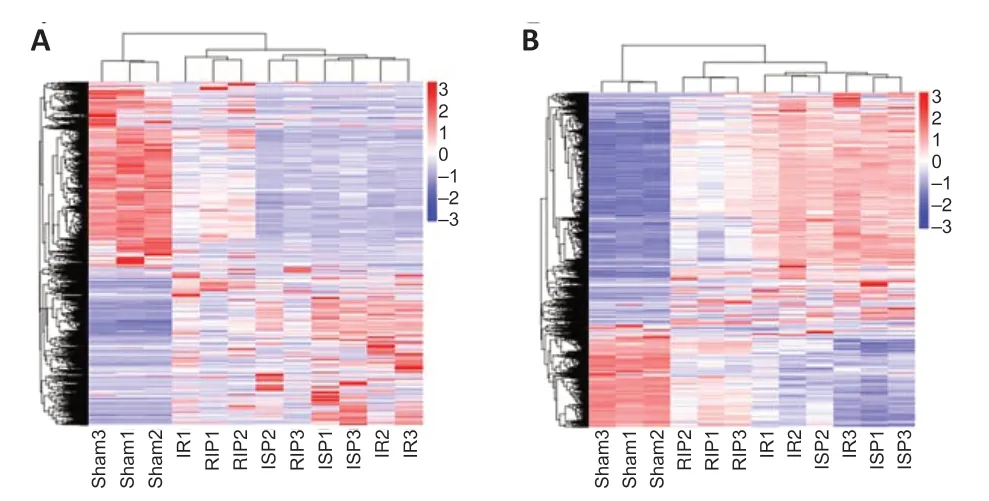
Figure 5|Hierarchical cluster analysis of differentially expressed long noncoding RNAs (lncRNAs) (A) and mRNAs (B).
Compared with the sham group, 665 differentially expressed lncRNAs and 407 mRNAs were identified in the IR group, which suggested extensive changes in lncRNA expression following cerebral IR injury. We identified 15 differentially expressed lncRNAs and 8 mRNAs that were identified in both the ISP and RIP groups when compared with the IR group (Figure 6AandB). We speculate that these 15 lncRNAs may play an important role in ischemic postconditioning. There are 51 common differentialy expressed lncRNAs in the IR vs. sham and RIPvs. IR groups. There are 38 and 2774 unique differentialy expressed lncRNAs in the RIPvs. IR groups and IRvs. sham groups respectively (Figure 6C).
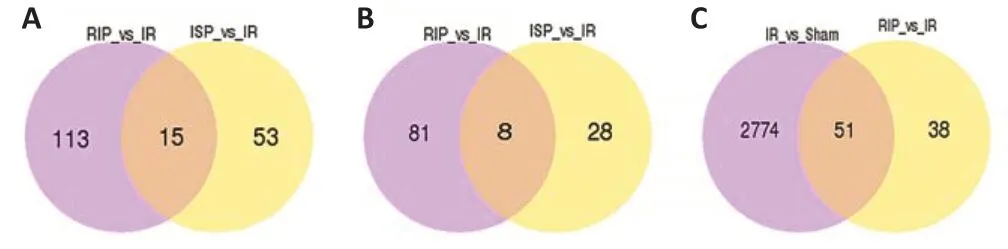
Figure 6|The number of common and unique differentially expressed transcripts among four groups.
The target gene enrichment analyses of differentially expressed lncRNAs incisroles
We examined protein-coding genes located 100 kb upstream and downstream of lncRNAs and performed functional enrichment analysis on identified potential mRNA targets to predict the primary functions of identified lncRNAs. The mRNAs identified as potential lncRNA targets within a distance of 100 kb are shown for each group inAdditional Table 1.
GO analysis was used to predict the functions of potential genes targeted by differentially expressed lncRNAs incisroles after ischemic stroke. The 20 most significantly enriched GO terms for the upregulated and downregulated lncRNAs in each group were related to the cellular components, biological processes, and molecular functions displayed inFigure 7. We conducted pathway analysis using the KEGG database. We identified 178, 155, 200, and 222 KEGG pathways in the IRvs. sham, ISPvs. IR, RIPvs. IR, and ISPvs. RIP comparisons, respectively (Additional Tables 2-5). The 20 KEGG pathways most associated with upregulated and downregulated lncRNAs after ischemic conditioning in ischemic stroke rats are shown inFigure 8. Significantly upregulated mRNAs were related to chemokines, mitogenactivated protein kinases, and tetrahydrofolate, whereas the significantly downregulated mRNAs after ischemic injury were involved in the synaptic vesicle cycle, cell adhesion molecules, inositol phosphate metabolism, and the AMPactivated protein kinase and Wnt signaling pathways, among others. After ischemic postconditioning, the significantly upregulated mRNAs were involved in metabolic pathways, the PI3K/Akt pathway, and the cell cycle, among others, whereas the significantly downregulated mRNAs were involved in the Notch, transforming growth factor-beta, and AMP-activated protein kinase signaling pathways.
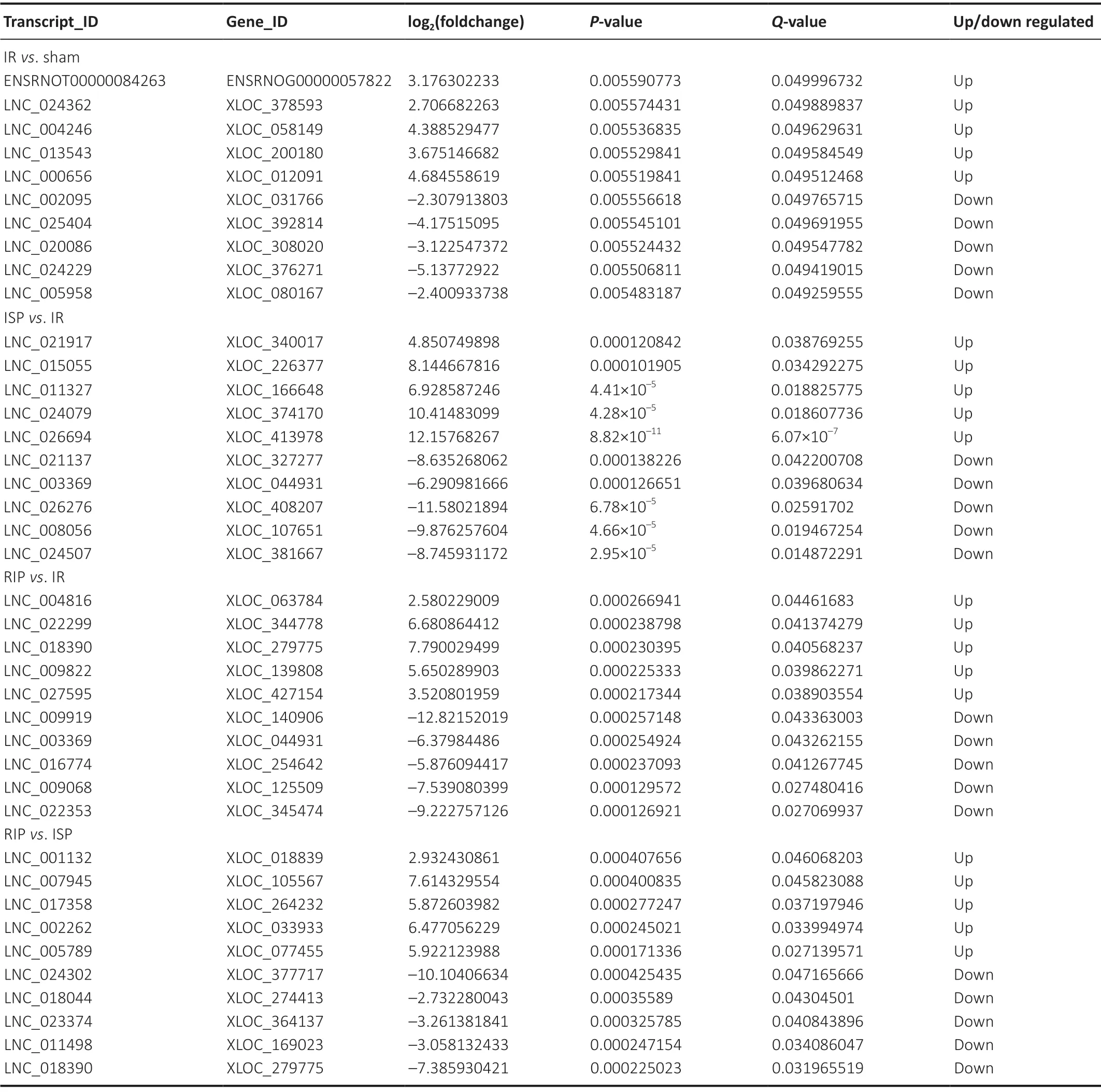
Table 2 |The five most upregulated and downregulated lncRNAs in cerebral tissue of IR rats treated with RIP and ISP
Construction of an LncRNA-mRNA co-expression network after ischemic postconditioning in ischemic stroke rats
According to the correlation between differentially expressed lncRNAs and theircis-acting target mRNAs, lncRNA and mRNA co-expression networks were constructed to identify the underlying molecular mechanisms of differentially expressed lncRNAs in ischemic stroke. We obtained 2616, 68, 128, and 172 differentially expressed lncRNAs and 2825, 36, 89, and 109cis-targeted mRNAs in the IRvs. sham, ISPvs. IR, RIPvs. IR, and RIPvs. ISP groups, respectively. We further focused on 15 lncRNAs that were significantly differentially expressed between the ISP and IR groups and between the RIP and IR groups for the co-expression network analysis (Figure 9). LncRNA 003369 targeted 15 mRNAs; lncRNA 000857 targeted 12 mRNAs; lncRNA 017702 targeted nine mRNAs; lncRNA 014770 targeted five mRNAs; lncRNA 000164 targeted seven mRNAs; lncRNA 023605 targeted eight mRNAs; lncRNA 023605 targeted eight mRNAs; lncRNAs 026276, 010495, and 018231 each targeted two mRNA; and lncRNAs 014827, 015397, and 000871 each targeted one mRNA.
Validation of differentially expressed lncRNAs after ischemic postconditioning in ischemic stroke rats
A total of 15 differentially expressed lncRNAs were identified in the comparison between the ISPvs. IR and RIP vs. IR groups (Figure 6A). Of these, we selected six lncRNAs that were significantly upregulated or downregulated in the post-ischemic adaptation groups (ISPvs. IR and RIPvs. IR groups) for verification using quantitative real-time reverse transcription-PCR. The overall expression levels of lncRNAs 018231, 000587, 014770, 014827, 017702, and 000871 were significantly downregulated in the IR group compared with those in the sham group (P< 0.01 inFigure 10A-C,E, andF;P> 0.05 inFigure 10D). The overall expression levels of lncRNAs 018231, 000587, 014770, 014827, 017702, and 000871 were significantly upregulated in the RIP and ISP groups compared with those in the IR group (P< 0.01 inFigure 10A-C,EandF;P< 0.05 inFigure 10D), consistent with the RNA sequencing data. The network suggests that these differentially expressed lncRNAs may regulate their corresponding target mRNAs during both ischemic injury and ischemic postconditioning.
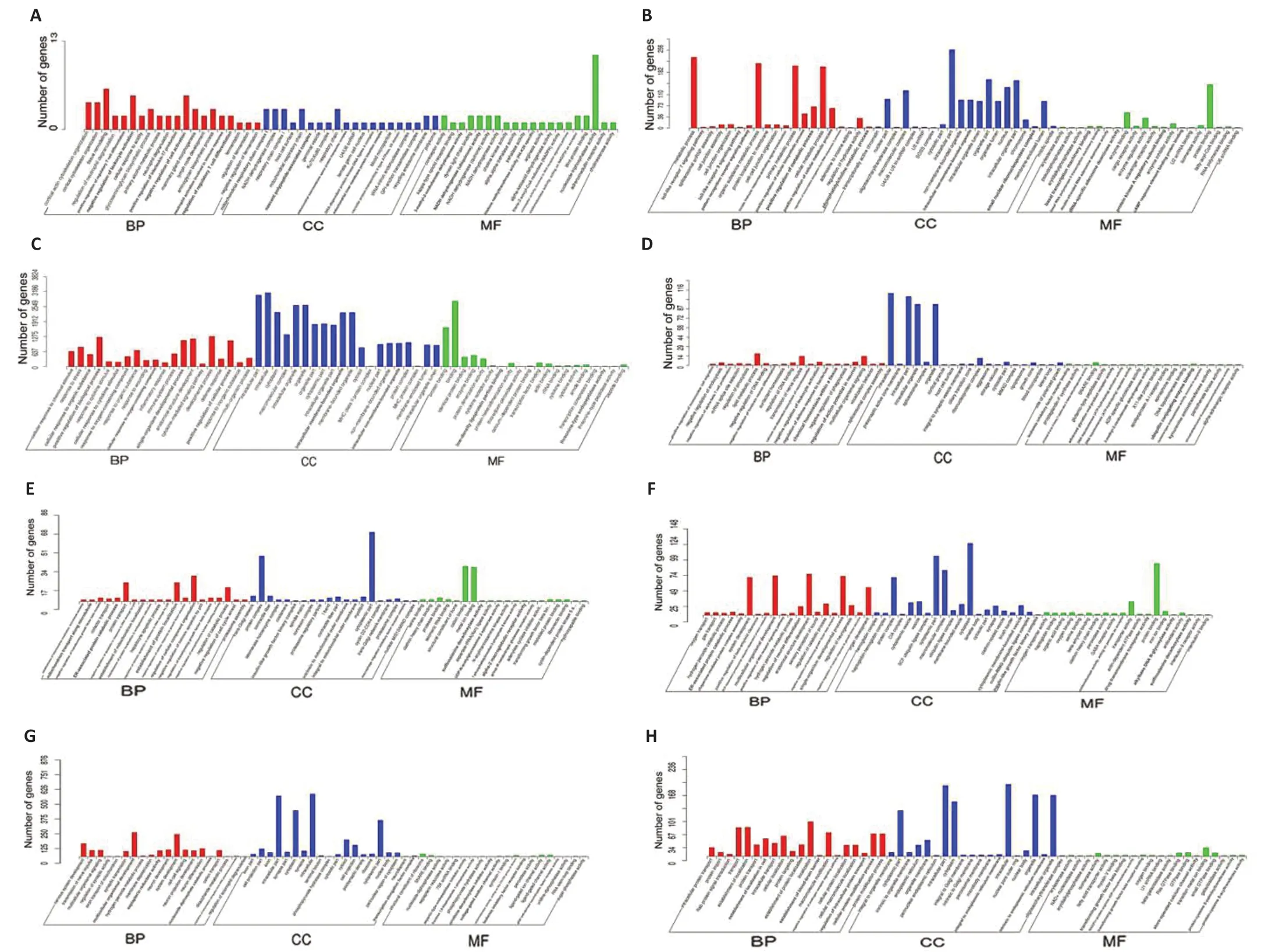
Figure 7|Gene Ontology (GO) analysis of significantly upregulated and downregulated mRNAs in the brain of IR rat with ISP and RIP treatment.

Figure 8|Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of differentially expressed mRNAs in cerebral tissue of IR rats with ISP and RIP treatment.

Figure 9|Co-expression networks of all differentially expressed long noncoding RNAs (lncRNAs) and mRNAs in the cerebral tissue of IR rats with ISP and RIP treatment.
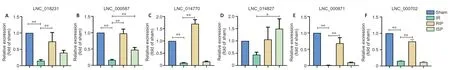
Figure 10| Validation of candidate long noncoding RNAs (lncRNAs) in the brain tissue of IR rats with ISP and RIP treatment.
Discussion
Previous studies have suggested that ischemic postconditioning has cerebral protective effects in various cerebral ischemic stroke models (Zhao, 2009; Xie et al., 2018; Li et al., 2021). Ischemic postconditioning can effectively reduce the size of the infarct area and cerebral edema, improve cerebral circulation, and relieve inflammation, reperfusion injury, and neural cell apoptosis (Liu et al., 2014a, b). Recent studies have shown that ischemic postconditioning is associated with many neuroprotective mechanisms, such as regulating the expression of neurotrophic factors (Ramagiri and Taliyan, 2017), neurovascular network-based ischemic tolerance (Lehotský et al., 2009; Deng et al., 2014), subcellular organelle-based ischemic tolerance (Lai et al., 2014; Pignataro et al., 2014), synaptic signaling-based ischemic tolerance (Bu et al., 2014; Jiang et al., 2015), protein degradation systemsbased ischemic tolerance (Della-Morte et al., 2012; Narayanan et al., 2013), and the regulation of anti-inflammatory, antiapoptotic, and anti-oxidative pathways (Dharap et al., 2012; Wei et al., 2016). These studies demonstrate that the neuroprotective mechanisms of ischemic postconditioning are multifaceted (Zhao, 2009; Xie et al., 2018; Li et al., 2021). However, systematic research exploring the neuroprotective mechanisms underlying the effects of ischemic postconditioning after stroke at the transcriptional level and the identification of the key biological processes, cellular components, molecular functions, and signaling pathways involved in these protective effects are lacking.
Studies have reported changes in the expression characteristics of noncoding RNAs after stroke; these noncoding RNAs regulate important cellular events in ischemic stroke through a variety of mechanisms (Zhao et al., 2015; Karner et al., 2020; Miao et al., 2020). Recent studies have shown that focal cerebral ischemia in adult rats extensively alter the lncRNA expression profiles of the brain during acute reperfusion, and altered lncRNAs may play critical roles in epigenetic changes after stroke (Dhami et al., 2013; Liu et al., 2018a; Shin et al., 2020). Bioinformatics analysis showed that among the significantly differentially expressed lncRNAs identified after stroke, 90% of sequences were homologous, indicating that hypoxic-ischemic injury altered the expression profile of brain lncRNAs in newborn rats (Hori et al., 2012). In this study, we found that ischemic postconditioning after ischemic stroke significantly altered the cerebral lncRNA expression profiles. Hierarchical clustering analysis showed systematic variations in the expression of lncRNAs and proteincoding RNAs among different groups in the ischemic brain. LncRNA expression patterns were similar among different samples within the same groups, indicating that lncRNAs in the same group may participate in similar biological processes and suggesting the functional conservation of lncRNAs (Zhao et al., 2015).
The KEGG enrichment analysis in the present study showed that significantly upregulated or downregulated lncRNAs after ischemic postconditioning in ischemic stroke rats were associated with the PI3K/Akt pathway. The PI3K/Akt pathway can regulate cell survival and growth by inhibiting apoptosis following cerebral IR injury (Ren et al., 2016). Moreover, we found that the lncRNAs that were significantly altered after IR injury were enriched in metabolic pathways, indicating that metabolic and cellular pathologies may be altered after ischemic postconditioning in ischemic stroke. Several studies have investigated the metabolic changes that occur during acute ischemic strokes of varying severities and found that excitotoxicity is the initial cellular-level insult mechanism associated with cerebral ischemia (Buga et al., 2012). Excitotoxicity is triggered by the failure to maintain metabolic homeostasis, resulting in the secretion of metabolites, including glutamate, glycine, D-serine, and polyamines (Xu et al., 2017).
LncRNA-mRNA regulatory networks might provide new insights into the molecular mechanisms underlying the therapeutic effects of ischemic postconditioning. The coexpression network suggested that lncRNAs may be involved in the regulation of corresponding target mRNAs in the IR, ISP, and RIP groups after ischemic stroke. Bioinformatics analysis indicated that the differentially expressed lncRNAs identified after ischemic postconditioning might be associated with inflammation, neuroactive ligand-receptor interactions, calcium signaling, and antigen processing- and presentationrelated pathways. We examined 15 lncRNAs and eight mRNAs that were differentially expressed between the ISP and IR groups and the RIP and IR groups in the co-expression network. We speculate that these 15 lncRNAs and eight mRNAs may play important roles in ischemic postconditioning. The expression of lncRNAs was highly correlated with the expression of neighboring mRNAs, which suggested that lncRNAs may exert their functions through these predicted mRNA targets (Zhao et al., 2015; Ren et al., 2016). We searched for coding genes located within 100 kb upstream and downstream of lncRNAs as potential target genes and used these genes to predict the functions of lncRNAs. We found that some lncRNAs, such as lncRNA 003369, lncRNA 017702, and lncRNA 014770, were associated with more than five mRNA targets. GO term and KEGG pathway analyses indicated that these lncRNAs might respond to ischemic postconditioning through different mechanisms after ischemic stroke, such as anti-inflammatory reactions (Chen et al., 2018), apoptosis (Pignataro et al., 2013; Esposito et al., 2018; Nichols et al., 2018), neuroactive ligand-receptor interactions (Li et al., 2021), oxidative stress (Pyfrom et al., 2020), calcium signaling (Vassallo et al., 2016; Pyfrom et al., 2020), and the cAMP response element binding protein/brain-derived neurotrophic factor signaling pathway (Lipovich et al., 2012). These findings suggest that changes in lncRNA expression profiles may be associated with ischemic stroke, and the neuroprotection afforded by ischemic postconditioning may involve the regulation of differentially expressed lncRNA expression. A previous study showed that cerebral lncRNAs were significantly altered after stroke (Duan et al., 2019), which supports our results. We further found that lncRNA 017702 was upregulated after ischemic postconditioning, according to the quantitative real-time reverse transcription-PCR and RNA sequencing results, and lncRNA 017702 was associated with nine target genes in the co-expression network, most of which were involved in the inflammatory response that causes brain edema. These results indicated that these genes might function in the regulation of the inflammatory response during ischemic postconditioning after stroke. A previous study found that the lncRNA Malat1 plays anti-apoptotic and antiinflammatory roles in the brain microvasculature, reducing ischemic cerebral vascular and parenchymal damage (Zhang et al., 2017). We speculated that lncRNA 017702 expression was upregulated, and pro-inflammatory gene expression was downregulated, following the RIP intervention, which reduced the degree of cerebral edema in rats with ischemic stroke. In support of this speculation, a previous study found that some lncRNAs that are upregulated after stroke in rats were identified to function in ischemic stroke through the inhibition of endothelial cell death and inflammation, such as the lncRNA Malat1 (Zhang et al., 2017).
This study has two limitations. First, we did not use more than three behavioral scoring methods to evaluate neurological deficits in tMCAO rats. The TTC staining images showed ischemic changes in brain tissue that are consistent with ischemic changes in the region of the middle cerebral artery. Therefore, we successfully established a rat tMCAO model. In future studies, we will use three behavioral scoring methods: the Zea Longa method, with a 5-point scale; the Garcia method, with an 18-point scale (Longa et al., 1989; Garcia et al., 1995); and modified neurologic severity scores (Zhou et al., 2011), combined with cerebral blood flow monitoring in rats to confirm the success of tMCAO model establishment. Although the cerebral tissue from the infarct area of each group was used to perform RNA sequencing in this study, the ischemic penumbra would provide more meaningful results because saving the ischemic penumbra is a key aim of ischemic stroke treatment. Therefore, our follow-up studies will use ischemic penumbra tissue from the rat brain to conduct RNA sequencing and explore the postconditioning protective mechanisms of the brain.
In conclusion, we investigated the expression profiles of the lncRNAs in the IR, ISP, and RIP groups after stroke. Studying the expression patterns of these RNAs at the overall gene expression level in cerebral IR injury and ischemic postconditioning after stroke is critical to developing new strategies for the treatment of ischemic stroke. Bioinformatics analyses revealed a complex lncRNA profile and significantly enriched GO terms and KEGG pathways associated with ischemic postconditioning after stroke. Nine candidate lncRNAs were identified by constructing lncRNA expression profiles and lncRNA-mRNA co-expression network analysis. These lncRNAs may be involved in the neurological protective effects associated with ischemic postconditioning in ischemic stroke. Our subsequent studies will focus on whether these specific lncRNAs can be targeted to prevent IR injury or promote angiogenesis and neuronal regeneration in animal models of ischemic stroke. Our study is the first study to provide a comprehensive, temporal description of the molecular events contributing to the pathogenesis of ischemic stroke in rats and uncovered functional RNA regulatory networks associated with ischemic stroke. These findings demonstrated that ischemic postconditioning methods might be used as effective interventions for cerebral ischemic injury, providing a theoretical basis for future clinical applications.
Author contributions:Study design: LYL, JHG; experiment implementation: WM, CYL, SJZ, CHZ; data analysis: WM, CYL, SJZ, CHZ, JWY, ZW, GDW, JL, WL, KPL, YL, XKZ, JJL; manuscript draft: WM, CYL, SJZ. All authors read and approved the final manuscript.
Conflicts of interest:The authors declare no conflicts of interest.
Financial support:This study was supported by the National Natural Science Foundation of China, No. 31560295 (to LYL); the Yunnan Applied Basic Research Projects of China, Nos. 2018FE001(-016) (to WM), 2018FE001(-163) (to LYL); and the Research Innovation Team of Yunnan Province of China, No. 2019HC022 (to LYL). The funding sources had no role in study conception and design, data analysis or interpretation, paper writing or deciding to submit this paper for publication.
Institutional review board statement:The study was approved by the Animal Experiment Ethics Committee of Kunming Medical University (approval No. KMMU2018018) in January 2018.
Copyright license agreement:The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement:The data used during this study are available from the corresponding author on reasonable request.
Plagiarism check:Checked twice by iThenticate.
Peer review:Externally peer reviewed.
Open access statement:This is an open access journal, and articlesare distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
Additional files:
Additional Figure 1: The schematic diagram about postconditioning intervenes in focal ischemic stroke rat.
Additional Figure 2: Effect of ISP and RIP on the neurological function deficit score of IR rats.
Additional Table 1: Long noncoding RNA target mRNA analysis in cis role.
Additional Table 2: KEGG pathway enrichment in IR vs. Sham group.
Additional Table 3: KEGG pathway enrichment in ISP vs. IR group.
Additional Table 4: KEGG pathway enrichment in RIP vs. IR group.
Additional Table 5: KEGG pathway enrichment in ISP vs. RIP group.
- 中国神经再生研究(英文版)的其它文章
- The importance of fasciculation and elongation protein zeta-1 in neural circuit establishment and neurological disorders
- Promoting axon regeneration in the central nervous system by increasing PI3-kinase signaling
- Microglial voltage-gated proton channel Hv1 in spinal cord injury
- Liposome based drug delivery as a potential treatment option for Alzheimer’s disease
- Retinal regeneration requires dynamic Notch signaling
- All roads lead to Rome — a review of the potential mechanisms by which exerkines exhibit neuroprotective effects in Alzheimer’s disease

