Investigation of kinetics in bioaugmentation of crude oil via highthroughput sequencing: Enzymatic activities, bacterial community composition and functions
Yong-Rui Pi , Mu-Tai Bao
a School of Ocean, Yantai University, Yantai, 264005, China
b Key Laboratory of Marine Chemistry Theory and Technology, Ministry of Education/Institute for Advanced Ocean Study, Ocean University of China,Qingdao, 266100, China
Keywords:Crude oil Bioaugmentation Kinetics Enzyme activities Microbial functions
ABSTRACT The enzymes and the characteristics of the community of the petroleum-degrading bacteria play a crucial role in the crude oil biodegradation.The prediction of kinetics of the key groups of hydrocarbons in crude oil was important to evaluate the bioremediation speed and constant.Most of the n-alkanes(C9-C29)were degraded in 25 days,and the average degradation rates of C18~C27 higher than 100 μg g-1d-1.The hopanes, such as H30, had a biodegradation rate more than 10 μg g-1d-1. The related enzymes activities changed along with the crude oil biodegradation, especially dehydrogenase. The 16S rRNA gene amplicon sequencing revealed that Proteobacteria, Firmcutes, Bacteroidetes, Actinobacteria, Acidobacteria were the main petroleum hydrocarbon degraders during the crude oil biodegradation,and the top two highest relative abundance of the genera were Alcaligenes and Acinetobacter. Acinetobacter presented positive correlation to biodegradation of n-alkanes and PAHs. Based on COG analysis, the largest group involved in the general function was amino acid transport and metabolism.The functional categories of bacterial communities were mainly focused on the carbohydrate and amino acid metabolism, xenobiotics biodegradation and metabolism, membrane transport, and so on. Overall, these findings highlight the potential guideline for more adequate monitoring of microbial degradation of crude oil.
1. Introduction
Oil spill poses significant impacts to the coastal environment,economics and human health.From 1970 to 2020,it is reported that more than 6 million tonnes of oil have been discharged into the marine environment over 466 large spills(>700 tonnes).Although no large spill was recorded,oil lost to the environment in 2020 was approximately 1000 tonnes (http://www.itopf.com/knowledgeresources/data-statistics/statistics/). The largest oil spill recorded in the US history was the Deepwater Horizon oil spill, releasing ~5 million barrels of crude oil (Quigg et al., 2021). The oil spill on the surface water in the open sea undergoes many physical, chemical and biological processes (Bacosa et al., 2020). Most of the components of the oil could ultimately be metabolized by hydrocarbon-degrading microorganisms as their carbon source,and they can acquire energy from the degradation of spilled oils(Kostka et al.,2011).Bioremediation was considered to be the most suitable method to deal with oil spills, which could be converted into CO2and H2O through mineralization(Pi et al.,2017;Gonz′alez-Gaya et al., 2019; Rodrigues et al., 2020; Xue et al., 2021). In the Deepwater Horizon oil spill, microorganisms, especially bacteria,played a key role and made great contribution to the residual oil,and 22%of the total spilled oil had been metabolized(Wang et al.,2013).
The biodegradation of spilled oil is influenced by different factors, such as pollutant characteristics, microorganisms and environmental conditions(Varjani,2017),it is important to evaluate the bioremediation speed and constant.Many previous kinetic studies focused on the bacterial growth and spilled oil consumption using models like Logistic, Tessier, Monod's and Contois (Sakthipriya et al., 2018; de Oliveira et al., 2013). However, few investigations have accurately predicted the kinetics of the key groups of hydrocarbon components of the crude oil.
The enzymes were crucial for crude oil biodegradation, were reported as environmentally friendly and cost effective (Mishra et al., 2014). They were beneficial for microbiota to getting adapted to the specific environmental conditions and the bioremediation of oil will be enhanced to several folds (Abatenh et al., 2017;Vaishnavia et al., 2021). The catalytic oxidation of organic substances by dehydrogenase which belongs to oxidoreductases, is achieved by separating electrons from the substrate and binding them with protons (Piotrowska-Cyplik et al., 2013). The biodegradation activity of the bacteria can be estimated by the presence of the enzyme dehydrogenase, providing information on their hydrocarbon removal efficiency. The hydrogenases were reported to be involved in the pathways of alkyl-CoA oxidation and the Wood-Ljungdahl pathway(Liu et al.,2020).Catalase is capable to detoxify H2O2to water(Sowani et al.,2020)and highly active oxygen,which is used by microorganisms in oil degradation(Wu et al.,2014).Wu et al.(2014)also confirmed that the hydrolysis of the amide group was catalyzed by urease, which could generate NH4+from organic nitrogen in the nitrogen cycle. The following factors might play a positive role on the activity of urease,such as the organic material content, microorganisms,total nitrogen and available phosphorus.It is particularly necessary to measure the enzymes involved in the crude oil degradation (Peixoto et al., 2011).
Microorganisms, especially hydrocarbon degrading bacteria,such as Oleispira, Thalassolituus, Oleiphilus and Alcanivorax, play a significant role in marine oil spill bioremediation process (Wang and Shao, 2014). While the abundance of the community shifts along the biodegradation going on.High-throughput sequencing is a biotechnology widely used in bioremediation of oil spill or polycyclic aromatic hydrocarbon (Kappell et al., 2014) and wastewater treatment (Guo et al., 2015) that provides great theoretical and practical significance to study the relationship between microorganisms and their environment.
Therefore, the objectives of this study were to: (1) investigate the degradation kinetics of the key groups of hydrocarbons of crude oil by an exogenous bacterial consortium; (2) quantify the key enzymes activities during the crude oil biodegradation;(3)explore the community composition of defined bacterial consortia by high throughput sequencing and predict the functions of the microbial community; (4) and explore the correlation between crude oil biodegradation and microbial communities.
2. Materials and methods
2.1. Chemicals and samples
All chemicals used were analytic grade without any purification.The crude oil was from Haierzhan oil well of the Shengli oilfield,and the physical property was as follows: 22.2 mPa s (viscosity,50°C,50 rpm),23.0°C(freezing point),0.8552 g cm-3(density).The component of the Haierzhan crude oil was as follows, 62.3% aliphatics,22.7%aromatics,9.5%resins and 0.9%asphaltenes(Pi et al.,2016).
The hydrocarbon-degrading strains were isolated from crude oil contaminated water and sediments (Loushan River, 36°12′N,120°20′E). Firstly, the mixture of the crude oil contaminated seawater and sediments were cultured in the LB medium at 30°C for 48h. Then 10 ml of the culture was transferred into a crude oil medium. Following cultured in a shaker at 150 rpm at 30°C for a week,20 ml culture was transferred into a fresh crude oil medium.After four circles, 20 ml crude oil medium culture was transferred into a LB medium. The density of the strains was measured by UV-visible Spectrophotometer at 600 nm. When OD600of the enriched culture was higher than 1.2(Fig.S1a),the consortia were harvested as crude oil degraders. The relationship between the density of the strains and OD600was shown in Fig. S1b.
The LB medium was used to enrich the hydrocarbon-degrading consortia,containing 3.0 g beef extract,10 g peptone and 5.0 g NaCl in 1L distilled water.The crude oil medium,which was referred to the literature with 2.0 g crude oil per liter (Obuekwe et al., 2009),was used to isolated the hydrocarbon-degrading consortia.The pH was adjusted to 7.0-7.2 with 1.0 M NaOH and 1.0 M HCl prior to sterilization. All the mediums were sterilized under 121°C for 20 min before used. Seawaters were collected from the coastal of Qingdao with the salinity of 32.5 ‰ and the dissolved oxygen of 9.5 mg/L.The seawater was filtered with the 0.45 mm membrane to remove the plankton and particulates before used.
2.2. Biodegradation of crude oil in flasks
Five Erlenmeyer 300 mL were prepared to evaluate biodegradation rate, nutrients change, enzyme activity and microbial community composition. The flasks were filled with 200 mL seawater amended with 0.4 g crude oil (2.0 g L-1) and some nutrients of N and P.10 mL LSH-series culture (OD600= 1.2) was added into the flasks. The growth curve of LSH-series in LB medium and the relationship between OD600and the bacteria density were shown in the supplementary. The flask without adding inoculation of hydrocarbon-degrading strains (natural seawater)was used as the control one. All the flasks were shaken continuously at 120 rpm,25°C and oil samples were collected after 1,3,7,15,25,40 and 60 days. The bacteria were also collected at these points for enzymes analysis and high-throughput sequencing.
The culture fluid was extracted twice with 50 mL of n-hexane for 20 min, the organic phase was emigrated to analyze the total petroleum hydrocarbons by GC-FID (Pi et al., 2017). The concentrations of the aromatic hydrocarbons, steranes and terpenes before and after biodegradation were analyzed by GC-MS(Pi et al.,2017).The details for the programs of GC and GC-MS were presented in the supporting information.
2.3. Measurement of enzymes activities
Once the crude oil was extracted, the aqueous phase was centrifuged at 8000 rpm for 10 min to collect the consortia,and the enzymes assay was taken out from the supernatant. A modified spectrophotometric method was used to measure the dehydrogenase activity(Wu et al.,2014).The catalase activity is performed as the rate of substrate oxidation of H2O2(Sowani et al.,2020).Urease activity was monitored by a modified indophenol reaction(Kandeler et al.,1988).The details for the measuring of the enzyme activities were included in the supporting information.All units of the enzymes activity were defined as U, and the details for U are presented in the supplementary.
2.4. Microbial diversity and functions analysis
According to the manufacturer's instructions,genomic DNA was extracted with the DNA extraction kit(Cwbio,China).The V4 region of the 16S rRNA gene was amplified using bacterial primers 515F and 806R constructed by 454 high-through sequencing.Qiagen Gel Extraction Kit (Qiagen, Germany) were used to purify all PCR products,then the PCR products were sequenced on Illumina MiSeq 250 platform.
Raw reads were processed by Uparse software (Edgar, 2013) to remove the low-quality sequences, and the operational taxonomic units (OTUs) were assigned with similarity ≥97%. The sequencing raw data was submitted to NCBI through Sequence Read Archive,and the SRA accession was PRJNA615237 with SRR11428947-SRR11428960. The software package of phylogenetic investigation of communities by reconstruction of unobserved states(PICRUSt2)was employed to predict the metabolic functions of the bacterial communities (Langille et al., 2013).
2.5. Biodegradation kinetic analysis
The biodegradation kinetic of the key groups of hydrocarbons was analyzed by the first-order kinetics (Zahed et al., 2010; Suja et al., 2014; Kheirkhah et al., 2020). The ratio of the concentration of the key groups of hydrocarbons to hopane was applied to calculate the microbial degradation. The key groups of hydrocarbons were divided into total hydrocarbon compounds (THC/hopane), total saturated hydrocarbon (n-C9~n-C38) (TSH/hopane),and total polycyclic aromatic hydrocarbons (TPAHs/hopane). The following first order model was used for the comparison (Bragg et al.,1994).

k refers to the biodegradation rate and t1/2is the time when initial concentration halved.
2.6. Statistical analyzes
All data in this study were the mean values (±SD) of three or more than three (six for 16S rRNA) duplicate samples. All of the statistical analyses were conducted using SPSS software (version 18.0, Chicago, IL, USA). A value of P < 0.05 was accepted as the criterion for statistical significance.The homogeneity of variance of all data was tested by Levene's test and the normality of residuals was tested by Shapiro-Wilk test. The one-way ANOVA was employed to conduct the multiple group comparison. * 0.01
3. Results and discussion
3.1. Biodegradation kinetics of key groups in crude oil
3.1.1. Biodegradation of crude oil
The concentration of n-alkanes was analyzed by GC-FID, while the aromatic hydrocarbons, steranes and terpenes were analyzed by GC-MS. The hydrocarbon compounds and abbreviations were listed in Table S1.The concentrations distribution of n-alkanes and PAHs were shown in the Fig.1. A large property of the n-alkanes(C9-C29)were degraded in 25 days,and the long n-alkanes(≥C30)were degraded in the next 35 days.It was very interesting that the concentration of the PAHs increased in the first 25 days,and some of them peaked at day 25.At this stage,the hydrocarbon-degrading bacteria were focused to “eat” n-alkanes, which were more“delicious”than PAHs.The total petroleum hydrocarbons declined,while the PAHs had almost no change,leading to a increase of PAHs presented as μg PAH per g oil. The PAHs with fewer substituents could be metabolized easily.
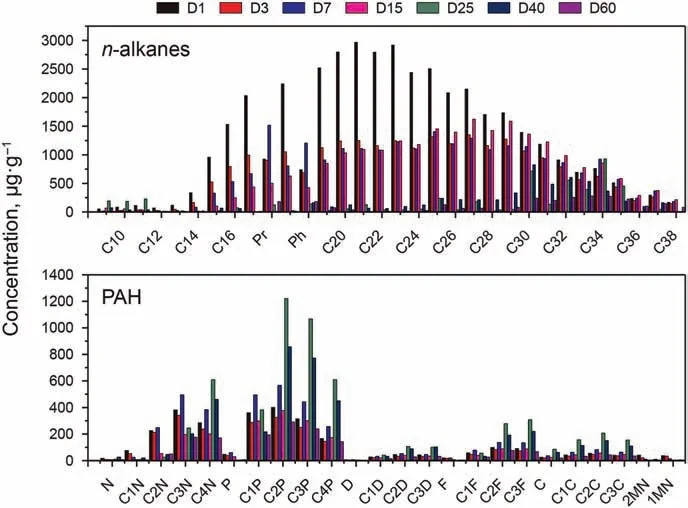
Fig. 1. The concentrations change of SH and PAH (five kinds) during the biodegradation.
3.1.2. Biodegradation kinetics of crude oil
The average biodegradation rate of n-alkanes, PAHs, hopanes,steranes and terpenes during the bioremediation of 60 days was presented in Fig. 2. Shorter n-alkanes (C9-C14) were almost completely degraded at ambient temperature (Fig. 2a). The media and long n-alkanes from C18to C27were susceptible to biodegradation and it was determined that the average degradation rates of n-alkanes seemed to be larger than 100 μg g-1d-1,with the highest degradation rate at above 100 μg g-1d-1for C21and C23(Fig.2a).The biodegradation rates of longer n-alkanes C29~C34were much lower than C18~C27, where the highest biodegradation rate was less than 100 μg g-1d-1(Fig.2a).From these results,it can be determined that the microbial community was effective to metabolize long n-alkanes C18~C27. Fig. 2b shows the average biodegradation rates of five targeted alkylated PAH homologues. Naphthalene, phenanthrene, as well as their alkyl homologues had higher degradation rate than that of dibenzothiophene, fluorene and chrysene. It indicates that there is a possibility that these compounds can be easily metabolized.It was obviously that the biodegradation rate of substituted PAHs was much higher than that of the non-substituted PAHs. The highest rate of degradation observed of C1P reached to 12 μg g-1d-1. As shown in Fig. 2c, hopanes were also degraded slightly, with the degradation rates ranging 0.5-10 μg g-1d-1.Obviously, H30had the highest biodegradation rate among all the hopanes homologues. The average biodegradation rates of some steranes and terpanes compounds were also tested and the results were shown in Fig. 2d. Most of the steranes and terpanes compounds were much persistent, while the average biodegradation rate of SES10(Table S1) was 1.2 μg g-1∙d-1.
Regressions were performed on the linearized (logarithmic)form of the Eq. (1). Fig. 3 presents plots of linear models for THC/hopane, TSH/hopane, TPAHs/hopane. The control showed no significant change during the 60-day experiment. For the three key groups, the slope of the lines had high significance. The biodegradation process was fitted well with the first-order model.
To quantify the biodegradation efficiency of various hydrocarbon components,the biodegradation rate constant(k)and half-life(t1/2) based on the first-order kinetic model were calculated(Table 1).The k values for THC/hopane,TSH/hopane,TPAHs/hopane were 0.330, 0.0489 and 0.0159 d-1, and the half-lives (t1/2) were 21.0,14.2 and 43.5d, respectively.
Kinetic is a fundamental tool to evaluate the equilibrium constant, the reaction speed as well as the half-time of the contaminants in petroleum hydrocarbon microbial remediation studies.The first order kinetics was employed to evaluate, model and analyze the kinetics for crude oil microbial degradation, and the half-life times for crude oil at 100,500,1000 and 2000 mg L-1were 31,40,50 and 75 days,respectively(Zahed et al.,2010).The results were quite similar to ours in this paper,however we have analyzed the degradation of key group of hydrocarbons.The half-life time of THC/hopane was 21 days, much shorter than the above 75 days when the crude oil concentration was 2000 mg L-1.A technology of continuous supply of inorganic nutrients was employed to investigate the microbial degradation of harbor sediments polluted by petroleum hydrocarbons,and the estimated value for k was 0.39 d-1(Beolchinia et al.,2010).Agarry et al.(2013)evaluated the kinetic of lubricating motor oil biodegradation in soil(25-200 mg kg-1)using the three different remediation treatments, natural attenuation,biostimulation and bioaugmentation, and the values of k during different biodegradation process were 0.015, 0.033, and 0.030 d-1,the values of t1/2were 46.2,21,and 23 days,respectively.In a field biostimulation of the intertidal sediments, the maximum rate of CO2production peaked at 18.40±1.04 μ mol CO2∙g-1wet sediment/day. When N and P were insufficient, Ksfor crude oil was only 4.52 ± 1.51 mg oil∙g-1wet sediment (Singh et al., 2014).
3.2. Enzymes activities
Dehydrogenases catalyze the oxidation-reduction reaction using a powerful coenzyme system scuh as NAD+/NADP+or flavin such as FAD,FMN as an electron acceptor.With the help of alcohol dehydrogenase, alcohol could be conversed to aldehyde/ketone with the reduction of NAD(P)+to NAD(P)H (Phale et al., 2019).Existing in aerobic organisms, catalase is an antioxidant enzyme,which could catalyze H2O2into H2O and O2in an energy-efficient manner in the cells exposed to environmental stress (Haider et al., 2021). Urease is a cytoplasmic enzyme, catalyzing the hydrolysis of urea to ammonium and CO2. The activities of dehydrogenase, catalase and urease during the biodegradation process were presented in Fig. 4. The activity of dehydrogenase increased quickly in the first 3 d,peaking at more than 150 U.Similarly,during a microbial degradation of the diesel oil, the highest activity of enzyme dehydrogenase was achieved at 4 d (Vaishnavia et al.,2021). Then, the dehydrogenase activity continued to decrease and reached a very low value in the last 20 d. The activity of dehydrogenase in the control was significantly lower than that in the biodegradation groups, indicating dehydrogenation plays an important role in the degradation of petroleum hydrocarbons,especially the early biodegradation stage of the petroleum hydrocarbons, such as n-alkanes. The catalase activity maintained steadily and had a slight increase at 40 d. Sowani (2020) found catalase activity in diesel oil treatment significantly higher than that in control groups. Vaishnavia (2021) found that the catalase activity had noted increase at the end of the incubation.Urease can promote hydrolyzation of urea-like substrates, such as protein,which is vitally important in mineralization of N (Huang et al.,2012). The urease activity did not show significant change during the biodegradation, while it had a slight rise at 60 d.
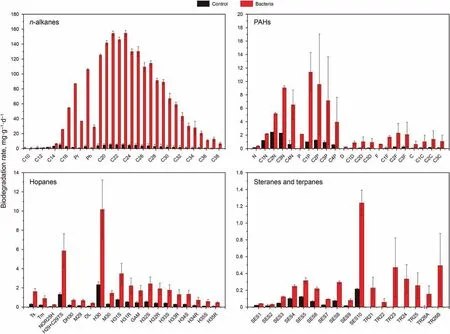
Fig. 2. The average biodegradation rates of the hydrocarbon compounds. a n-alkanes hydrocarbons. b Five target alkylated PAHs. c Hopanes. d Target steranes and terpenes. The hydrocarbon compounds and abbreviations were listed in Table S1.
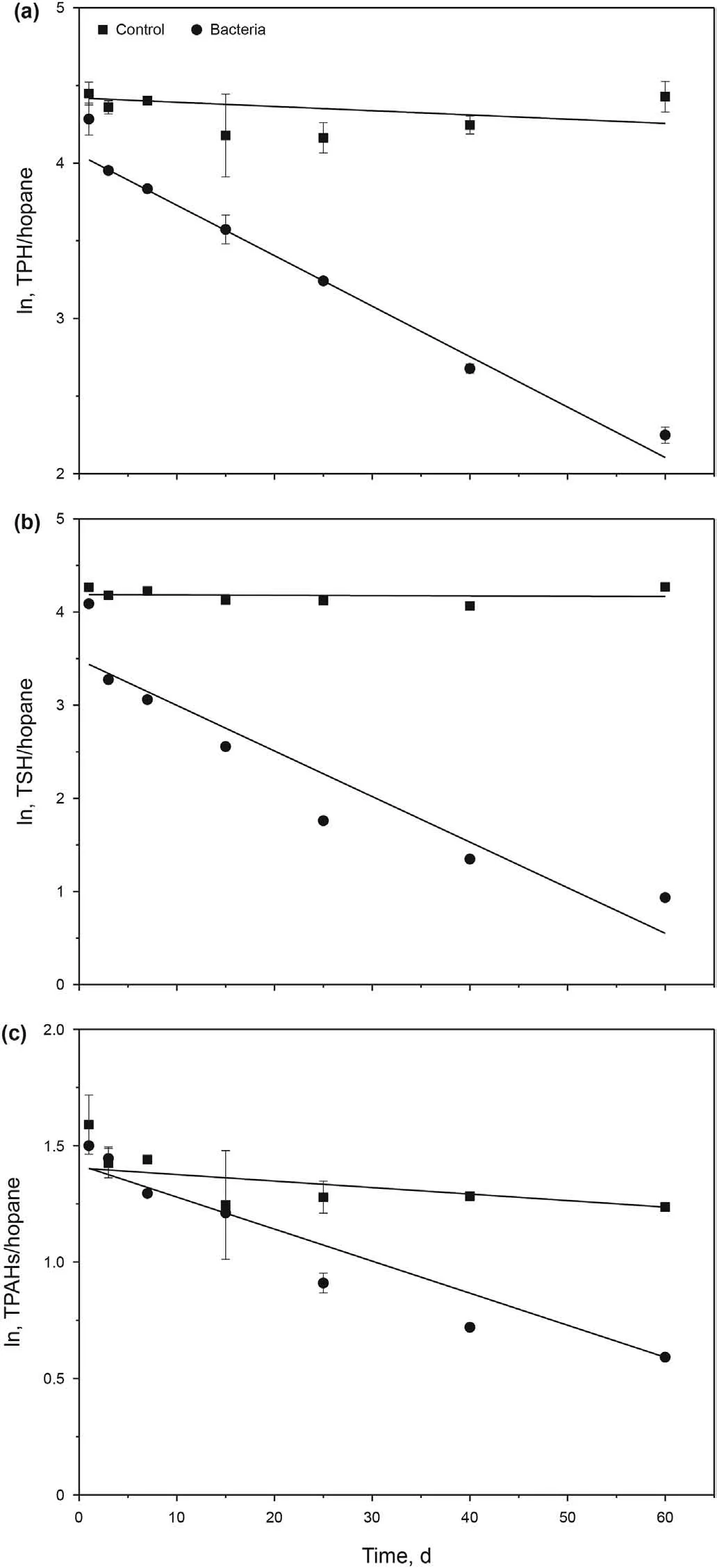
Fig.3. The linearized(logarithmic)form of biodegradation kinetic of the key groups of hydrocarbons.a The total hydrocarbon compounds (THC/hopane) detected by GC-MS.b The total saturated hydrocarbon(n-C9~n-C38)(TSH/hopane)measured with GC-FID.c The total polycyclic aromatic hydrocarbons (TPAHs/hopane) measured by GC-MS.
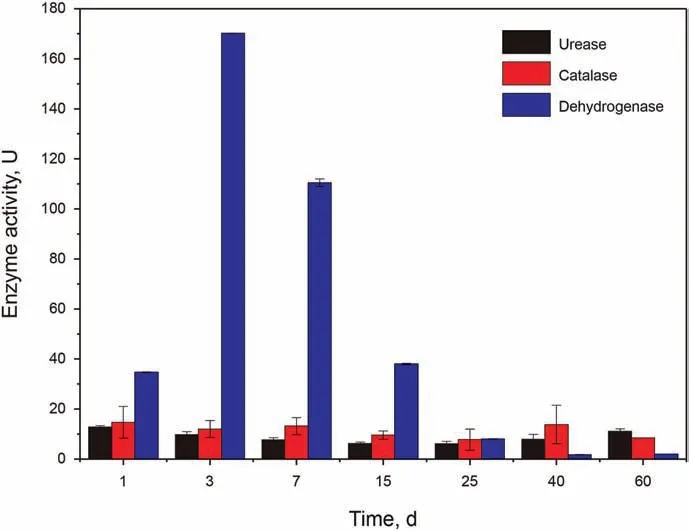
Fig. 4. The activities of urease, catalase and dehydrogenase measured during the biodegradation process along with time.
The top 30 high relative abundance of enzymes predicted by PICRUSt analysis ware shown in Fig. 5. The two most abundant enzymes were DNA-directed DNA polymerase (EC [2.7.7.7]) and histidine kinase (EC [2.7.13.3]). The dehydrogenase includes NADH:ubiquinone reductase (H(+)-translocating) (EC [1.6.5.3]), 3-oxoacyl-[acyl-carrier-protein] reductase (EC [1.1.1.100]),cytochrome-c oxidase (EC [1.9.3.1]), alcohol dehydrogenase (EC[1.1.1.1]), aldehyde dehydrogenase (NAD (+)) (EC [1.2.1.3]) and NAD(P)(+) transhydrogenase (Re/Si-specific)(EC [1.6.1.2]). Except these enzymes, the relative abundance of some other dehydrogenases was quite high,such as dihydrolipoyl dehydrogenase(EC[1.8.1.4]),medium-chain acyl-CoA dehydrogenase(EC[1.3.8.7]),formate dehydrogenase (EC [1.2.1.2]), dihydroxy-acid dehydratase(EC [4.2.1.9]), and 3-hydroxyisobutyrate dehydrogenase (EC[1.1.1.31]) (supplementary). The enzymes, such as NADH: ubiquinone reductase (H(+)-translocating) (EC [1.6.5.3]), could transfer H+during the biodegradation. Histidine kinase (EC [2.7.13.3])played a key role in the bacterial chemotaxis. Cytochrome P450 enzymes implicated into the degradation of xenobiotic compoundswere also detected (supplementary).Cytochrome P450, which is a terminal monooxygenase,is capable to metabolize endogenous and exogenous compounds. It could regulate the cytokines and temperature,due to its location of the cell's endoplasmic reticulum and mitochondrial inner membrane (Xue et al., 2021).

Table 1 Kinetic parameters for biodegradation of various hydrocarbons components.
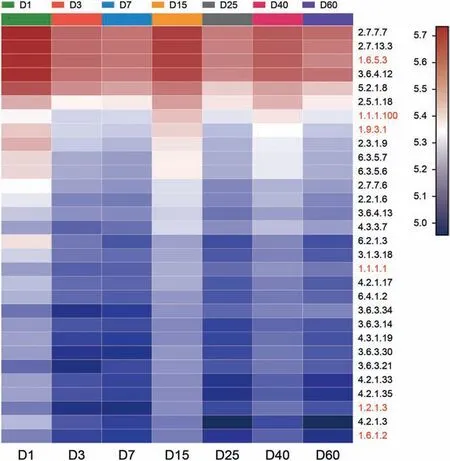
Fig. 5. Heatmap profiles showing the enzymes of bacterial communities as predicted by PICRUSt analysis. Rows represent the 30 enzymes, and the color intensity in the heatmap represents the relative abundance of the enzymes.
3.3. Bacterial community structure analysis
The microbiota was characterized by partial 16S rRNA gene sequencing obtained from DNA extracted from the bacteria harvested after biodegradation. The microbial communities at different biodegradation days were analyzed. According to the pyrosequencing of oil samples, the total data were as follows:effective sequences: 637736; Phylum: 31; Class: 85; Order: 179;Family:276;Genus:443;Species:645;OTU:898(Table S2).These sequences were assigned into 177-617 OTUs at a 97% similarity.The Shannon indexes had a peak at 25d, and Simpson indexes gradually increased then reduced, suggesting the microbial diversity increased from 1d to 7d, the decreased. High Good's coverage indexes(≥0.996)illustrated that the microbial diversity of samples could be covered by the obtained sequence libraries (Liu et al.,2015).
The hydrocarbon-degrading bacteria on the phylum level was major in Proteobacteria, Firmcutes, Bacteroidetes, Actinobacteria,Acidobacteria, the total relative abundance of the five phyla were more than 99.7% among all samples (Fig. 6). The abundances of Proteobacteria were decreased from 93.6%at 1 d to 83.0 at 60 d.It was very interesting that Firmcutes presented a decreasing tendency in the relative abundance from 5.4% at 1d to 1.4%, then increased to 14% at 60 d. Bacteroidetes showed an increase tendency from 0.54% to 5.3% among the first 3 d, then decreased to 1.4 at 60 d.The relative abundance of Actinobacteria increased from 0.25% at 1d to 1.1% at 40d. Acidobacteria presented a peak at 25 d with the relative abundance of 1.1%.
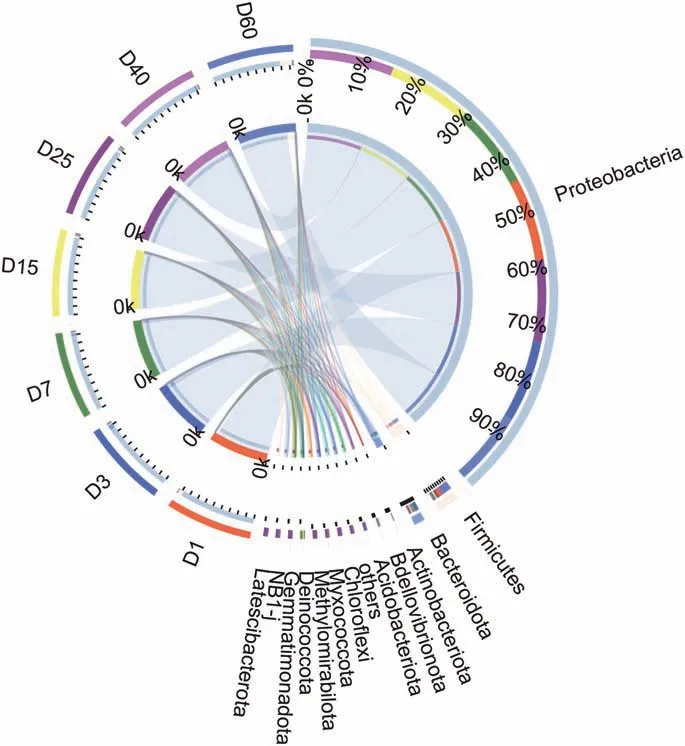
Fig. 6. The circos diagram of the relationship between samples and species on the phylum level.The small semicircle(left half circle)represents the species composition in the sample,the color of the outer ribbon represents the group from which it comes,the color of the inner ribbon represents the species, and the length represents the relative abundance of the species in the corresponding sample. The greater half circle(right half circle)represents the distribution proportion of species in different samples at the phylum level, the outer band represents species, the inner band represents different groups,and the length represents the distribution proportion of the sample in a certain species.
The hydrocarbon compounds degrading microorganisms are among the most extensively microbial communities in applied and environmental microbiology. Particularly, Proteobacteria(Kleindienst et al., 2015) including Alcanivorax, Cycloclasticus,Marinobacter, Oleiphilus, Oleispira, Planomicrobium and Thalassolituus (Head et al., 2006), have been recognized as the relative abundance of widespread specialized hydrocarbon-degrading bacteria of genera. In our study, the Haierzhan crude oil performed a distinct effect on the relative abundance of bacterial communities known as oil degraders(Head et al.,2006)during the biodegradation process, with the most abundant taxa Proteobacteria.
To analyze the key bacteria during the crude oil biodegradation,the species differences in all samples at the genus level were presented using one-way ANOVA(Fig.7).The significant differences of species at the genus level were Pseudochrobactrum, Ochrobactrum,Stenotrophomonas, Brevundimonas and unclassfied_f_Alphaproteobacteria.The top two highest relative abundance of the genera were Alcaligenes and Acinetobacter during the crude oil biodegradtion.Pseudochrobactrum is a powerful genus with the capability of degrading phenols and halogenated aromatics (Liu et al., 2020).In our study, Pseudochrobactrum, members of the Firmicutes, correlated positively along with the crude oil biodegradation,suggesting that it played a major role in crude oil remediation. Alcaligenes,known as a typical hydrocarbon-degrading genus,could degrade a wide range of xenobiotics, including aliphatic and polyaromatic hydrocarbons (Kahla et al., 2021). Acinetobacter play an indispensable role in the biodegradation of petroleum-hydrocarbon pollutants (Phulpoto et al., 2021). Except this, Acinetobacter was reported carrying with tetracycline-resistance genes and the quinolone-resistance genes, resulting in great potential resistance to multiple classes of antibiotics(Fang et al.,2018;Jang et al.,2021).The antibiotics above carry many benzene ring structures, which were similar in structure to PAHs. The antibiotics genes might be helpful for the PAHs degradation through co-metabolic degradation.

Fig. 7. Species differences in all samples on the genius level using one-way ANOVA. Data are presented as mean ± SD. * 0.01 < P ≤0.05, ** 0.001 < P ≤0.01, ***P ≤0.001.

Fig. 8. Clusters of orthologous groups (COG) function classification of the gut microbiota by PICRUSt analysis.

Fig.9. Heatmap profiles showing the functional categories(KEGG level 3)of bacterial communities as predicted by PICRUSt analysis.Rows represent the 50 KEGG Orthology (KO)functions, columns represent the 7 samples, and the color intensity in the heatmap represents the relative abundance of the functional genes.
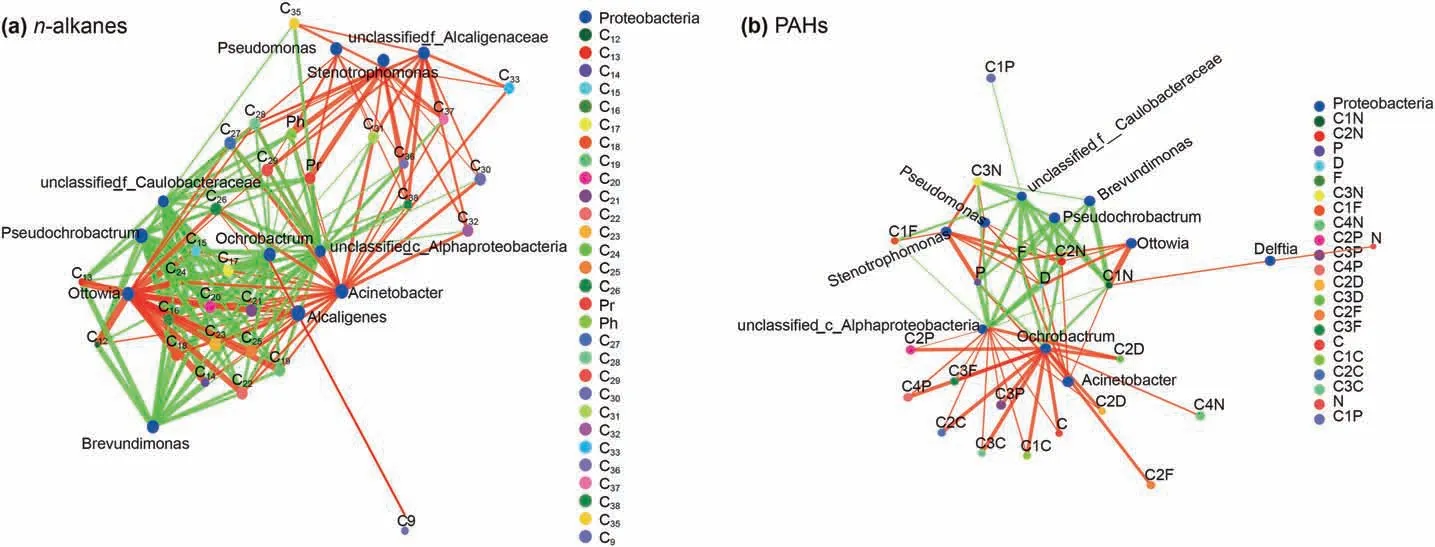
Fig.10. The species correlation network diagram between crude oil biodegradation and microbial community.The microbial species at genus level with P<0.05 are displayed in the figure.The size of nodes in the figure indicates the abundance of species, and different colors indicate different species. The red lines indicate positive correlation and green ones indicate negative correlation. The thickness of the line indicates the correlation coefficient. The thicker the line, the higher the correlation between species. The more lines, the closer the connection between nodes is.
3.4. Functional prediction of the microbial community
Cluster of orthologous Groups (COG) analysis showed that all microbial genes of the samples were classified into 23 categories assigned by COG (Fig. 8). In this classification, except the function unknown, the largest group involved in the general function was amino acid transport and metabolism (E), which was followed by energy production and conversion(C),inorganic ion transport and metabolism (P), transcription (K), Translation, ribosomal structure and biogenesis (J), Cell wall/membrane/envelope biogenesis (M),Carbohydrate transport and metabolism (G), lipid transport and metabolism(I),Coenzyme transport and metabolism(H)and signal transduction mechanisms (T).
The functional categories (KEGG level 3) of bacterial communities as predicted by PICRUSt analysis through a heatmap(Fig.9).The ABC transporter belonging to the membrane transport was no doubt the largest pathway. Following the ABC transporter,biosynthesis of amino acids and carbon metabolism were the important pathways of metabolism. The two-component system was important to the signal transduction. Quorum sensing was a critical function for cellular community with prokaryotes. The highest relative abundance of genes related to carbohydrate metabolism,including metabolism of glyoxylate and dicarboxylate,pyruvate, propanoate, butanoate, glycolysis, gluconeogenesis,amino sugar and nucleotide sugar. The gene with so high relative abundance indicated that the microbial community worked hardly to mineralize the crude oil. Except carbohydrate metabolism, the genes of amino acid metabolism accounted high of the total genes,including the genes to metabolize glycine,serine,threonine,caline,leucine, isoleucine, cysteine, methionine, phenylalanine, alanine,aspartate,glutamate,arginine and proline,phenylalanine,histidine and lysine. Also, the microbiota could metabolize the lipids, vitamins, and nucleotide. During the biodegradation of crude oil, a mass of lipid acid could be produced as the pH values were lower after the biodegradation than that of before. The high relative abundance of genes for lipid transport and metabolism was a great of significance for the crude oil biodegradation. The pathway of benzoate degradation was important for petroleum hydrocarbon degradation. Except benzoate degradation, the genes related to other xenobiotics biodegradation and metabolism were found,such as aminobenzoate,styrene,steroid,chloroalkane and chloroalkene,toluene, xylene, ethylbenzene, nitrotoluene, naphthalene, chlorocyclohexane and chlorobenzene, fluorobenzoate, polycyclic aromatic hydrocarbon, bisphenol and even atrazine (supplementary).Except the oxidative phosphorylation, methane and sulfur metabolism was the main function of energy metabolism. Bacteria secretion system was of great significance for membrane transport,which was much helpful for the microbiota to access to environmental information.Bacteria chemotaxis,which has a key enzyme of histidine kinase, played an important role to cell motility.
3.5. Correlation between crude oil biodegradation and microbial communities
The correlation between crude oil biodegradation and microbial communities was presented in Fig. 10. Based on Fig. 10a,Acinetobacter was responsible for all n-alkanes biodegradation.Alcaligenes and Ottowia presented positive correlation to biodegradation of the short and medium long chain alkanes (≤C29). The
Pseudomonas and Stenotrophomonas were the main driven biodegradation force for the long chain alkanes (≥C27). While unclassified_c_Alphaproteobacteria, Ochrobactrum, Pseudochrobactrum and Brevundimonas presented negative correlation to biodegradation of alkanes.Interestingly,some microalgae were also detected during the biodegradation of alkanes, except unclassified_f_Alcaligenaceae, the microalgae were not beneficial for the alkanes biodegradation.
Unlike the alkanes biodegradation, Ochrobactrum and unclassified_c_Alphaproteobacteria presented positive correlation to biodegradation of branched chain PAHs. Brevundimonas, Pseudochrobactrum and unclassified_f_Caulobacteraceae show negative correlation to biodegradation of D, F and P. On the contrary, Acinetobacter, ottowia, Pseudomonas and Stenotrophomonas were helpful for D, F and P biodegradation. Delftia was just responsible for N and C1N biodegradation (Fig.10b).
4. Conclusion
This study monitored the kinetic of the key hydrocarbon groups,enzymes and microbial community structure and functions during the crude oil biodegradation in 60 d. The biodegradation of different hydrocarbon compounds of the crude oil followed well with the first-order kinetic model. The related enzymes activities changed along with the crude oil biodegradation, especially dehydrogenase. It was highlighted that Proteobacteria, Firmcutes, Bacteroidetes, Actinobacteria and Acidobacteria were domain phylum during the crude oil biodegradation. The highest abundance of genus was Acinetobacter and it presented positive correlation to biodegradation of n-alkanes and PAHs. The most significant difference gene was Pseudochrobactrum among the whole biodegradation and it did not show any help for biodegradation of n-alkanes and PAHs. The genes of amino acid transport and metabolism has the highest relative abundance based on the COG analysis. The main microbial functions were carbohydrate metabolism, membrane transport, signal transduction, cell motility and xenobiotics biodegradation and metabolism based on KEGG analysis. These findings would provide a technology platform for the utilization of natural microbiome,especially for the in-situ remediation of crude oil or petroleum hydrocarbon contamination.
Acknowledgement
This research was funded by the Shandong Provincial Natural Science Foundation [Grant number: ZR2018MD018]; Yantai University Doctoral Start-up Foundation[Grant number:HX2018B32];Shandong Key Laboratory of Marine Ecological Restoration (Grant number: 201919). The authors declare no competing financial interests. The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.petsci.2022.01.022.
- Petroleum Science的其它文章
- A fast space-time-domain Gaussian beam migration approach using the dominant frequency approximation
- Predicting gas-bearing distribution using DNN based on multicomponent seismic data: Quality evaluation using structural and fracture factors
- Reflection-based traveltime and waveform inversion with secondorder optimization
- Determination of dynamic capillary effect on two-phase flow in porous media: A perspective from various methods
- Settling behavior of spherical particles in eccentric annulus filled with viscous inelastic fluid
- Laboratory investigation on hydraulic fracture propagation in sandstone-mudstone-shale layers

