Risk assessment of gene flow in rice mediated by the housefly Musca domestica
PU De-Qiang, LIU Jia-Fu, REN Shao-Peng, GAO Ming-Qing, YANG Fan,SHI Min, YE Gong-Yin, WEI Shu-Jun, CHEN Xue-Xin,*
(1. Ministry of Agriculture and Rural Affairs Key Laboratory for Molecular Biology of Crop Insects and Pathogens,Institute of Insect Sciences, Zhejiang University, Hangzhou 310058, China; 2. Institute of Plant Protection,Sichuan Academy of Agricultural Sciences, Chengdu 610066, China; 3. Institute of Plant and EnvironmentalProtection, Beijing Academy of Agriculture and Forestry Sciences, Beijing 100097, China)
Abstract: 【Aim】 Ecological risk assessment of transgenic crops is a necessary step before large-scale planting. There are hundreds of species of flower visiting insects in rice (Oryza sativa), including the housefly (Musca domestica). This study aims to assess the potential risk of rice gene flow mediated by flower-visiting M. domestica. 【Methods】 We conducted field experiments with the genetically modified (GM) rice lines B1, B6 and G8-7 as the pollen donors, the parental non-GM rice lines Jiazao 935 and Wuyunjing 7 as the pollen recipients, and M. domestica as the pollinator in Huajiachi and Changxing testing base of Zhejiang University in 2010. All harvested rice offspring seeds were planted in the laboratory and the plants were used for the detection of transgenic hybrids by the hygromycin B and glyphosate treatments, and then the surviving plants were further detected for hygromycin- and glyphosate-resistant genes by PCR to test the gene flow of GM rice mediated by M. domestica. 【Results】 The results based on the examination of over 216 500 geminated seeds from the three GM rice lines in the two testing bases in Zhejiang showed that there were few hybrids, indicating low frequencies (0-0.64%) of transgene flow from GM to non-GM rice at close spacing both in plots with and without M. domestica. 【Conclusion】 It is concluded that the frequency of transgene flow from GM to non-GM rice mediated by M. domestica is low and M. domestica does not increase the risk of gene flow of rice.
Key words: Oryza sativa; Musca domestica; gene flow; genetically modified crop; hygromycin-resistant gene; glyphosate
1 INTRODUCTION
Rice is one of the most important food crops in the world, especially in Asia, and feeds over half of the global population (Stone, 2008). Scientists have dedicated considerable efforts to enhancing the quality, reproduction and pest defense capacity of rice. Genes from the bacteriumBacillusthuringiensis(Bt) that code for insecticidal crystal (Cry) proteins were engineered into plants and the first insect-resistant genetically modified (GM) plants were developed in the mid-1980s (Vaecketal., 1987), with the rapid development of transgenic biotechnology, soon after, several transgenic rice varieties have also been developed (Chenetal., 2011), many of them have been entering into farm-scale preproduction trials. However, the release of these transgenic crops along with the risk of unwanted gene flow, creates concerns about biosafety worldwide (Ellstrand, 2001; Snow, 2002). Gene flow always has happened (Burkeetal., 2003; Chapmanetal., 2006) and is difficult for our capacity to predict ecological impacts of introduced species accurately (Wolfenbarger and Phifer, 2000), it is a crucial component of risk assessment and thus an important consideration in biosafety and trade policies (Huangetal., 2005; Luetal., 2006) of transgenic crops.
Oryzasativais thought to self-pollinate often before the flower opens, lowering the likelihood of pollen spread by wind or insects (Vogel, 2006), therefore, the gene flow frequency of rice is very low (<1.0%) (Oka, 1988). There were many studies focusing on wind-mediated gene flow of anemophilous crops such as rice (Messegueretal., 2004; Rongetal., 2004, 2005, 2007, 2010; Jiaetal., 2007), maize (Halseyetal., 2005; Gustafsonetal., 2006; Sanvidoetal., 2008), wheat (Gatfordetal., 2006; Riebenetal., 2011) and bentgrass (Reichmanetal., 2006), the important mechanism of wind in pollen transmission of these self-pollinated plants was revealed, and the management suggestions in production were put forward. However, gene escape due to flower-visiting insects of these plants has received surprisingly little attention. In fact, rice can produce a large amount of pollen during flowering, and there are many insects in fields, rice pollen may attach to the body of flower-visiting insects, which will cause the long-distance transmission of pollen and the escape of rice genes (Free and Williams, 1972).
In order to clarify the species of rice flower visiting insects, we conducted a two-year study that sampled locations were distributed in the rice growing regions of China and found more than 500 species of flower-visiting insects, of which approximately half of the insects collected had rice pollen on their bodies, including those famous pollinators such as tens of species of bees (Apoidea) and hoverflies (Syrphidae)(Puetal., 2014). Moreover, there were more than 100 species of higher Diptera (Tachinidae, Sarcophagidae, Ephydridae, Muscidae, Stratiomyidae and Dolichopodidae) in the paddy field during flower periods. The housefly,Muscadomestica, was founded in all of the rice growing regions of China and loaded more than ten rice pollen grains for each one (Puetal., 2014). Housefly is not considered as a regular flower-visiting insect and its role in plant pollination is rarely studied, especially in rice and other gramineous crops. However, a large number of them visiting rice flowers lead us to question if they may be able to mediate gene flow in rice. To determine the gene flow in GM rice in relation to flies, we designed cage experiments to examine the frequencies of housefly-mediated gene flow in field, involving three GM rice lines and their non-GM parental counterparts. The objectives of this study are to answer the following question: whether flies such as housefly increase the risk of gene flow of rice?
2 MATERIALS AND METHODS
2.1 Plant materials
The Bt rice lines B1 and B6 contain a syntheticcry1Abgene under the control of the maize ubiquitin promoter and linked in tandem withgus(encoding β-glucuronidase),hph(encoding hygromycin phosphotransferase) andnpt(encoding neomycin phosphotransferase) genes (Chengetal., 1998), their non-GM parental line is Jiazao 935. The Bt rice lines effectively control the target lepidopteran species including rice stem borers (Yeetal., 2001) and leaf-folders (Yeetal., 2003), under field conditions.
Another GM rice G8-7 contains the synthetic fusion genecry1Ab-vip3DAand a glyphosate-resistant geneG170 both under the control of a maize ubiquitin promoter, its non-GM parental line is Wuyunjing 7 (unpublished material).
2.2 Housefly
A colony ofM.domesticawas maintained in insect cages of Institute of Insect Science, Zhejiang University, healthy adults with the same size and no more than 2 days of eclosion were selected for field test.
2.3 Field experimental design
Experiment A (April-July 2010, Hangzhou): Insect-resistant GM rice B1 and its non-GM parental variety Jiaozao 935 (JZ) were planted in six plots in one straight line (Fig. 1). Each plot was 5 m×2 m, 0.6 m between plots, and the area ratio of B1∶non-GM rice Jiazao 935 was 2∶2. Rice plants were individually transplanted with distances of 1.0 m between the GM and non-GM subplots, 0.2 m between rows, and 0.2 m between hills within rows. During the flowering stage, each plot was covered with one closed cage (5 m×2 m×2 m high) with 0.5 mm×0.5 mm openings, three plots were placed without houseflies while the other three ones were placed with 140 houseflies respectively, and the same numbers of houseflies were added in each plot one week later to maintain the numbers of houseflies. Experiment B (April-August, 2010, Hangzhou): Insect-resistant GM rice B6 and its non-GM parental variety Jiaozao 935 (JZ) were designed in the same way as experiment B except the area ratio of B6∶non-GM rice was 3∶5, houseflies were placed in six plots in the same way as experiment A. Experiment C (July-November, 2010, Changxing): Insect- and glyphosate-resistant GM rice G8-7 and its parental variety Wuyunjing 7 were designed and the houseflies placed in the same way as experiment B.
Fig. 1 Field layout of the transgene flow experiments in Hangzhou in 2010
2.4 Seed harvest
At seed maturity, only seeds from the non-GM subplots were harvested, 1/3 of the entire row of non-GM rice plants were harvested at different distance intervals of 0.5, 1.0, 1.5, 2.0, 2.5 and 3.0 m (experiment A) and 0.5, 1.0, 1.5, 2.0, 2.5, 3.0 and 3.5 m (experiments B and C) from the GM rice subplots. All harvested seeds were used for the detection of transgenic hybrids.
2.5 Identification of transgenic seedlings
After drying, the seeds were soaked in fresh water for 3 d at 38℃ and placed in a seed germination dish at 38℃ for 1-2 d. The germinated seeds from the parental line Jiazao 935 of B1 and B6 were cultured in dishes containing a nutrient solution (Yoshidaetal., 1971), in addition to 50 mg/L hygromycin B (Bio Basic Inc., Markham Ontario, Canada) for 5-7 d in an illuminated growth chamber at 28-30℃. The surviving individuals were considered as transgenic plants containing the hygromycin-resistant gene. To confirm the hybrids detected by hygromycin B, 10% of the surviving seedlings from the hygromycin B treatment were randomly selected for polymerase chain reaction (PCR) detection with specific primer pairs designed for the hygromycin-resistant gene (hph) (Table 1), and the insect-resistantBtgene (cry1Ab) (Table 1). Total genomic DNA was extracted from leaf samples of individual seedlings following the method described by Dellaportaetal. (1983). A denaturation period of 5 min at 94℃ was followed by 35 cycles of 30 s at 94℃, 30 s at 56℃ and 80 s at 72℃, and then 5 min at 72℃ for final extension. Reactions were performed in a volume of 15 μL containing 1×buffer, 1.675 mmol/L MgCl2, 0.5 μmol/L primer [TaKaRa Biotechnology (Dalian) Co., Ltd., Dalian, China], 200 μmol/L dNTP, c. 50 ng genomic DNA, and 1 UrTaqpolymerase (TaKaRa Inc.). The PCR products were distinguished by electrophoresis using 1.0% agarose gel.
The seeds from the G8-7 parental line Wuyunjing 7 were soaked in fresh water for 3 d at 38℃ and placed in a seed germination dish at 38℃ for 1-2 d. The germinated ones were cultured in dishes containing the same nutrient solution as above for 6-7 d in an illuminated growth chamber at 28-30℃. The seedlings were sprayed with a solution of glyphosate (Roundup, Monsanto, USA) at the concentration of 2.05 g/L and cultured in the same way for 5-7 d. The surviving individuals were transgenic plants containing the glyphosate-resistant gene. To confirm the hybrids detected by the glyphosate, 10% of the surviving seedlings from the glyphosate treatment were randomly selected for PCR detection with specific primer pairs designed for the glyphosate-resistant gene (G170)(Table 1). Total genomic DNA was extracted from leaf samples of individual seedlings following the method described by Dellaportaetal.(1983). A denaturation period of 5 min at 94℃ was followed by 35 cycles of 40 s at 94℃, 40 s at 60℃ and 80 s at 72℃, and then 5 min at 72℃ for final extension. Reactions were performed in a volume of 15 μL containing 1×buffer, 1.675 mmol/L MgCl2, 0.5 μmol/L primer [TaKaRa Biotechnology (Dalian) Co., Ltd., Dalian, China], 200 μmol/L dNTP, c. 50 ng genomic DNA, and 1 ULaTaqpolymerase (TaKaRa Inc.). The PCR products were distinguished by electrophoresis using 1.0% agarose gel.

Table 1 Primers used in this study
2.6 Determination of transgene flow
For each row, more than 1 000 germinated seeds were examined, and the frequency of transgene flow (Fgf) was calculated as the number of transgenic seedlings divided by the total number of seedlings examined. The statistical analyses were all performed using the Wilcoxon two-sample test (http:∥www.fon.hum.uva.nl/Service/Statistics/Wilcoxon_Test.html), and the significant level (α) of 0.05 was taken into consideration.
3 RESULTS
The transgene-specific PCR examination confirmed that hybrids identified by the hygromycin B and glyphosate treatment were true hybrids generated from transgene flow (Fig. 2). This indicates that the methods of the hygromycin B and glyphosate screening for hybrids applied in the present study are reliable.

Fig. 2 Determination of hybrids containing the hygromycin- and glyphosate-resistant genes resulted from transgene flow
The frequencies of transgene flow from the three Bt transgenic rice lines to their adjacent non-transgenic counterparts were generally very low. This result was based on the screening of a total of 216 565 seeds collected from two experimental sites in Hangzhou and Changxing. For B1 and its non-GM parental variety Jiazao 935, in no housefly plots, all seeds harvested from 14 subplots were detected and hybrids in 12 subplots were found, and the highest frequency of transgene flow was 0.64% at 1.0 m from B1 to Jiazao 935; in housefly plots, all seeds harvested from 15 subplots were detected and hybrids in 7 subplots were found, and the highest frequency of transgene flow was 0.19% at 1.0 m from B1 to Jiazao 935 (Table 2). For B6 and its non-GM parental variety Jiazao 935, in no housefly plots, all seeds harvested from 18 subplots were detected and hybrids in 8 subplots were confirmed, and the highest frequency of transgene flow was 0.20% at 1.0 m from B6 to Jiazao 935; in housefly plots, all seeds harvested from 18 subplots were detected and hybrids in 5 subplots were confirmed, and the highest frequency of transgene flow was 0.10% at 1.0 m from B6 to Jiazao 935 (Table 3). As to G8-7 to its non-GM parental variety Wuyunjing 7, in no housefly plots, all seeds harvested from 18 subplots were detected and hybrids in 2 subplots were verified, and the highest frequency of transgene flow was 0.25% at 3.5 m from G8-7 to Wuyunjing 7; in housefly plots, all seeds harvested from 18 subplots were detected and hybrids in 1 subplot were verified, and the frequency of transgene flow was 0.09% at 2.5 m from G8-7 to Wuyunjing 7 (Table 4). These data indicated that the frequencies of transgene flow from the GM rice lines to their adjacently planted non-GM counterparts never exceeded 1.0% in any of the experimental plots with different mixed-planting proportions in this study, and in all the three experiments, the transgene flow in housefly plots were not significantly different from those in no housefly plots.

Table 2 Frequencies of transgene flow from GM rice B1 to its non-GM parental variety Jiazao 935

Table 3 Frequencies of transgene flow from GM rice B6 to its non-GM parental variety Jiazao 935
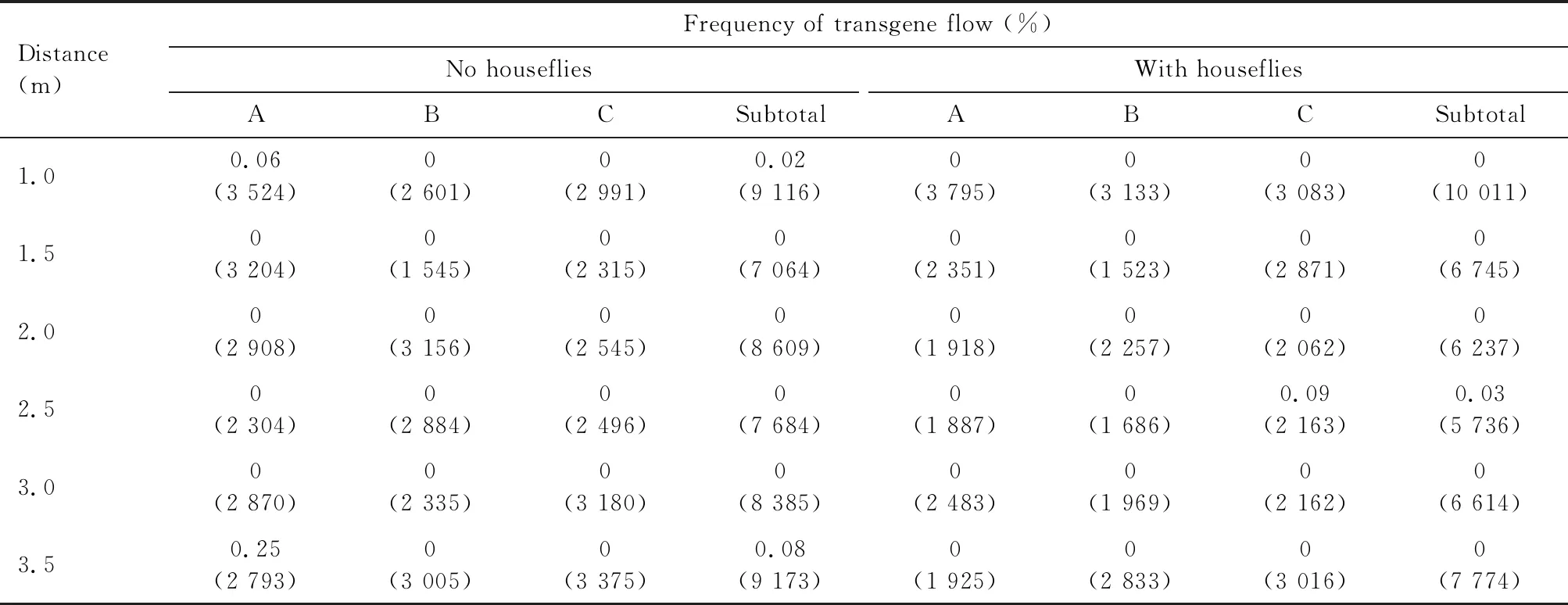
Table 4 Frequencies of transgene flow from GM rice G8-7 to its non-GM parental variety Wuyunjing 7
The Wilcoxon two-sample test confirmed no significant differences in frequencies of transgene flow between the plots treated with houseflies and without houseflies (P>0.05), meaning that even the number of houseflies more than ten times compared with that in the field would not increase the gene flow frequencies of rice significantly.
4 DISCUSSION
Rice has a large amount of pollen and flower-visiting insects, in addition to the famous pollinators such as bees and hoverflies, there were more than 100 species of higher Diptera visiting rice flowers during the blooming period, and tens of rice pollen grains loaded on their bodies by our previous study (Puetal., 2014). However, results from this experiment revealed very low frequencies of transgene flow from the three GM rice lines to their non-GM counterparts at extremely close spacing, even in the ‘extreme scenario’ with more than 10 houseflies per square meter, the frequencies of transgene flow were always below 1.0%-the strictest threshold that is adopted to determine ‘transgene contamination’ in the international trade of cereals. This result is similar to the reported frequencies of crop-to-crop and crop-to-weed rice gene flow by different authors (Messegueretal., 2001, 2004; Bashiretal., 2004; Chenetal., 2004). The gene flow frequencies were as low as those have been reported previously (Rongetal., 2005, 2007), and were like those of other wind-pollinated plants such as maize (Sanvidoetal., 2008), wheat (Gatfordetal., 2006) and bentgrass (Reichmanetal., 2006).
It is important to emphasize that the frequency of hybrids from our experiments was determined based on the observation of >216 500 geminated seeds collected from the transgene flow experimental fields at two sites using hygromycin- and glyphosate- resistant genes as the selection markers (confirmed by PCR of the transgenes). The data can well represent the actual transgene outflow from the three GM rice lines to their non-GM counterparts with a relatively high confidence. This method facilitates the massive scale of experiments that can provide a convincing assessment of likelihood of transgene outflow in rice fields. The frequencies of transgene flow determined from this experiment were all based on the GM and non-GM rice individuals cultivated adjacently with 20 cm between the hills in a row and between the rows. This means that the frequencies of transgene flow measured from GM to non-GM rice were nearly at the ‘zero distance’, because the canopy size of a rice plant is usually about the same as or larger than an area of 20 cm×20 cm and rice panicles of different individuals were connected to each other in the experiment. Additionally, the houseflies in the treated plots were several times compared with those in the same area of the rice field where at most 5-7 houseflies were present per square meters in a natural condition. Therefore, the frequencies of transgene flow from GM to non-GM rice in this study are the extreme (or maximum) situation.
From the results shown by our study, we can speculate that the individuals of Muscidae, Sarcophagidae, Sciomyzidae and Tachinidae may not increase the risk of gene flow of rice in rice fields because there are few numbers of rice pollen grains loaded by these insects, and without obvious flower visiting characteristics. However, as agroforests can provide important food and nesting resources for pollinators that translate into taxonomically and functionally diverse pollinator communities as well as stable pollinator visitation networks (Hassaetal., 2018), more research should be conducted in those rice fields surrounded by nectar source and nest for flies. In addition, there are many other pollinators such as bees (Apoidea), hoverflies (Syrphidae) and blowflies (Calliphoridae) by previous study (Puetal., 2014). Accordingly, for an accurate assessment of pollen-mediated transgene flow from GM to non-GM rice varieties, we should take all possible pollinators, such as those of bees, blowflies and hoverflies into consideration. Therefore, we should conduct more studies about rice gene flow mediated by potential pollinators.
ACKNOWLEDGEMENTSThe authors thank Prof. MO Jian-Chu for providing houseflies, Prof. SHEN Zhi-Cheng (Institute of Insect Sciences, Zhejiang University) for providing GM rice G8-7 and non-GM variety, and GOU Shou-Peng, CHEN Yi-Kai, CHEN Zheng, JIANG Jian, QIAO Chen-Yun and LIN Chao-Yang (Institute of Insect Sciences, Zhejiang University) for their assistance in harvesting rice and detecting hybrids.

