Molecular characterization and expression patterns of nuclear androgen receptors in the ovoviviparous black rock fish Sebastes schlegelii
Shaojing Yan, Jiahui Chen, Likang Lyu, Xiaojie Wang, Yijia Yao, Haishen Wen, Xin Qi
Key Laboratory of Mariculture, Ministry of Education, Ocean University of China, Qingdao, 266110, China
Keywords:
Androgen receptor
Gene cloning
Tissue expression
In situ hybridization
Sebastes schlegelii
A B S T R A C T
nARs are ligand-activated transcription factors associated with gonadal development and reproductive regulation. In this study, two nuclear androgen receptor genes (ara and arb) were cloned from an ovoviviparous teleost,black rock fish (Sebastes schlegelii). The phylogenetic analysis of nARs showed that nARα and nARβ clustered into teleost nARα and nARβ, respectively, but differed from the nARs of tetrapods. Four module domains of the nuclear receptor superfamily are present in the black rock fish nARs, an N-terminal domain (NTD), a DNA-binding domain (DBD), a hinge region (HR) and a ligand-binding domain (LBD). Among the four domains,only the DBD and LBD of nARα and nARβ are relatively conserved. Tissue distribution analysis revealed that ara was mainly expressed in the gonad, muscle, intestine and kidney in males and in female, while arb was mainly detected in the gonad, followed by the intestine, kidney and head kidney. During the gonadal development process, the expression levels of ara and arb were significantly decreased from the regenerating to late stages of spermatogenesis and significantly increased in the degeneration stage in the testis (P<0.05). In the ovary, the expression levels of ara and arb were not significantly different in different stages. In situ hybridization revealed that in black rock fish, both ara and arb transcripts were localized to the Sertoli cells of the testis in black rock fish.In general, the present study is the first to characterize and analyse the expression of nuclear androgen receptors,including ara and arb, in black rock fish.
1.Introduction
Androgens are a class of steroid hormones involved in several physiological processes, including sexual maturation, sexual differentiation, and spermatogenesis (Hiort, 2002). Their physiological effects are mainly mediated by the nuclear androgen receptors (nARs), which are ligand-dependent transcription factors that belong to the nuclear receptor superfamily (Schuppe et al., 2020). nARs are mainly distributed in the cytoplasm and bind to cytoskeletal proteins, heat shock protein 70 and heat shock protein 90. In the presence of their ligands, nARs change conformation and release heat shock proteins. They then enter the nucleus as transcription factors that bind to the androgen response elements in target gene promoters to regulate transcription (Del Rosso et al., 2020).
Only oneargene copy is present in amphibians, birds, and mammals(Douard et al., 2008), but most teleosts have two copies due to the unique process of genome duplication during evolution. The first successful cloning of theargene from teleosts was carried out with Japanese eels (Anguilla japonica) in 1999 (Ikeuchi et al., 1999; Todo et al.,1999). Thereafter, the coding sequences ofargenes have been characterized from fishes in Cypriniformes, Perciformes, Siluriformes and other orders (Huang et al., 2011; Jørgensen et al., 2007; Pu et al., 2013).Similar to other nuclear receptors, nAR is composed of four modular domains: an N-terminal domain (NTD), a DNA-binding domain (DBD), a hinge region (HR) and a ligand-binding domain (LBD) (Gelmann, 2002).
Black rock fish (Sebastes schlegelii) is an important commercial marine species which is mainly distributed in the coastal waters of China, Japan and Korea. Black rock fish are important fish bred in cage cultures and have the characteristics of fast growth and low temperature resistance(Zhang et al., 2020). Black rock fish exhibit a special reproductive strategy in which the gonadal development of males and females is not synchronized. In males, spermatogenesis begins in late July, and sperm mature in November. The mature male sperm are emitted into the ovary of the female through its modified urogenital papillae. Sperm are stored in the ovarian cavity after mating (Wang et al., 2021). Females undergo ovoviviparous reproduction, and the ovary matures in April of the following year. Females normally mate multiple times with different males from late November to early December and then store the sperm for more than four months until fertilization. The embryo developsin situin the ovary until parturition (Liu et al., 2019). Two of the important steps of this process, mating and parturition, are quite crucial in aquaculture. Mating behaviour significantly affects the storage of sperm,which in turn affects the number of offspring produced (Cardozo et al.,2020). On the other hand, the pregnant fish are easily affected by changes in the environment or stimulation caused by fright, leading to a preterm parturition or dystocia. In mammals, the androgen participates in both reproductive steps, and plays important regulatory roles.Accordingly, to investigate the role of androgen in the mating and parturition, in the present study, we cloned the coding sequences of botharsubtypes, and then, we analysed the protein domains, evolutionary relationships and mRNA expression patterns ofaraandarbduring different gonadal developmental stages and three perinatal time points,namely, before, during and after the delivery of fries, in black rock fish.
2.Materials and methods
2.1.Ethics statement
All the animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee of Ocean University of China. The field studies did not involve endangered or protected species.The experimental fish were anaesthetized with 100 ng/ml 3-aminobenzoate methanesulfonic acid (MS-222) to minimize pain before sacrifice and treatment.
2.2.Fish and sampling
The fish used in the experiment came from the aquaculture population maintained in marine cages offshore of Rushan, Shandong, China(36.92 N, 121.54 E). A total of 43 adult black rock fish were selected randomly for the experiment (over 3 years old, body weight: 1.0 ±0.4 kg, body length: 24.3 ±1.3 cm). All the animals were sampled after being housed for 2 days in indoor cement pools with a culvert system.Whole tissue samples including heart, liver, spleen, stomach, kidney,head kidney, intestine, gills, muscle, skin, brain, pituitary gland, testis and ovary samples, were collected from three males and three females in September 2020 and October 2020, respectively. Three testis samples from each stage were collected in July 2020, September 2020, December 2020, and January 2021, which corresponded to the regenerating (stage II), early spermatogenesis (stage III), and late spermatogenesis (stage Ⅳ)stages. Three ovary samples from each stage were collected in July 2020,October 2020, December 2020, and March 2021, which corresponded to the oogenesis (stage II), previtellogenesis (stage III), and vitellogenesis(stage Ⅳ) stages. Thirteen female individuals were sampled at time points of 24 h before delivery (n =5), during delivery (n =3), and 24 h after delivery (n =5) in April 2021. Parts of the testis samples collected during the degeneration (stage Ⅴ) stage were fixed in 4% paraformaldehyde for thein situhybridization.
2.3.Total RNA extraction and reverse transcription
Total RNA was extracted from testis, ovary and other tissues using TRIzol reagent (Invitrogen, America) according to the reagent instructions. RNA quantity and purity were assessed by a Biodropsis BD-1000 nucleic acid analyser (OSTC, China) and electrophoresis using a 1% agarose gel. cDNA was prepared using the Prime ScriptTM RT real Time kit with gDNA Eraser (Perfect Real Time) (TAKARA, Japan) according to the reagent instructions.
2.4.Molecular cloning and sequence analysis
Based on the genomic and transcriptome data, the open reading frames (ORFs) of thearaandarbgenes were predicted. Primers for cloning foraraandarbwere designed using Primer5 software. All the primers used in the present study are listed in Table 1. The 2 ×Phanta Max Master Mix (Dye Plus) (Vazyme, China) was used for cloning and gonad cDNA was used as a template. The PCR product was purified and cloned into the pEASY-T1 vector (TransGen Biotech, China) for subsequent sequencing.

Table 1Primers sequences used for ORF cloning, ISH and qPCR.
2.5.Sequence and phylogenetic analysis
ClustalX 2.1 and ESPript 3.0 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi) were used to perform multiple alignments of the amino acid sequences based on the AR amino acid sequences of several species. A phylogenetic tree was reconstructed from the multiple alignments of the amino acid sequences with the neighbour-joining method using MEGAX. The values on the trees represent bootstrap scores of 1,000 iterations,indicating the credibility of each branch. SWISS-Model (https://swi ssmodel.expasy.org/) was used to model the AR tertiary structure.
2.6.Quantitative real-time PCR
Thearaandarbexpression levels in the ovary, testis and other different tissues of black rock fish were measured by quantitative realtime PCR (qPCR). According to the reagent instructions, a ChamQTM SYBR® Color qPCR Master Mix (High Rox Premixed) kit (Vazyme,China) was used for qPCR. The qPCR program was as follows: 95 ℃ for 30 s; 40 cycles of 95 ℃ for 10 s and 60 ℃ for 30 s. The threshold cycle(CT) values were measured for each sample, and 18S rRNA was used as the reference gene. qPCR was run in triplicate to con firm the results.
2.7.In situ hybridization (ISH)
According to the reagent instructions, the 2 × Phanta® Max Master Mix (Dye Plus) kit (Vazyme, China) was used for PCR, the testis cDNA of testis was used as a template. According to the reagent instructions, the DIG RNA Labeling Kit (SP6/T7) (Roche, Switzerland) was used to synthesize antisense or sense probes for the in vitro transcription ofaraandarbmRNA.
Testes collected from black rock fish were fixed in 4% paraformaldehyde (in PBS, pH 7.4) for 4–6 h, embedded in paraffin and sectioned at thicknesses of 6–7 μm. The tissue sections were then deparaffinized, rehydrated, and successively immersed in the following RNase-free solutions successively: 0.1 M HCl (10 min) and 10 μg/mL proteinase K (in PBS) (37 ℃, 2 min). The tissue sections were then prehybridized at 42 ℃ for 1 h, and then hybridized with DIG-labelled riboprobes diluted in hybridization buffer at 58 ℃ overnight in a wet box. After overnight hybridization (12–16 h), the sections were washed in preheated 2 × SSC, 1 × SSC, 0.2 × SSC, and 0.1 × SSC (55 ℃, 15–30 min for each step). According to the reagent instructions, alkalinephosphatase-conjugated anti-DIG Fab fragments (1:400 dilution) and nitro-blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP) substrate (Roche, Switzerland) were used to detect the DIG-labelled probes.
2.8.Statistical analysis
All the data are shown as the mean ±standard error of the mean(SEM). Data analysis was performed by independent-samplest-test and one-way ANOVA followed by Tukey’s and Sidak’s multiple range tests.Differences were considered significant atP < 0.05. All the statistical procedures were performed using SPSS 19.0 software (SPSS, Chicago, IL,USA), and all the graphs were generated with GraphPad Prism 9(GraphPad Software, USA).
3.Results
3.1.Molecular cloning and characterization of ara and arb
Twoargenes (araandarb) were identified in black rock fish. The ORF ofarais 1,941 bp long and encodes a precursor protein of 646 amino acids (accession number: OL406398, Supplementary 1), while the ORF ofarbis 2,364 bp long and encodes a 766 amino acids precursor protein(accession number: OL406399, Supplementary 2).In silico,the physicochemical properties of the AR proteins were determined as follows:the molecular weights of ARα and ARβ were 71,477.19 Da and 85,622.92 Da, respectively, and the theoretical isoelectric points of ARα and ARβ were 6.47 and 6.19, respectively.
Based on the amino acid alignment, the vertebrate AR proteins were determined to contain 4 modular domains: an NTD, a DBD, an HR and an LBD (Fig. 1A). The overall amino acid identity of the two ARs of black rock fish was 63.22%, and the sequence identities of the NTD, DBD and LBD were 61.54%, 74.70% and 69.39%, respectively. The amino acid sequence of black rock fish ARα was homologous to Nile tilapia ARα(68.88%), zebra fish ARα (63.22%), human ARα (57.21%), and mouse ARα (60.05%). Furthermore, black rock fish ARβ was found to be homologous to Nile tilapia ARβ (70.29%), zebra fish ARβ (58.21%), human ARβ (65.27%), and mouse ARβ (66.27%) (Fig. 1A). The 3D structures of the two AR proteins were constructed (Fig. 1B and C). The phylogenetic analysis of vertebrate ARs was performed. As shown in Fig. 2, ARα and ARβ of black rock fish were clustered with teleost ARα and ARβ,respectively, and differed from the ARs of other vertebrates.
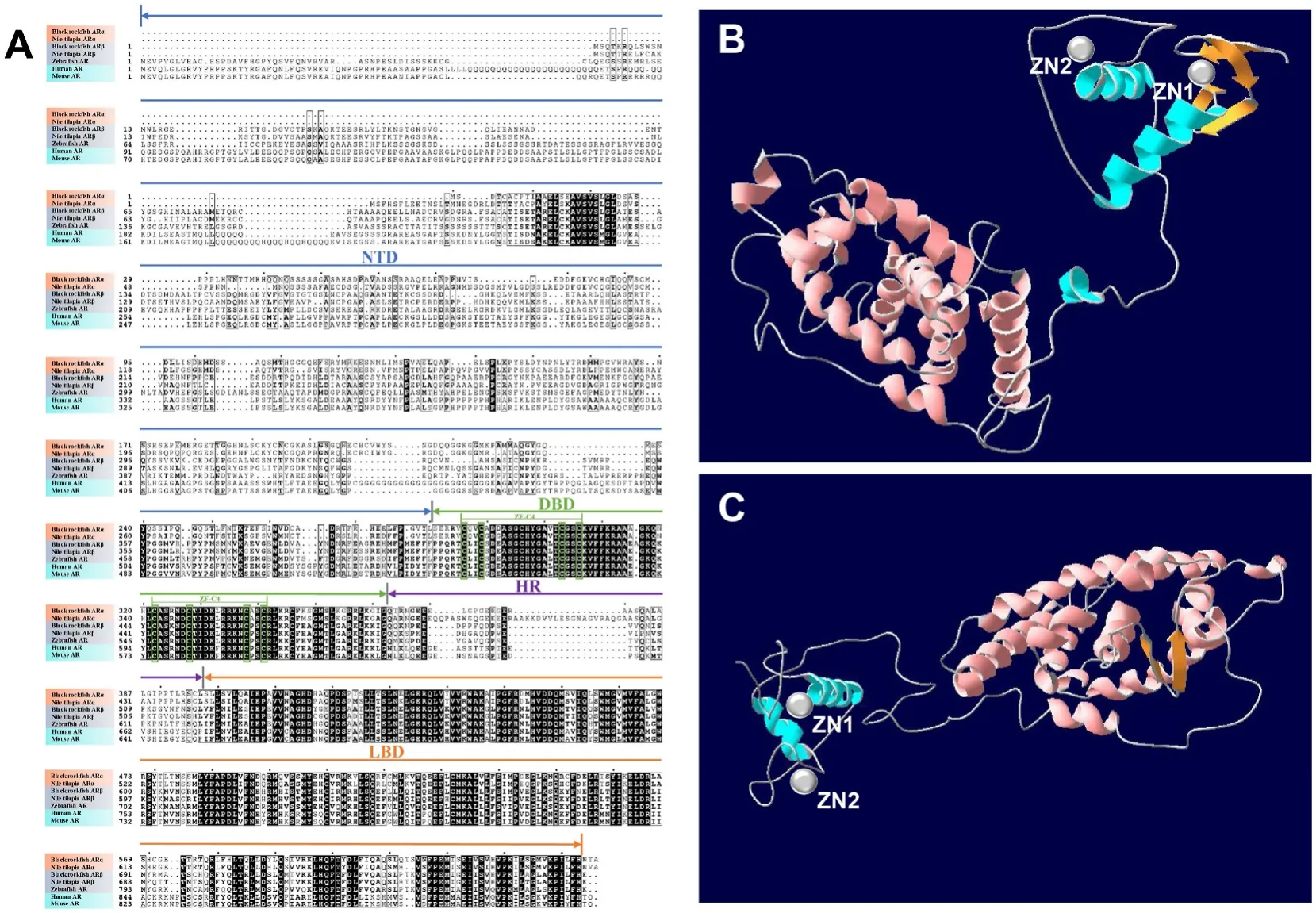
Fig. 1.Multiple comparisons of the amino acid sequences of the amino acid sequences of black rock fish ARα and ARβ, Nile tilapia ARα and ARβ, zebra fish, human,and mouse. Black rock fish ARα NTD (1–280), DBD (281–362), HR (363–397), and LBD (398–643); black rock fish ARβ NTD (1–403), DBD (404–486), HR (487–520),and LBD (521–765); Nile tilapia ARα NTD (1–300), DBD (301–382), HR (383–441), and LBD (442–685); Nile tilapia ARβ NTD (1–401), DBD (402–483), HR(484–517), and LBD (518–762); zebra fish AR NTD (1–506), DBD (507–588), HR (589–621), and LBD (622–867); human AR NTD (1–554), DBD (555–636), HR(635–672), and LBD (673–918); mouse AR NTD (1–533), DBD (534–615), HR (616–651), and LBD (652–897). The 3D protein structures of ARα (B) and ARβ (C) of black rock fish were constructed by the SWISS-MODEL online tool. ZN: zinc finger, NTD: N-terminal domain, DBD: DNA-binding domain, HR: hinge region, LBD:ligand-binding domain.
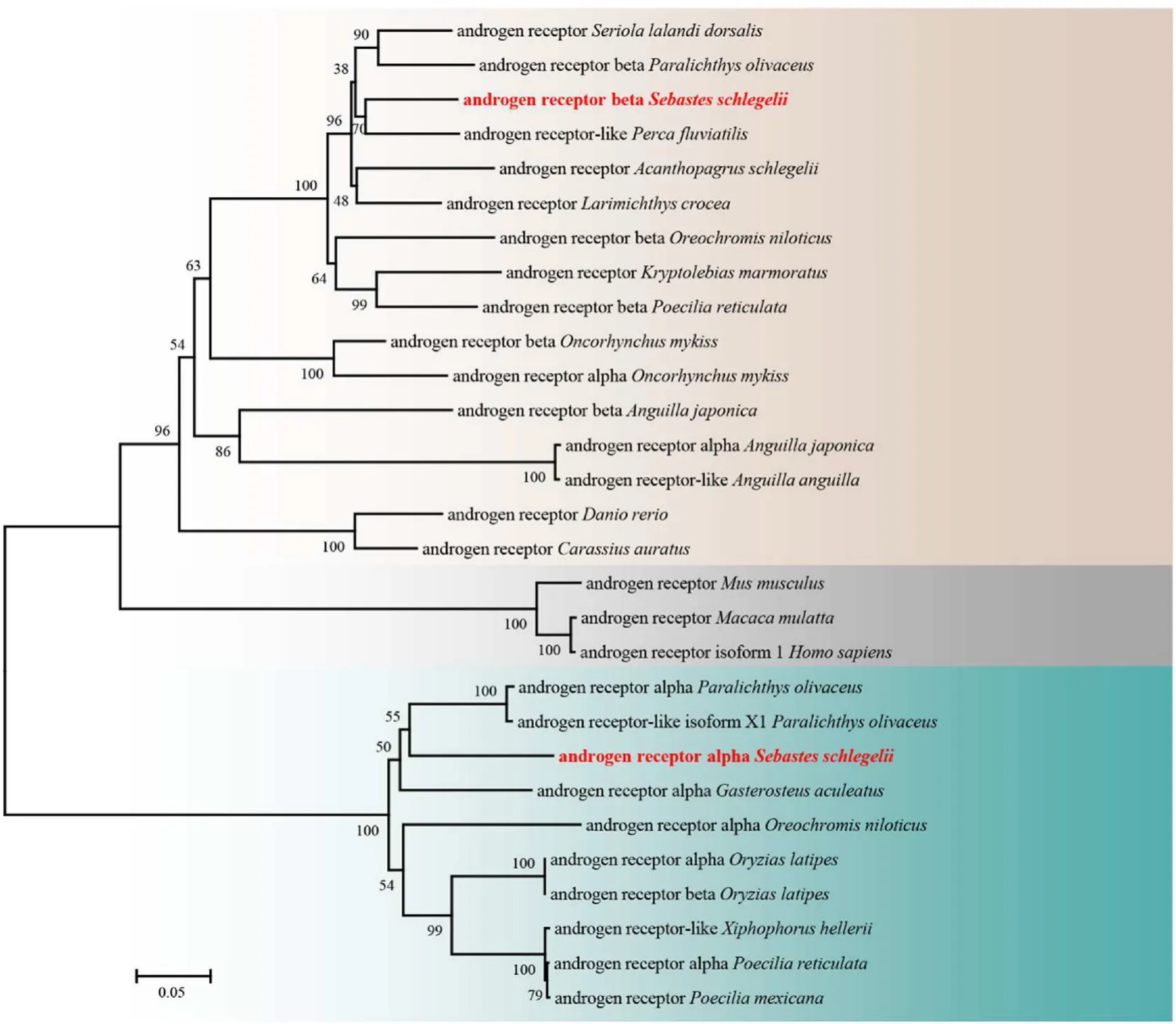
Fig. 2.A phylogenetic tree was constructed by MEGA 6 using the neighbour-joining method. The data were resampled with 1,000 bootstrap replicates. The GenBank accession numbers of the selected AR sequences are as follows: Seriola lalandi dorsalis (XP_023266520.1), Paralichthys olivaceus (XP_019968003.1), Perca fluviatilis(XP_039633381.1), Acanthopagrus schlegelii (AAO61694.1), Larimichthys crocea (NP_001290296.1), Oreochromis niloticus (BAB20082.1), Kryptolebias marmoratus(NP_001316288.1), Poecilia reticulata (XP_008425737.1), Oncorhynchus mykiss (NP_001117657.1), Oncorhynchus mykiss (NP_001117656.1), Anguilla japonica(BAA83805.1), Anguilla japonica (BAA75464.1), Anguilla anguilla (XP_035266767.1), Danio rerio (NP_001076592.1), Carassius auratus (AAM09278.1), Mus musculus(NP_038504.1), Macaca mulatta (NP_001028083.1), Homo sapiens (NP_000035.2), Paralichthys olivaceus (AGV29985.1), Paralichthys olivaceus (XP_019968003.1),Gasterosteus aculeatus (NP_001254620.1), Oreochromis niloticus (NP_001266542.1) Oryzias latipes (BAI22839.1), Oryzias latipes (NP_001116383.1), Xiphophorus hellerii(XP_032410588.1), Poecilia reticulata (XP_008417846.1), and Poecilia mexicana (XP_014860861.1).
3.2.Tissue distribution of ara and arb
qPCR was performed to measure the expression levels of bothargenes in different tissues of male and female rock fish. The results showed thatarawas highly expressed in various tissues and mainly expressed in the gonad, muscle, intestine and kidney in males and in female. The expression level ofarain the skin and brain was very low,and no expression was detected in the heart, spleen, stomach, or gill(Fig. 3A). The expression level ofarbwas mainly detected in the gonad,followed by the intestine, kidney and head kidney (Fig. 3B). In male liver, muscle, skin and gonad, the expression level ofarawas signi ficantly higher (P<0.05) than that in female (Fig. 3A). Correspondingly,the expression level ofarbin liver, stomach, intestine, kidney and gonad in male was significantly higher (P<0.05) than that in female (Fig. 3B).
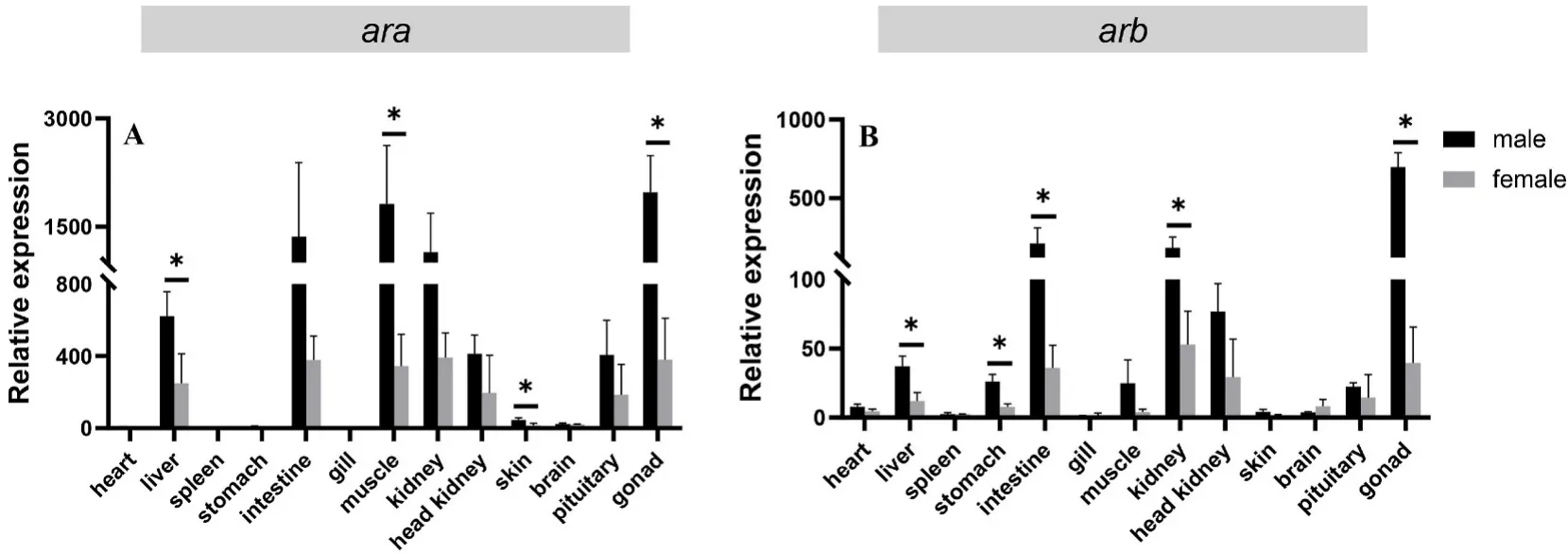
Fig. 3.Gene expression patterns of ara and arb in different tissues of male and female black rock fish. Relative expression levels of ara (A) and arb (B) in different organs (heart, liver, spleen, stomach, intestine, gill, muscle, kidney, head kidney, skin, brain, pituitary, testis, and ovary). The X axis indicates different tissues. The Y axis indicates the relative expression normalized to 18S RNA. The data are presented as the mean ±S.E.M. (n =3). The data were analysed by independent-samples ttest. Asterisks indicate significant differences (P < 0.05).
3.3.Expression of ara and arb in different gonad developmental stages and delivery periods
Based on a previous study (Lyu et al., 2021; Wang et al., 2021), in this study, the developmental stages of the testis and ovary were classified into 5 and 7 stages, respectively, in the present study. The different developmental stages of the testis include the original testis stage (stage I), regenerating (stage II), early spermatogenesis (stage III), late spermatogenesis (stage Ⅳ), and degeneration (stage Ⅴ) stages. The different developmental stages of the ovary include the original ovary stage (stage I), oogenesis (stage II), previtellogenesis (stage III), vitellogenesis (stageⅣ), oocyte maturation (stage Ⅴ), embryonic stage (gestation) (stage Ⅵ),and regressed (stage Ⅶ) stages.
The results showed thataraexpression was significantly downregulated in stage IV testes compared with stage II and stage V testes(P<0.05) (Fig. 4A). Moreover, the expression ofarbin stage V testis was significantly higher than that in other stages (P<0.05), and the expression ofarbin stage IV testis was significantly lower than that in other stages (P<0.05) (Fig. 4C). However, there were no significant differences in eitheraraorarbexpression during the different stages of ovary development (Fig. 4B, D). The expression levels ofaraandarbof 24 h after delivery were significantly lower than 24 h before delivery(P<0.05) (Fig. 5).
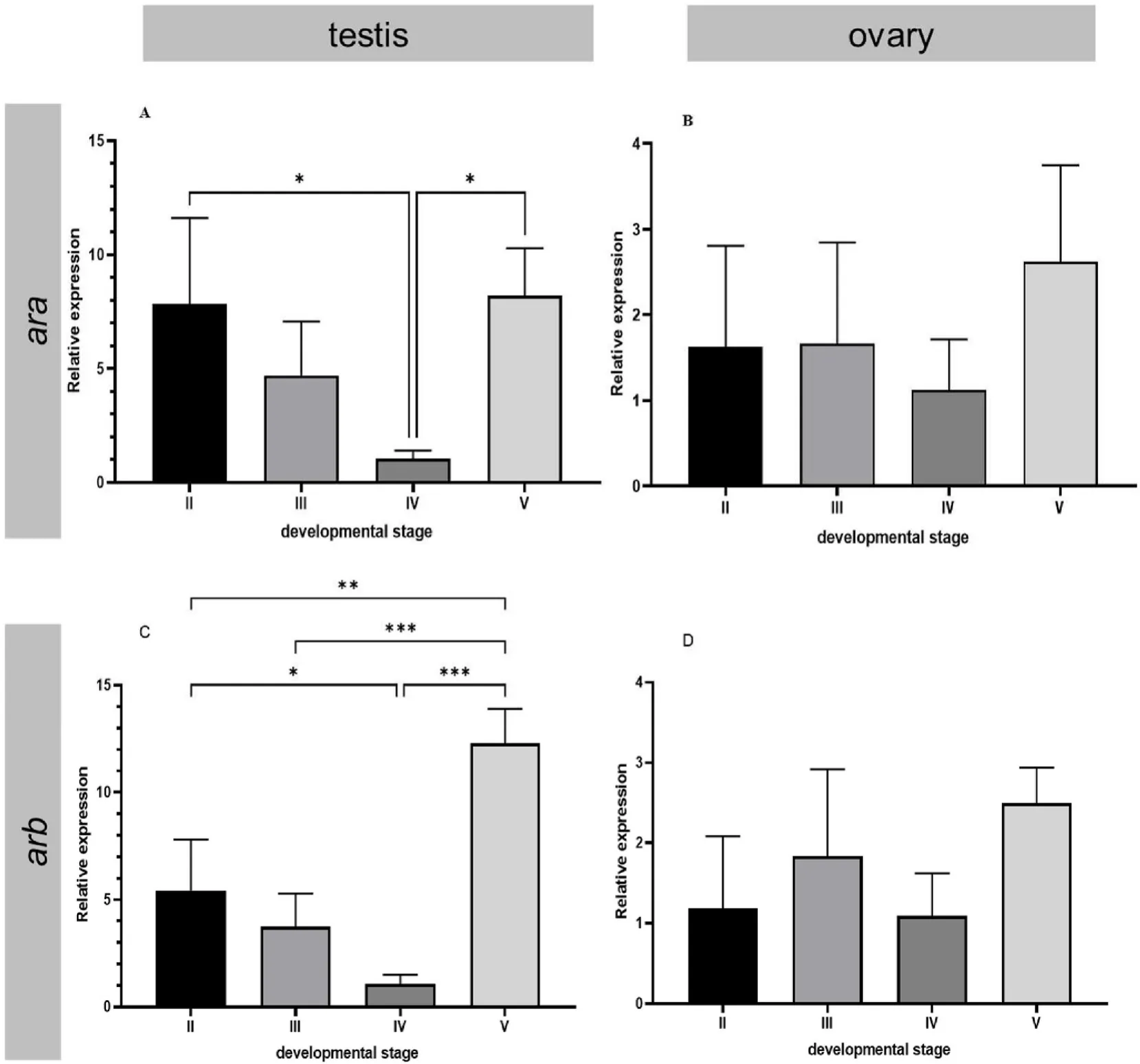
Fig. 4.Relative expression levels of ara and arb in the development stage of the testis and ovary in black rock fish. The different developmental stages of the testis include the regenerating (stage II), early spermatogenesis (stage III), late spermatogenesis (stage Ⅳ), and degeneration (stage Ⅴ) stages. The different developmental stages of the ovary include the oogenesis (stage II), previtellogenesis (stage III), vitellogenesis (stage Ⅳ), and oocyte maturation (stage Ⅴ) stages. The X axis indicates different developmental stages. The Y axis indicates the relative expression normalized to 18S RNA. The data are presented as the mean ±S.E.M. (n =3). The data were analysed by one-way ANOVA followed by Tukey analysis. One, two and three asterisks indicate significant differences (P < 0.05, 0.01 and 0.001, respectively).
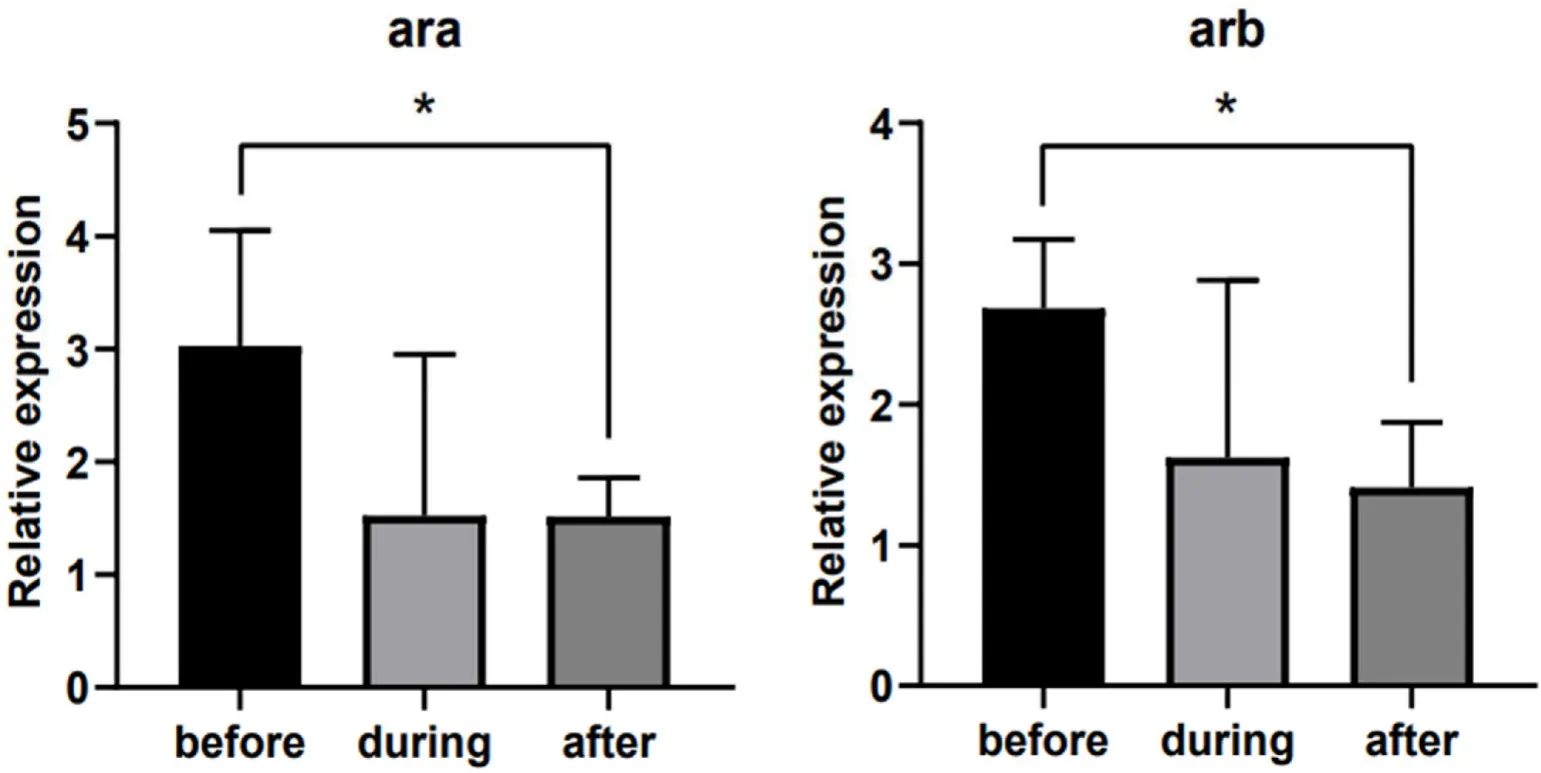
Fig. 5.Relative expression levels of ara and arb in black rock fish in different delivery periods. The different delivery periods included 24 h before delivery (before, n =5), during delivery (during, n =3), and 24 h after delivery (after, n =5). The X axis indicates different delivery periods. The Y axis indicates the relative expression normalized to 18S RNA. The data are presented as the mean ±S.E.M. The data were analysed by one-way ANOVA followed by Sidak analysis.Asterisks indicate significant differences (P< 0.05).
3.4.Localization of ara and arb in testis
According to the qPCR results, stage V testis was used foraraandarbmRNA localizationviaISH. The results showed that positive ofaraandarbsignals in the Sertoli cells which surrounding the lobular cavity(Fig. 6A, B, D, E) compared with the negative control (Fig. 6C, F).

Fig. 6.Localization of ara and arb mRNA in testis of black rock fish via in situ hybridization. Antisense probe signals of ara and arb presented in Sertoli cells (A, D),respectively. Positive signals in Sertoli cells are indicated by arrows (B, E). Negative signal with sense probes (C, F). Scale bars are shown in the corner of each figure.
4.Discussion
In the present study, twoarsubtype genes were identified in black rock fish, and these results were similar to the findings in rainbow trout(Oncorhynchus mykiss) (Takeo & Yamashita, 1999); Japanese eel (Ikeuchi et al., 1999); mosquito fish (Gambusia affinis) (Sone et al., 2005); and stickleback (Gasterosteus aculeatus) (Olsson et al., 2005), but different from the findings in zebra fish (Olsson et al., 2005), black sea bream(Acanthopagrus schlegelii) (Touhata et al., 1999) and large yellow croaker(Larimichthys crocea) (Pu et al., 2013), which have only one gene subtype. Similar to their orthologues, the black rock fish nARs contained all four domains, including an NTD, a highly conserved DBD, an HR and a C-terminal LBD. The NTD is the first effector region of AR and is largely responsible for transactivation (Gelmann, 2002); accordingly, the NTD is less conserved between species (Pu et al., 2013. The DBD and LBD were found to be highly conserved by protein functional domain comparison. The DBD consists of 2 or 3 α helices and 2 zinc fingers, and its main function is DNA binding (Thornton & Kelley, 1998). In addition,AR binds to androgen response elements (AREs) through its DBD,anchoring AR dimers to androgen response elements in gene promoters,thus causing effective gene transcription. The LBD is a ligand binding region with highly specific functions. The LBD of ARα and ARβ consists of 14 and 13 α structures, respectively. It can recruit SRC1 through the AF-2 modulo to improve the transcription output function of AR (Bevan et al., 1999; Brinkmann et al., 1999; Grad et al., 1999).
Phylogenetic analysis showed that ARα and ARβ of the black rock fish were clustered into two separate groups. Interestingly, species containing only one type of AR protein were separately clustered into the ARα or ARβ groups, separately. Among these species, Atlantic mollies (Poecilia mexicana) and green swordtail (Xiphophorus hellerii) were clustered into the ARα group, and gold fish (Carassius auratus), zebra fish and yellowtail amberjack (Seriola lalandi dorsalis) were clustered into the ARβ group.The loss of the second AR gene from these species could be caused by the whole genome duplication (Hossain et al., 2008), while the subtype which was lost may be varied during the evolution. Our data provided a different result from a previous study, in which all species with one AR subtype were clustered into the ARβ group(Zou et al., 2019). The results may differ because of the number of AR sequences used in building the phylogenetic tree. Indeed, the conclusion that the teleost ARβ type could be the canonical form, whereas ARα may be a duplicated copy generated later in evolution and lost in some fish species(Hossain et al., 2008),possibly lacks sufficient evidence.
The expression patterns ofaraandarbwere investigated by qPCR. In the liver and gonad, the male showed a significantly higher level of bothargenes expression than the female. It is also significant that the expression ofarain male muscle and skin is higher than the female counterpart. In addition,arbwas detected to be significantly higher in male stomach, intestine and kidney than female. These results showed similar expression patterns in zebra fish (Hossain et al., 2008). The distribution ofarais widespread and its highly expressed in important tissues. However, the range ofarbis relatively limited and its expression level is low. It may indicate that the different functions of the two gene subtypes. The results were consistent with those of olive flounder(Paralichthys olivaceus) (Zou et al., 2019), zebra fish (Hossain et al.,2008), and rainbow trout (Takeo & Yamashita, 1999). This coincidence indicates a conserved function of teleost AR. On the other hand, the different tissue expression patterns may be related to the different affinity of the androgen receptor for androgen. In kelp bass (Paralabrax clathratus), two nARs, termed as kbAR1 and kbAR2, possess different binding affinities for natural androgens. KbAR1 expressed in brain tissue has a high affinity for the T binding site, while kbAR2 expressed in ovarian cytoplasm has a higher affinity for dihydrotestosterone (DHT)(Sperry & Thomas, 1999). In Atlantic croaker (Micropogonias undulatus),AR2 has a broader affinity for androgens and a greater affinity than AR1 for structurally diverse androgens (Sperry & Thomas, 2000). Differences in affinity between ARα and ARβ for three androgen ligands have also been observed in Japanese eel (Ikeuchi et al., 1999). The different affinities of fish nARs for androgens suggest that nARs mediate different physiological roles in different tissues. In addition, one study identified two key substitutions in ARα in teleosts by site-directed mutagenesis and protein–ligand interaction prediction. These substitutions result in structural differences between ARα and ARβ, which may lead to different transcriptional activation responses in cells (Ogino et al.,2016).
In black rock fish testis, the mRNA expression of bothargenes in Leydig-like, Sertoli and peritubular cells is similar to the findings in fish,birds and mammals (de Waal et al., 2008; Dornas et al., 2008; Kotula et al., 2000; Leska et al., 2012; Liu et al., 2009). In zebra fish testis,arwas found to be expressed in support cells that are in contact with early spermatogonial cells (de Waal et al., 2008). AR knockout zebra fish have a low amount of mature sperm (Ogino et al., 2016). ARKO mice have fewer spermatocytes and produced fewer round sperm, suggesting that AR is required to support sperm meiosis support (Johnston et al., 2004;Walker, 2021). This further suggests that Sertoli cells are the target of androgen action. When AR is knocked out in Sertoli cells, the integrity of the blood-testosterone barrier is impaired, leading to the exposure of germ cells. Exposed germ cells are attacked by autoimmune responses,which is not conducive to sperm formation (Holdcraft & Braun, 2004;Walker, 2011; Chen et al., 2016). In addition, after specific AR gene knockout in the Sertoli cells in mouse testis, spermatogenesis was found to be arrested in the pachytene or diplotene primary spermatocyte state,and spermatocytes had serious DNA double-strand breaks, repair disorders, and abnormal chromosomal union (Chang et al., 2004; De Gendt et al., 2004, 2005). Based on the above research results of research on AR function in Sertoli cells, it can be hypothesized that ARs may play an important role in the regulation of the process of occurrence and release of black rock fish sperm.
During the gonad development in black rock fish, botharaandarbwere highly expressed in the degenerating testis and primary spermatogonia stages. Androgen stimulates the AR-mediated proliferation and differentiation of spermatogonia and plays important roles in the regulation of meiosis and the release of mature spermatozoa. Thearexpression patterns during spermatogenesis process varies in different species, as shown by previous studies. In European eel (Peñaranda et al.,2014), European seabass (Viñas & Piferrer, 2008) andSpinibarbus denticulatus(Liu et al., 2009), the expression of theargenes decreased significantly as testis development progressed but increased in the degeneration stage, which is consistent with our results. In sticklebacks,no variation inarexpression level was observed (Hoffmann et al., 2012).These results indicate that thearexpression pattern during the spermatogenesis varies in different species. On the other hand, knockout ofarin zebra fish decreased the amount of sperm, but did not affect the maturation process, indicating that AR plays an important role in the early process of spermatogenesis instead of during the maturation stage.Accordingly, it is reasonable that thearexpression decreased when the sperm matured. However, the function ofarin the testis degradation is still unknown.
In the ovary, thearexpression levels showed no significant variation during the oocyte development. However,argene expression varied in the parturition process, which is a specific process of ovoviviparous teleosts. Studies have shown that the fetus and placenta are additional sources of androgen during pregnancy is the fetus and placenta.Androgen from these sources can enter the maternal circulation, where their levels can be detected (Cantineau et al., 1985; Makieva et al.,2014). The mRNA levels ofarin female black rock fish are relatively high during the third trimester (before delivery) and then decrease during and after delivery. From this, we infer that the developing larvae in the ovary of black rock fish serve as additional sources of androgen, which enters the maternal circulation and bind to ARs. As delivery progresses,the androgen levels from these additional sources decrease, as doesarexpression. In mammals, androgens levels in maternal circulation increase throughout gestation, and this might regulate key processes during pregnancy and parturition; additionally, these increased androgen levels might be critical for cervical remodelling at term, in particular cervical ripening, via the regulation of cervical collagen fibril organization (Makieva et al., 2014). Additionally, a number of studies have highlighted potential roles for androgens in myometrial relaxation via nongenomic, AR-independent pathways that are critical for a pregnancy to reach full term (Makieva et al., 2016). However, due to limited conditions, the specific mechanism underlying the role ofargenes in parturition has not been studied.
In summary, botharaandarbwere identified in black rock fish, and their expression patterns were characterized in selected tissues and at different gonad development stages as well as during the parturition process. Our results indicate that AR plays important roles not only in the gonadal development but also in the mating behaviour and parturition in ovoviviparous teleosts.
Author contributions
This study was designed by HSW and XQ. SJY and JHC performed the experiment. SJY, LKL, XJW and YJY participated in the sample collection. SJY wrote the manuscript and XQ gave feedback and edited the article. All authors read and approved the final manuscript.
Acknowledgement
This study was supported by the National Key R&D Program of China(2018YFD0901204) and the National Natural Science Foundation of China (4197608).
Appendix A.Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aaf.2022.04.008.
 Aquaculture and Fisheries2022年5期
Aquaculture and Fisheries2022年5期
- Aquaculture and Fisheries的其它文章
- Impact of xenoestrogens on sex differentiation and reproduction in teleosts
- Sexual plasticity in bony fishes: Analyzing morphological to molecular changes of sex reversal
- Germ cell markers in fishes - A review
- Understanding the impact of stress on teleostean reproduction
- Molecular determinants regulating the release of the egg during ovulation:Perspectives in piscine models
- Reproductive farming technology in Japanese eel and chub mackerel
