OsbHLH59 involved in rice resistance to Nilaparvata lugens(Stål)by regulating the expression level of xylanase inhibitor protein OsXIP
LÜ Xueqi, XU Ying, HUANG Yingying, LIU Mingqi, WENG Xiaoyan* (. Institute of Plant Biology,College of Life Sciences, Zhejiang University, Hangzhou 30058, China; . College of Life Sciences, China Jiliang University,Hangzhou 3008,China)
Abstract Xylanase inhibitor protein(XIP)is considered to participate in plant defense.OsXIP was found to be a stressresponsive gene of Nilaparvata lugens (Stål) in the previous study. The transcription factor OsbHLH59 could be combined with the promoter of OsXIP in response to brown planthopper (BPH) treatment. In order to study whether OsbHLH59 was involved in regulating the expression of OsXIP and mediating rice resistance to BPH, OsbHLH59 overexpression transgenic lines were obtained by transgenic methods, and OsbHLH59 mutant lines were obtained by using CRISPR/Cas9 system. The results showed that the expression levels of xylanase inhibitor OsXIP enhanced accompanied with the increased overexpression level of transcription factor gene OsbHLH59.Through the determination of agronomic traits,it was found that the growth of OsbHLH59 overexpression lines and OsXIP overexpression lines was less affected by the BPH treatment compared with wild type(WT).When WT plants and transgenic plants were exposed to BPH,the feeding preference of BPH in OsbHLH59 overexpression and OsXIP overexpression lines reduced compared with that of WT.With the BPH treatment,higher expression levels of defense-related genes were found in OsbHLH59 overexpression and OsXIP overexpression lines.In addition,compared with WT,OsbHLH59 overexpression and OsXIP overexpression lines had stronger abilities to remove excess H2O2 and higher antioxidant enzyme activities.This study reveals that OsbHLH59 can activate the expression of OsXIP,and OsXIPparticipates in rice resistance to BPH.
Key words xylanase inhibitor protein;OsXIP;OsbHLH59;gene expression;plant defence
Xylanase inhibitor protein (XIP) was first discovered in 1997[1]. So far, three xylanase inhibitor proteins have been found in gramineous plants[2].They areTriticum aestivumxylanase inhibitor (TAXI)-type,XIP-type and thaumatin-like xylanase inhibitor(TLXI)-type xylanase inhibitors[3-5]. Xylanase inhibitor proteins in plants can effectively inhibit the activity of xylanase derived from microorganisms, but cannot inhibit the activity of xylanase derived from plants,so they are considered to participate in the defense of plants. Overexpression of xylanase inhibitor OsHIXIP reduced the feeding and oviposition preference of the rice brown planthopper (BPH) [Nilaparvata lugens(Stål)], thereby increasing the resistance to BPH[6]. PADILLA-HURTADO et al. found that xylanase inhibitor XIP-I in wheat reduced the loss of coffee production caused by the coffee berry borer[7].
OsXIP is a XIP-type xylanase inhibitor, which was first cloned by TOKUNAGA et al.[8]. Our previous research found that BPH treatment significantly increased the transcription level ofOsXIP, and furthermore illustrated that the expression ofOsXIPmight be regulated by transcription factor OsbHLH59 for that it could bind to the promoter of OsXIP in response to BPH[9]. However xylanase inhibitorOsXIPwas considered to participate in plant defense[2,9], but little was known about the effect of expression level ofOsbHLH59onOsXIPexpression.
Basic helix-loop-helix (bHLH) transcription factors are widely present in higher plants, participating in various signal transduction, anabolism, and play important roles in various biotic and abiotic stresses[10].YANG et al. demonstrated thatTabHLH1mediated wheat tolerance to osmotic stress by regulating abscisic acid(ABA)-related pathways[11].The bHLH transcription factor MYC2 interacted with theArabidopsisMED25 subunit to activate the expression of downstream defense genes to further increase the resistance ofArabidopsistoBotrytis cinerea[12]. WANG et al. found that OsHLH61-OsbHLH96 complex mediated rice resistance to BPH through the regulation ofpathogenesis-related(PR)genes[13].
In this study,OsbHLH59overexpression transgenic lines were obtained by transgenic methods, andOsHLH59mutant lines were obtained by using CRISPR/Cas9 system.We try to explore the relationship between the transcription factor geneOsbHLH59and the xylanase inhibitorOsXIP, and further illustrate whether transcription factor OsbHLH59 is involved in regulatingOsXIPexpression and mediates resistance to rice BPH.
1 Materials and methods
1.1 Plant materials and growth conditions
Nipponbare(Oryza sativacv.Nipponbare) was used as a wild type(WT)rice.OsXIP-1 was anOsXIPoverexpression rice line, o-59-1, o-59-2, o-59-3,and o-59-4 wereOsbHLH59overexpression trans‐genic rice lines, and c-59-1, c-59-2, c-59-3, and c-59-4 wereOsbHLH59mutant lines. According to the method of YOSHIDA et al.[14],the seeds were placed in a 28 ℃, 80% relative humidity, 16 h light/8 h dark greenhouse,and grew in the culture solution.
1.2 Plasmid construction of overexpression lines and rice transformation
UsingNipponbare(Oryza sativacv.Nipponbare)rice cDNA as a template, forward primer (5´-ATGGACGGAGGCGGAGACCCCGTT-3´) and reverse primer(5´-TTAAGCATTTGGTGGGGCCAA AGCT-3´) were used for polymerase chain reaction(PCR) to amplify the coded sequence (CDS) ofOsbHLH59(LOC_Os02g02480). TheOsbHLH59fragment was digested withBamHⅠandSacⅠ, and the product was cloned into a site in the downstream of 35S::sGFP (green fluorescent protein) promoter in pCAMBIA1300-sGFP vector. The 35S::OsbHLH59-sGFP constructs were verified by sequencing and then introduced intoAgrobacterium tumefaciensstrain EHA105 by electroporation. The calli derived from the mature rice (Oryza sativacv.Nipponbare)seeds were used for transformation[15]. The calli were selected by hygromycin-containing medium. The transgenic rice line obtained by the differentiation of the selected callus is T0.The T0rice lines were mature to harvest the T1generation rice seeds, and then planted and cultivated after PCR identification. The T2generation ofOsbHLH59overexpression rice line that was stable and homozygous was used for further physiological and biochemical research.
1.3 Construction of CRISPR/Cas9 plasmid vector and rice transformation
Enter the accession number of the geneOsbHLH59(LOC_Os02g02480)on RiceXPro(https://www.genome.arizona.edu/crispr/CRISPRsearch.html). Find the exon sequence and compare the CDS ofOsbHLH59to find a pair of exon sequences with a sequence difference of about 150 bp. Design the primers according to the template of XIE et al.[16].The primers used in plasmid construction were listed in Table 1.
Table 1 Primers used for plasmid construction
Use the plasmid of pGTR bacteria as a template for PCR. The ligated products were amplified by PCR and then the target fragments were purified by using the universal DNA purification kit [DP214,Tiangen Biotech (Beijing) Co., Ltd.]. The purified PCR products were digested withFokⅠ. TheFokⅠdigested fragments were ligated into theBsaⅠdigested pRGEB32 vector with T4 DNA ligase. The ligation product was transformed intoEscherichia coliDH5α, and after the construct was confirmed by sequencing, it was transformed into EHA105 by electroporation.
1.4 DNA extraction and PCR analysis
DNA was extracted from fresh leaves of rice by using the cetyltriethylammonium bromide(CTAB)method[17]. We designed a pair of primers to detect the presence of GFP during PCR analysis (GFP-F, 5´-ATGGTGAGCAAGGGCGAGGAGCTGT-3´; GFP-R,5´-TTACTTGTACAGCTCGTCCATGCCG-3´).
1.5 Real-time quantitative PCR (qRT-PCR)analysis
According to the manufacturer’s operating procedures, using RNA extraction kit (Code No. 9767,TaKaRa, Japan) to extract total RNA from rice leaves.cDNA synthesis kit (Code No. RR047A, TaKaRa,Japan)was used to synthesize cDNA from 1 μg of total RNA. qRT-PCR analysis of all genes was performed with gene-specific primers (Table 2), which was total of 40 cycles(95 ℃for 5 s;60 ℃for 30 s)[15].
Table 2 Primers used for qRT-PCR analysis
1.6 Determination of net photosynthetic rate
The net photosynthetic rate was measured by using the Li-6400 portable photosynthesis system(LI 6400XT, LI-COR, USA). During the measurement,the saturated light intensity was set to 1 500 μmol/(m2·s).The net photosynthetic rate of rice was measured at the tillering stage (grown after 80 d), heading stage(110 d)and filling stage(140 d)of rice to analyze the growth status of different rice lines[18].
1.7 BPH treatment
The fourth-instar BPH was selected. Put the 14-day-old rice into the rice incubator separately under the treatment of BPH. Suck the BPHs and pour them into a rice incubator (10 BPHs for every four rices), and then place them in the BPH cultivation room.
1) Determination of the mortality rate.Individual plant test was carried out by using 15 replicates of the susceptible variety TN1 and each line. When all TN1 plants died, the mortality rate of each line was recorded[19].
2) Determination of agronomic traits. The rice seedlings were taken out on the 0, 1, 2, 3 d after the BPH treatment, and the rice was placed in a rice nutrient solution and grew in a normal environment for 25 d. After that, the plant height and the number of leaves of different rice lines treated with BPH at different times were measured[18].
3) Determination of physiological and biochemical indexes.The rice seedlings were taken out on the 0,1, 2, 3, 4 d after the BPH treatment, and the physiological and biochemical indexes were measured.
1.8 BPH resistance test
In order to study the colonization behavior of BPH,WT plants and transgenic plants exposed to BPH in pairs were confined with plastic cylinders (5 cm in diameter, 30 cm in height, with small holes). Each cylinder received 10 fourth-instar BPHs. Count the number of BPHs on each plant at 1, 2, 4, 8, 12, 24, 36,and 48 h after the release of BPHs. The experiments were repeated five times[20].
1.9 Measurement of H2O2 content and antioxidase activity
Rice leaf tissue was taken on the 0, 1, 2, 3, and 4 d after the BPH treatment to determine the H2O2content and antioxidase activity.
The measurement of H2O2was carried out according to the method of JANA et al.[21].Grind 0.05 g leaf tissue into homogenate, and then add 3 mL sodium phosphate buffer (including 50 mmol/L KH2PO4, pH 6.5) containing 1 mmol/L hydroxylamine.After centrifugation for 25 min (6000 r/min, 4 ℃),the supernatant was collected. The extract was mixed with 0.1% (V/V) titanium chloride in 20% (V/V)H2SO4. Measure the absorbance at 410 nm and calculate the H2O2content.
Enzyme extraction was as following: the leaf tissue was ground into homogenate in sodium phosphate buffer (including 50 mmol/L Na2HPO4/NaH2PO4, pH 7.8). After centrifugation for 20 min(12 000 r/min, 4 ℃), the supernatant was collected for the analysis of antioxidase activity.
The activity of catalase (CAT) was measured by spectrophotometry[22]. Add 200 μL enzyme extract to the reaction mixture which containing 200 μL H2O2(100 mmol/L) and 1.5 mL phosphate buffer (50 mmol/L, pH 7.0) and adjust it to 3 mL with distilled water. Measure the absorbance at 240 nm and calculate the CAT activity.
The peroxidase (POD) activity was measured by the guaiacol method[23].Add 50 μL enzyme extract to a mixture which containing 1 mL phosphate buffer(50 mmol/L,pH 7.0),1 mL H2O2(0.3%)and 0.95 mL guaiacol (0.2%). Within 2 min after adding the enzyme extract, record the increase of absorbance at 470 nm and calculate the POD activity.
2 Results
2.1 Obtaining of OsbHLH59 overexpression lines and mutant lines
The independent transgenic rice lines were selected by PCR analysis using gene-specific primers for the hygromycin resistance gene (hyg) and the 35S-OsbHLH59sequence (Fig. 1). The expression levels ofOsbHLH59in 14-day-old seedlings of o-59-1, o-59-2, o-59-3, and o-59-4 overexpression lines were analyzed. Compared with WT plants, the expression levels ofOsbHLH59in o-59-1, o-59-2,o-59-3, o-59-4 overexpression lines were 9.11, 8.94,6.23 and 7.30 times,respectively(Fig.1A).
Compared with WT plants,the expression levels ofOsbHLH59in c-59-1,c-59-2,c-59-3,and c-59-4 mutant lines were 0.27, 0.36, 0.54 and 0.33 times,respectively (Fig. 1B). Higher expression levels ofOsXIPwere found in transgenic plants. The expression levels ofOsXIPin o-59-1,o-59-2 and OsXIP-1 were 6.56, 5.40 and 11.34 times of those in WT plants,respectively; andOsXIPexpression levels in c-59-1 and c-59-2 were 0.94 and 0.93 times of WT plants,respectively(Fig.1C).
OsbHLH59mutant plants were obtained by using CRISPR/Cas9 system. Sequence analysis results(Fig.1D)revealed that there were four mutation edits in the T1mutant lines:one large deletion mutation of 65 bp,one large deletion mutation of 34 bp, simultaneous one base insertion and one base deletion,and simultaneous three bases substitution and one base deletion.
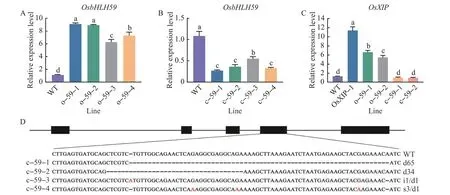
Fig.1 Identification of OsbHLH59 mutant rice and relative expression levels of OsbHLH59 and OsXIP in overexpression lines,mutant lines and WT plants
2.2 Photosynthetic characteristics of transgenic rices
The net photosynthetic rates ofOsbHLH59overexpression lines and mutant lines were measured during the growth and development of rice. In those,the net photosynthetic rates of rice leaves in all transgenic rice plants at the tillering stage, heading stage, filling stage showed trends of increasing first and then decreasing(Table 3).
There were no significant differences in the net photosynthetic rates of the four lines at the tillering stage, heading stage and filling stage (Table 3). The results indicated that the overexpression ofOsXIPandOsbHLH59and the knockout ofOsbHLH59did not adversely affect the growth and development of rices.
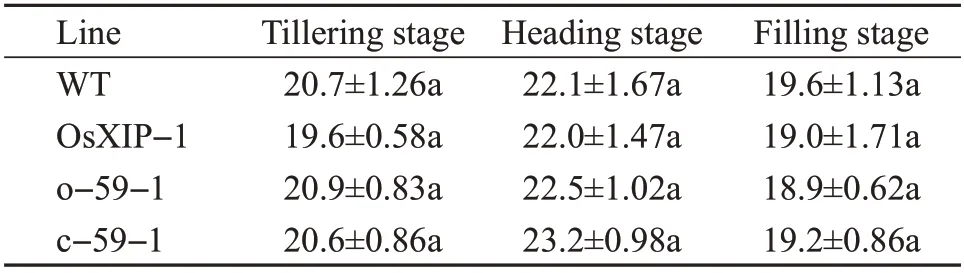
Table 3 Net photosynthetic rates of transgenic plants at different stages μmol/(m2·s)
2.3 Growth characteristics of OsbHLH59 overexpression plants and mutant plants after the BPH treatment
We selected the T2generation of each line and used individual tests to determine their response to BPH. When all TN1 plants died, the mortality rates ofOsbHLH59mutant lines were much higher than that ofOsbHLH59overexpression lines andOsXIPoverexpression line(Fig.2A).
The plant heights ofOsbHLH59overexpression lines and mutant lines on the 25th day were observed to analyze the growth of different rice plants after being infected by BPH (Fig. 2B). The data showed that as the treatment time of BPH increased, the plant height on the 25th day gradually decreased.At the same treatment time with BPH, the plant height ofOsXIPoverexpression line was the highest, followed byOsHLH59overexpression lines, WT, andOsHLH59mutant lines. The plant heights of o-59-1, o-59-2 and OsXIP-1 were 13.8%, 13.3% and 16.9% higher than that of WT, respectively, on the 3 d after the BPH treatment. The plant heights of c-59-1 and c-59-2 were 6.9% and 5.5% lower than that of WT,respectively. It showed thatOsXIPoverexpression line had the strongest resistance to BPH,OsHLH59overexpression lines were the second, andOsHLH59mutant lines had the weakest resistance to BPH.
The growth rates of new leaves ofOsbHLH59overexpression lines and mutant lines treated with BPH were obviously inhibited.OsXIPoverexpression line had the strongest resistance to BPH, whileOsHLH59mutant lines had the weakest resistance to BPH, which was the same as the result obtained in Fig.2C.
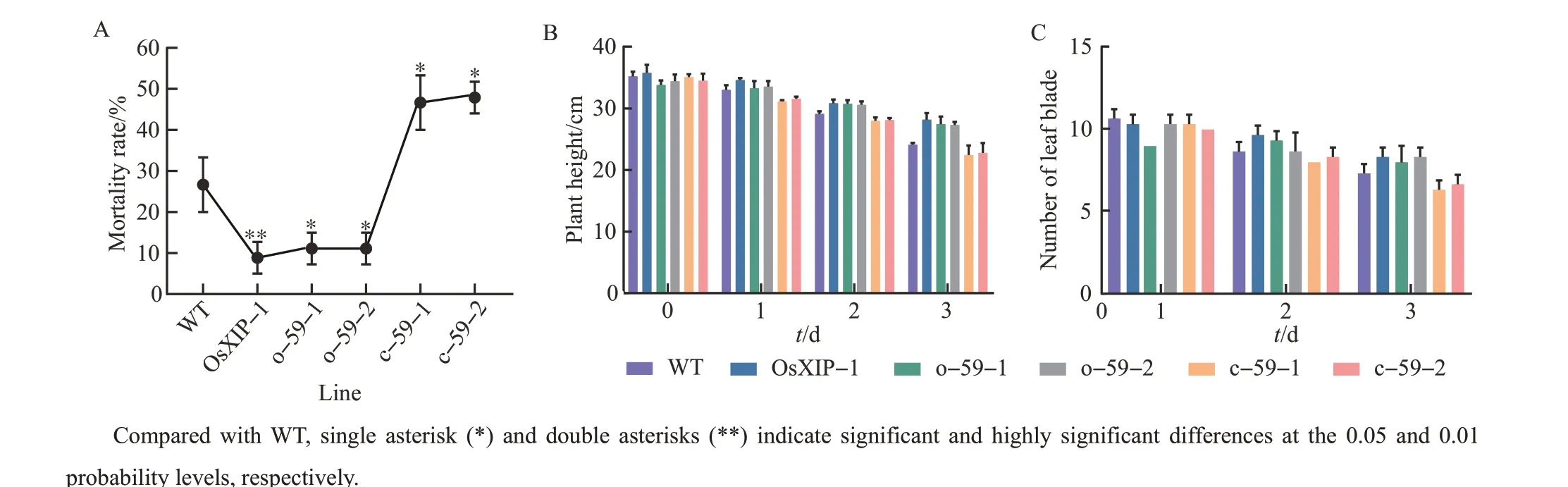
Fig.2 Growth characteristics of overexpression lines,mutant lines and WT after the BPH treatment
2.4 Overexpression of OsXIP and OsHLH59 positively regulating rice resistance to BPH
WhenOsXIPandOsbHLH59overexpression lines (OsXIP-1, o-59-1, o-59-2) and WT plants were exposed to a BPH colony,BPH preferred to feed on WT plants rather than overexpression lines. And theOsbHLH59mutant lines (c-59-1, c-59-2) were more favored by BPH than WT plants. These results indicated that overexpression ofOsXIPandOsHLH59positively regulated rice resistance to BPH(Fig.3A-E).
Different lines of 14 days old were infected with BPH, and other lines were observed when all the susceptible lines TN1 died. Compared with the WT,OsXIP(OsXIP-1) andOsHLH59overexpression lines (o-59-1, o-59-2) were less damaged by BPH,whileOsHLH59mutant lines (c-59-1, c-59-2) were more severely damaged(Fig.3F).
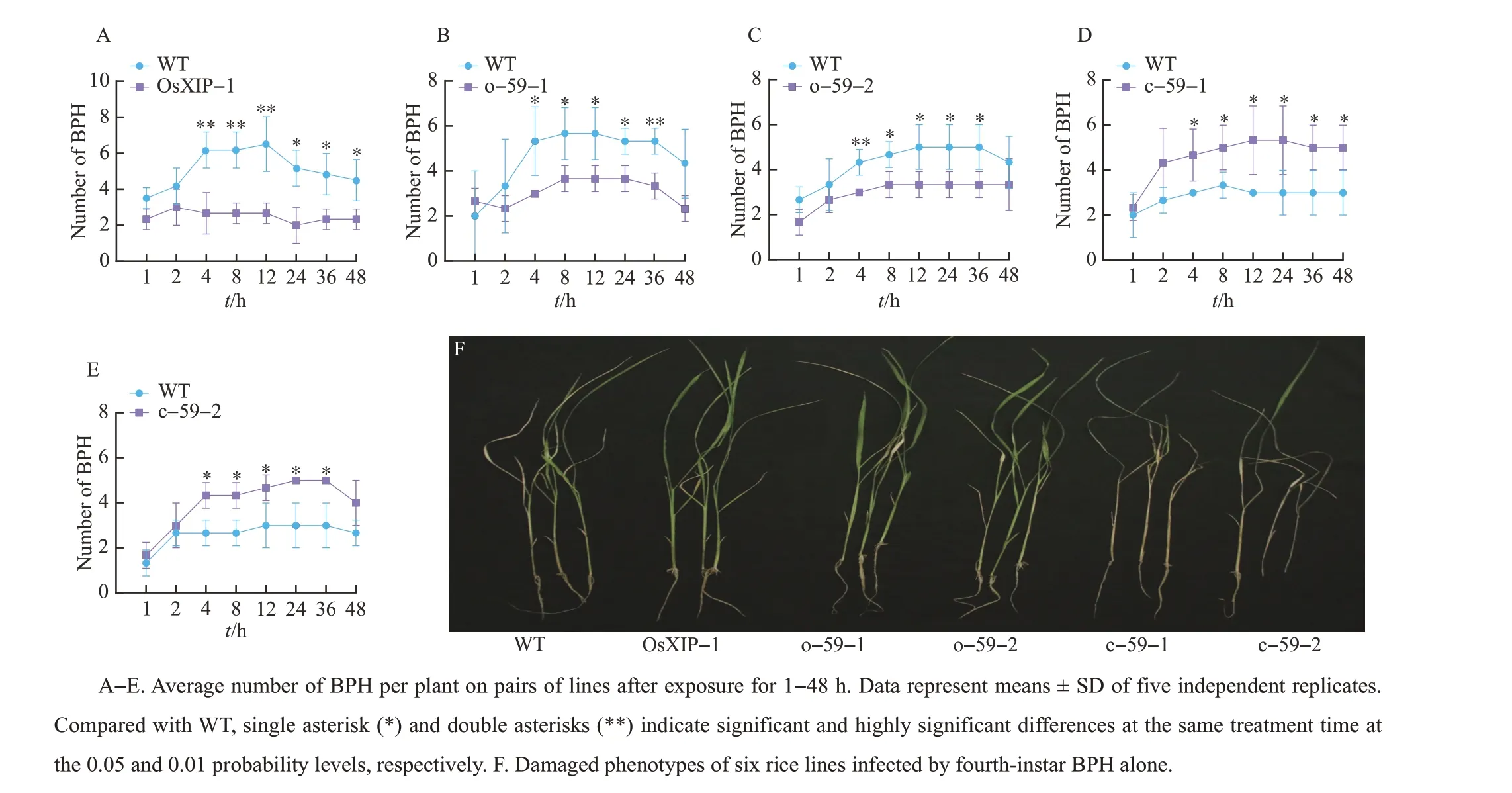
Fig.3 Overexpression of OsXIP and OsHLH59 positively regulating rice resistance to BPH
2.5 Relative expression levels of genes OsbHLH59 and OsXIP in different transgenic rice plants after the BPH treatment
After the BPH treatment, the expression levels ofOsbHLH59in theOsXIPandOsbHLH59overexpression lines increased significantly, and were much higher than that of WT (Fig. 4A).However, the expression level ofOsbHLH59inOsbHLH59mutant rice was too low to change significantly. Besides, BPH treatment induced different levels ofOsXIPexpressions in each line. The expression levels ofOsXIPin overexpression lines were higher than those of WT and two mutants(Fig.4B).
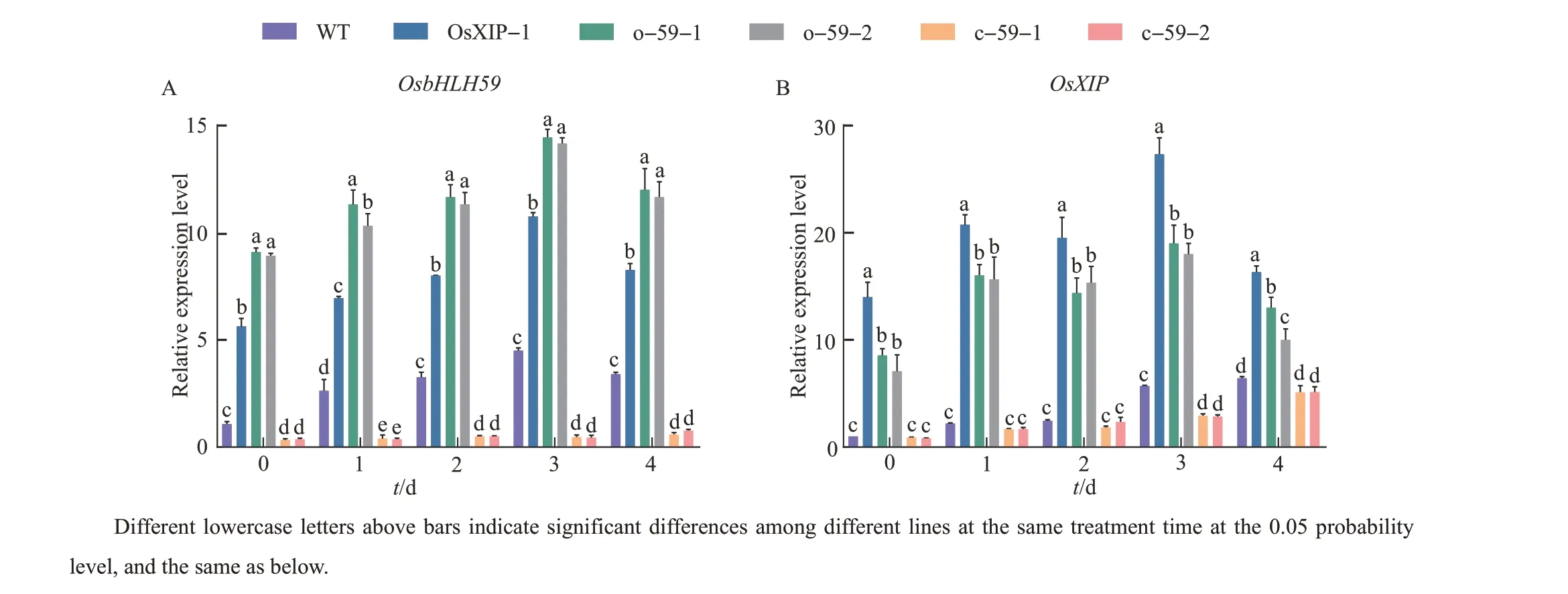
Fig.4 Relative expression of OsbHLH59 and OsXIP in the overexpression lines, mutant lines and WT after the BPH treatment
2.6 Expression patterns of defense genes in different transgenic rice plants after the BPH treatment
The expression patterns of defense-related genes inOsbHLH59overexpression lines and mutant lines treated with BPH were investigated.The expression levels of defense-related genes in the five lines infected by BPH were compared by qRT-PCR analysis (Fig. 5).After the BPH treatment,the expression levels ofOsPR1in different lines gradually increased with the extending treatment time. The expression levels ofOsPR1inOsXIPoverexpression line andOsbHLH59overexpression lines increased more significantly than WT andOsbHLH59mutant lines (Fig. 5A).The expression levels ofOsPR1bin WT did not change significantly but decline slowly inOsbHLH59mutant lines from 1 d to 3 d. The expression levels ofOsPR1binOsbHLH59overexpression lines andOsXIPoverexpression line were significantly increased(Fig. 5B). As forOsNH1, the expression levels ofOsNH1in WT andOsbHLH59mutant lines were similar and did not change significantly. The expression levels ofOsNH1inOsbHLH59overexpression lines andOsXIPoverexpression line increased significantly with the increase of the BPH treatment time (Fig. 5C). As shown in the Fig. 5D, the expression levels ofOsAOS2inOsXIPandOsbHLH59overexpression lines increased with the increase of treatment time. The expression levels ofOsAOS2in WT andOsbHLH59mutant lines continued to increase, but they were always lower than the overexpression lines. In summary,BPH treatment can affect the expression levels of all tested defense-related genes, andOsXIPandOsbHLH59overexpression lines showed higher expression levels of defense genes,as well as insect resistance.
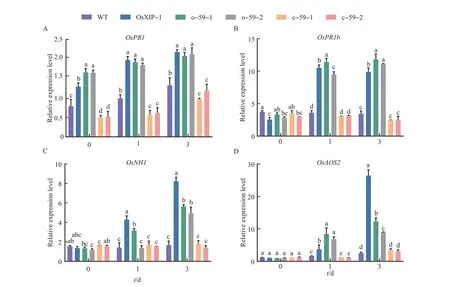
Fig.5 Relative expression levels of defense-related genes in the overexpression lines,mutant lines and WT after the BPH treatment
2.7 Analysis of H2O2 content and antioxidant enzyme activity in rice after the BPH treatment
The H2O2content and the activities of antioxidant protective enzymes CAT and POD in rice were measured on the 0, 1, 2, 3, and 4 d after the BPH treatment to study the physiological and biochemical changes ofOsbHLH59overexpression and mutant lines. In general, after being infested by BPH, the H2O2content, CAT and POD activities of the six rice lines all increased to varying degrees.The measuring results of H2O2content showed that with the treatment time increasing, the H2O2contents in WT andOsbHLH59mutant lines continued to increase,whileOsXIPandOsbHLH59overexpression lines increased, but the increase rates were relatively slow(Fig. 6A). After 3 d of treatment, the H2O2contents inOsXIPandOsbHLH59overexpression lines were still slightly lowered than other lines. CAT (Fig. 6B) and POD(Fig.6C)activity assay results showed that after being infested by BPH, the CAT and POD activities of the four lines all increased to varying degrees.Compared with WT andOsbHLH59mutant rices,the activities of CAT and POD inOsXIPandOsbHLH59overexpression lines increased faster,with higher activities and longer durations.
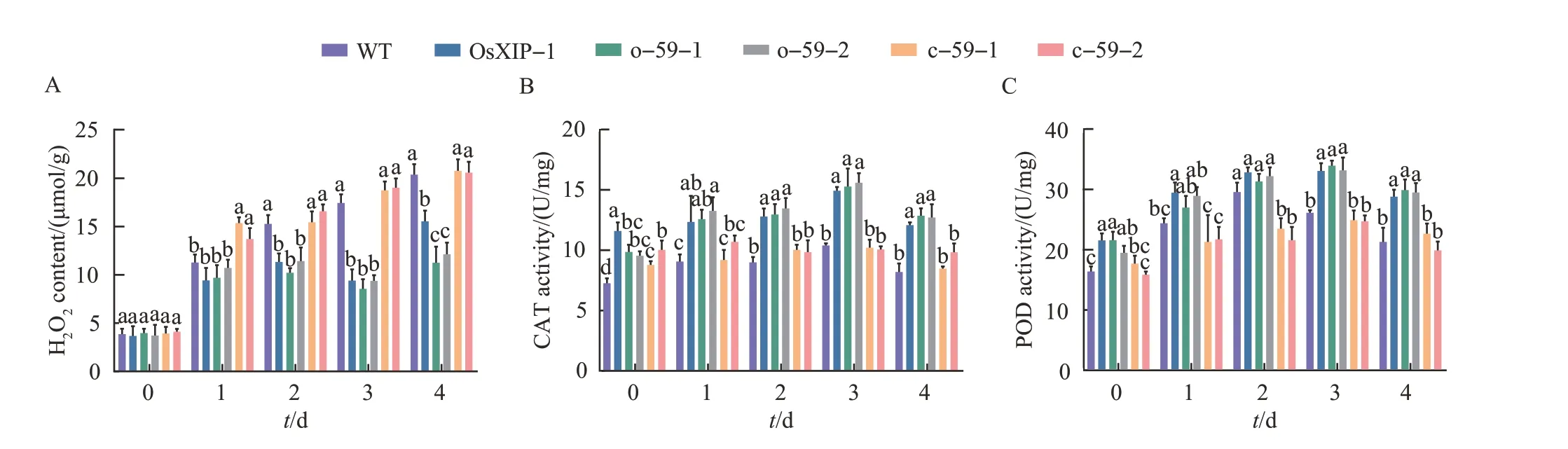
Fig.6 Changes in H2O2 contents and antioxidant enzyme activities in overexpression lines, mutant lines and WT after the BPH treatment
3 Discussion
3.1 OsbHLH59 transcription factor gene had transcriptional activation effect on rice xylanase inhibitor OsXIP
OsXIPencoding XIP-type xylanase inhibitor is a stress-responsive gene in rice. TOKUNAGA et al.reported that the expression level ofOsXIPwas increased when rice seeds were treated by citrate,sodium chloride, exogenous ascorbic acid (ASA) and wounding[8]. Herbivore infestation, wounding and methyl jasmonate (MeJA) treatments enhanced mRNA levels and protein levels of OsXIP[9]. Using yeast one-hybrid (Y1H) screening and chromatin immunoprecipitation (ChIP)-PCR analysis, we found thatOsbHLH59could bind to the promoter ofOsXIP,and regulated its expression level in rice plants to response herbivore stress[9].
In this study, we obtainedOsbHLH59overexpression transgenic rice plants and mutant lines through transgenic technology. The results showed that the overexpression ofOsbHLH59significantly increased the expression level ofOsXIPin rice, while the knockout ofOsbHLH59had little effect on the expression level ofOsXIP.
There are many bHLH transcription factors, and they play important regulatory roles in plant growth,development, morphogenesis and stress resistance[24-26].The bHLH transcription factor was documented to regulate the response of plants to stress by regulating the expression levels of downstream stress-responsive genes. XU et al. found that overexpression ofVabHLH1orVvbHLH1could increase the resistance of grapes to low temperature by inducingcold regulated(COR) genes[27]. In this experiment,OsbHLH59transcription factor gene had an effect on the expression ofOsXIP.
3.2 OsbHLH59 enhanced the insect resistance of rice by regulating the expression of OsXIP
In our study, the overexpression ofOsbHLH59enhanced the expression level ofOsXIP, which inOsbHLH59overexpression lines was higher than that in WT. After the BPH treatment, the expression levels ofOsXIPandOsbHLH59in theOsbHLH59overexpression andOsXIPoverexpression lines increased significantly,and showed the same trend.The mortality rates ofOsbHLH59overexpression andOsXIPoverexpression lines were significantly lower than WT after the BPH treatment. For the results of agronomic traits (plant height and number of leaf blades), it could be seen that the effect of BPH treatment onOsbHLH59overexpression andOsXIPoverexpression lines was much lighter than that of WT, while the effect onOsbHLH59mutant lines was more serious than that of WT. Correspondingly, the experiments showed thatOsbHLH59overexpression andOsXIPoverexpression lines might enhance the resistance of rice to BPH by reducing the feeding preference of BPH.
As a signal molecule, H2O2can induce plants’response to stress[28]. In our experiment, the H2O2content in the six different lines all increased after the BPH treatment. With the accumulation of H2O2in the plants, antioxidant enzyme activity also increased after the BPH treatment. Antioxidant enzymes can clear excess H2O2which could cause oxidative damage to plant cells[29]. In this study,compared with WT, high activities of CAT and POD were found in bothOsbHLH59overexpression andOsXIPoverexpression lines, indicating thatOsbHLH59andOsXIPoverexpression lines had stronger abilities to eliminate excess H2O2compared with WT.
OsPR1andOsPR1barePRgenes, closely related to plant defense[30]. In this study, after the BPH treatment, higher expression levels ofPRgenes were found inOsbHLH59overexpression andOsXIPoverexpression lines compared with WT.Studies have reported that salicylic acid (SA) and jasmonic acid (JA) are important signal molecules in plants and participate in the signal transduction of stress-responsive expression[31]. TOKUNAGA et al.found thatOsXIPwas induced by SA and MeJA treatments, indicating thatOsXIPmay participate in plant defense through SA and JA pathways[8].OsNH1is a homolog of genes in the SA-mediated defense system ofArabidopsis.OsAOS2is the gene of key enzyme in JA synthesis pathway[32]. After the BPH treatment,OsbHLH59overexpression andOsXIPoverexpression lines also showed higher expression levels ofOsNH1andOsAOS2compared with WT,while theOsbHLH59mutant lines showed lower expression levels ofOsNH1andOsAOS2compared with WT. This indicated that theOsbHLH59transcription factor gene may regulate the expression levels ofOsXIPand some defense-related genes(OsPR1,OsPR1b,OsNH1,OsAOS2), andOsbHLH59overexpression andOsXIPoverexpression lines may increase rice resistance to BPH through JA and SA signaling pathways[8].
4 Conclusions
In summary,OsbHLH59overexpression andOsXIPoverexpression transgenic plants had higher resistance to BPH. Overexpression of transcription factor geneOsbHLH59might enhance the expression ofOsXIPand other defense-related genes (OsPR1,
OsPR1b, OsNH1, OsAOS2), and mediate rice resistance to BPH.

