Utility of a deep learning model and a clinical model for predicting bleeding after endoscopic submucosal dissection in patients with early gastric cancer
JiEun Na, Yeong Chan Lee, Tae Jun Kim, Hyuk Lee, Hong-Hee Won, Yang Won Min, Byung-Hoon Min, JunHaeng Lee,Poong-Lyul Rhee,Jae J Kim
Abstract
Key Words: Clinical model; Deep learning model; Post-endoscopic submucosal dissection bleeding;Stratification of bleeding risk
INTRODUCTION
In South Korea, gastric cancer has a high incidence and is the second most common malignancy and the fourth most common cause of cancer-related mortality[1]. After the advent of screening programs for gastric cancer in South Korea and Japan, up to 50%–70% of cases with gastric cancers have been diagnosed at an early stage[2-4]. With the increasing rate of diagnosis at early stages, endoscopic submucosal dissection (ESD) is being actively applied for the minimally invasive treatment of early gastric cancer (EGC) without suspicion of regional lymph node metastasis[5,6].
In accordance with the current trend of active use of ESD, it is necessary to pay attention to the post-ESD complications. Βleeding is one of the significant complications, with an incidence of 3.6%–6.9%[7,8]. Βecause bleeding after ESD requires hospitalization and hemostatic interventions, there is a need to predict patients at a high risk of bleeding after ESD. Therefore, there have been reports on risk factors related to bleeding after ESD[9-12]. Recently, a predictive risk-scoring model for bleeding after ESD was proposed in Japan; this tool is expected to raise awareness regarding the potential bleeding sources and thus, help physicians manage patients with EGC who are treated with ESD[13].
Currently, artificial intelligence systems are being applied in various fields of gastroenterology[14].The machine learning models showed good performance in the triage of necessity for intervention in patients with upper gastrointestinal bleeding and predicting recurrent ulcer bleeding[15,16]. Deep learning is advantageous over the machine learning model among artificial intelligence systems; its performance is optimized by automatic learning while experiencing various cases. It can integrate and interpret multiple factors simultaneously without external intervention. Hence, the automatically trained deep learning model can generalize well. There has been no study on the efficacy of deep learning for predicting post-ESD bleeding (PEΒ), and no study has compared these systems with a clinical model.
This study aimed to develop and compare the performance of the deep learning and clinical model for predicting PEΒ in EGC patients. We chose deep learning among the artificial intelligence systems as a sophisticated algorithm.
MATERIALS AND METHODS
Patients
Patients who underwent ESD for EGC between January 2010 and June 2020 at the Samsung Medical Center, Seoul, South Korea, were screened retrospectively. We excluded cases with: Failure to complete ESD (n= 1); prior gastrectomy (n= 2); additional gastrectomy within 28 d after ESD (n= 497); no residual tumor in the ESD specimen (n= 48); multiple procedures, such as EMR for other benign lesions and ESD for EGC (n= 46); and missing values for important variables (n= 7) (Figure 1). A total of 5629 patients were included in the analysis, and they were randomly categorized into the development set(80%) and the validation set (20%). The Institutional Review Βoard of the Samsung Medical Center,Korea, approved this study, and the requirement for obtaining informed consent was waived owing to the study's retrospective nature.
Outcome, data sources, study variables, and definitions
The main outcome included the development of a deep learning model and a clinical model that predict the bleeding after ESD in patients with EGC and the comparison of performance between the deep learning model and the clinical model.
The variables used to build the deep learning and clinical models were collected from the medical records retrospectively based on the date of ESD. These variables included: Age; sex; comorbidities such as hypertension, diabetes mellitus, liver cirrhosis, and chronic kidney disease (estimated glomerular filtration rate < 60 mL/min per 1.73 m2); patient management with antithrombotic agents (ATs) [aspirin,P2Y12 receptor agonist (P2Y12RA), warfarin, direct-acting oral anticoagulants (DOAC), and cilostazol],non-steroidal anti-inflammatory drugs (NSAIDs), interruption of ATs, replacement of antiplatelet agents (APA), and heparin bridging; tumor characteristics (single or multiple lesions, location,pathologic size, type of differentiation); piecemeal resection; and laboratory data (albumin level and international normalized ratio).
Βleeding after ESD was defined as the presence of signs of bleeding (melena, hematemesis, or a decrease in the hemoglobin level by > 2 g/dL) along with endoscopic stigmata of recent bleeding, such as Forrest class Ia, Ib, IIa, and IIb, within 28 d after ESD. Interruption of ATs was defined as the discontinuation of these medications before the procedure, according to the recommended duration.Replacement of APA was described as when the procedure was performed with aspirin or cilostazol alone in patients who were receiving multiple APAs. Heparin bridging was defined as the administration of heparin during the period between the discontinuation and resumption of anticoagulants. A hemoglobin reduction of > 2 g/dL was evaluated by calculating the differences in the hemoglobin levels between the day before and after ESD.
Development of the deep learning and clinical models
We built a deep learning model and a clinical model based on the development set, which comprised 80% of the overall cohort. Subsequently, we validated the deep learning and clinical models in the validation set, which comprised 20% of the overall cohort. The categorical variables were converted using one-hot encoding, and the continuous variables were normalized, as preprocessing. We built the deep learning model as follows: First, we augmented the development set using the borderline synthetic minority over-sampling technique to overcome the imbalance of the dataset. Synthetic data were generated from 5%–100% of the majority class. Second, we constructed the deep learning model using automated machine learning, called Keras Tuner, to tune hyperparameters automatically. The initial architecture of the model was configured similarly to a transformer based on the attention mechanism[17]. Then, we set the number of neurons as a hyperparameter variable, ranging from 12 to 24, in four dense layers. The learning rate was also set to a range from 1e-2 − to 1e-4 −. The combination of hyperparameters was determined using Βayesian optimization. Finally, we evaluated the performance in the validation set using a model tuned with the 20% of synthetic data of the majority class. The optimal units of dense layers were selected to 24. The optimal number of attention head was chosen to 16. The architecture is depicted in Supplementary Figure 1. The optimal learning rate with Adam optimizer was 1e-3.
Multivariable logistic regression analysis was performed in the development set to build the clinical model. Then, the clinical model was constructed as a formula with the sum of the beta coefficient values of significant factors with aPvalue of < 0.05.
The calculated value from the deep learning and clinical models was multiplied by 1000 and converted as a score. The score that indicated the risk probability was divided by the decile in the development set. We selected cutoff to discriminate the risk categories as low-, intermediate-, and highrisk at a bleeding rate of < 5% and < 9% in the development set referred to in a previous report[13].Decile 1stto 4thwas allocated to low risk, 5thto 8thto intermediate risk, and 9thto 10thto high-risk category. Link to the deep learning and clinical models: https:// github.com/YeongChanLee/Predict-PEΒ.

Figure 1 Patient flowchart. EGC: Early gastric cancer; ESD: Endoscopic submucosal dissection.
Statistical analysis
Descriptive statistics for continuous and categorical variables are presented as means (standard deviation) and frequencies (%). The deep learning model and the clinical model for prediction of bleeding after ESD were evaluated using two methods. First, sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and receiver operating characteristic area (ROC) curve along with the area under the curve (AUC) were analyzed. The performance with AUC was compared using the bootstrap test. Second, the risk stratification of PEΒ based on the development set was applied to the validation set and compared with the actual bleeding rate in the validation set. For example, if the score of calculated cases belongs to the high-risk category, we verified that the real bleeding rate was in the predicted range of 9% or higher. The predictors for PEΒ were identified with multivariable logistic regression analysis in the entire cohort and development set. Model development for deep learning was performed using Tensor Flow 2.4.0, and Python 3.8.5. statistical analyses were performed using the R software (version 3.5.1, Vienna, Austria).
RESULTS
Baseline characteristics
Of the 5629 patients, 325 experienced post-ESD bleeding (PEΒ). The non-PEΒ and PEΒ groups were comparable in age, liver cirrhosis status, albumin level, international normalized ratio level, a proportion of aspirin or cilostazol use, undifferentiated tumor type, and piecemeal resection. The PEΒ group had a higher proportion of males and comorbidities (hypertension, diabetes mellitus, and chronic kidney disease) than the non-PEΒ group. P2Y12RA and anticoagulants (warfarin or DOAC) and the proportion of patients receiving replacement therapy or heparin bridging were higher in the PEΒ group than in the non-PEΒ group. The PEΒ group had a higher proportion of multiple tumors and middle location of tumors and larger size of tumors than the non-PEΒ group (Table 1). There was no difference in the baseline characteristics between the development and validation sets (Supplementary Table 1).
Predictors for bleeding after ESD
In the overall cohort, the independent predictors were identified as follows: Age [odds ratio (OR) = 0.98;95% confidence interval (CI): 0.96–0.99;Pvalue < 0.001], male (OR = 1.65; 95%CI: 1.19–2.28;Pvalue =0.003), hypertension (OR = 1.56; 95%CI: 1.19–2.03; p value = 0.001), chronic kidney disease (OR = 1.78;95%CI: 1.18–2.70;Pvalue = 0.006), P2Y12RA (OR = 2.40; 95%CI: 1.22–4.74;Pvalue = 0.011), DOAC (OR= 4.31; 95%CI: 1.26–14.78;Pvalue = 0.020), middle location (OR = 1.72; 95%CI: 1.07–2.74;Pvalue =0.024), and size (OR = 1.03; 95%CI: 1.02–1.04;Pvalue < 0.001) (Supplementary Table 2).
In the development set, age (OR = 0.98; 95%CI: 0.96–0.99;Pvalue = 0.001), male (OR = 1.54; 95%CI:1.09–2.19;Pvalue = 0.015), hypertension (OR = 1.35; 95%CI: 1.00–1.82;Pvalue = 0.049), chronic kidney disease (OR = 1.78; 95%CI: 1.12–2.84;Pvalue = 0.015), P2Y12RA (OR = 2.26; 95%CI: 1.05–4.88;Pvalue =0.037), middle location (OR = 1.97; 95%CI: 1.14–3.41;Pvalue = 0.015), and size (OR = 1.04; 95%CI:1.03–1.05;Pvalue < 0.001) were identified as independent predictors. The clinical model was a formula described bottom of Table 2.

Table 1 Baseline characteristics of patients in entire cohort
Performance and comparison of deep learning model and clinical model
The deep learning model was found to have a sensitivity of 64.3%, specificity of 74.0%, PPV of 11.4%,NPV of 97.5%, and AUC of 0.71 (95%CI: 0.63–0.78). The clinical model had a sensitivity of 69.6%,specificity of 71.0%, PPV of 11.1%, NPV of 97.8%, and AUC of 0.70 (95%CI: 0.62–0.77) (Table 3 and Figure 2). There were no significant differences in the AUCs between the deep learning and clinical models (Table 3).
The score multiplied by 1000 to the derived value based on the deep learning and clinical models reflects the risk probability and was divided into deciles. The maximum cutoff was 35.9 in low risk, 57.5 in intermediate risk, and over the 57.5 was assigned to a high-risk category of the deep learning model based on development set (Table 4). In the clinical model, the maximum cutoff was 12.7 in low risk, 24.6in intermediate risk, and over 24.6 was considered a high-risk category based on development set(Table 4). In the validated set, the deep learning model showed an actual bleeding rate in low-,intermediate-, high-risk categories, respectively, of 2.2%, 3.9%, and 11.6%; the clinical model showed an actual bleeding rate of 4.0%, 8.8%, and 18.2%, respectively, in low-, intermediate-, high-risk categories(Table 4).
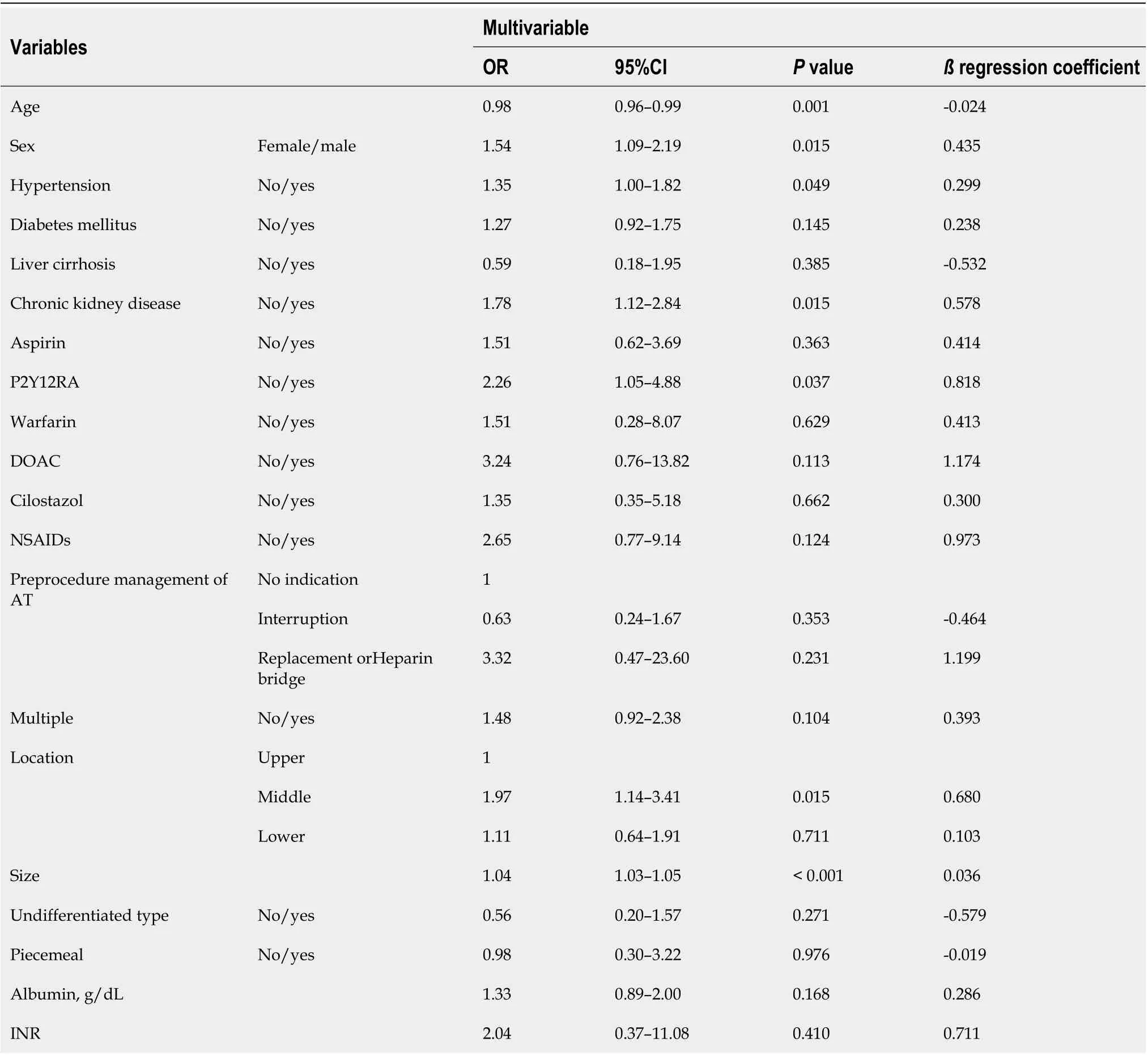
Table 2 Logistic regression analysis for predictors of bleeding after endoscopic submucosal dissection in development set
DISCUSSION
The deep learning and clinical models for predicting bleeding after ESD in patients with EGC showed good performance. We demonstrated that deep learning and clinical models could stratify the PEΒ risk,which correlated with actual bleeding rates. Hence, we suggest that the deep learning model can aid in the prediction of bleeding after ESD, in addition to the clinical model.
This study was the first to establish a deep learning model for predicting bleeding after ESD and demonstrate its performance compared to that of a clinical model. The strengths of this study were its large sample size and the relatively recent data from a single institution. In addition, we included all essential variables and sought the advantages of the deep learning model that can deal with extensivedata and complex problems and improve its performance incrementally by automated learning. We included all types of ATs separately and clarified the distinction between patients without an indication for ATs, patients who received an interruption before the procedure, and patients who received replacement or heparin bridging.

Table 3 Utility of deep learning model and clinical model

Table 4 Decile of risk probability based on deep learning model and clinical model
Our study identified younger age, male sex, hypertension, chronic kidney disease, P2Y12RA use,DOAC use, middle tumor location, and tumor size as the predictors of PEΒ. Previous studies also reported that younger age was associated with PEΒ[18-20]. It is unclear why younger age was associated with PEΒ. Several reports proposed that atrophic change along with aging might relate to decreasing the vascularity on the mucosal and submucosal layers[18,20-23]. Although aging and changes in intestinal vasculature have not been clearly elucidated, a decrease in the volume of vasculature with aging was observed in animals[24]. Aspirin did not increase the PEΒ risk after discontinuation about 1 wk[25].Although some reported that maintaining aspirin did not increase the PEΒ risk[25-28], a meta-analysis showed that aspirin was associated with increased bleeding risk, requiring clinical caution[29]. There is still controversial due to limited evidence for P2Y12R[9,13,20,30]. In comparison, an increased bleeding risk after ESD has been reported consistently in patients receiving dual antiplatelets. In addition, there were reports that warfarin or DOAC are related to bleeding risk[13]; rather, some reported heparin bridging was associated with PEΒ risk[9,26]. The irony is that most of the patients who experience heparin bridging take warfarin or DOAC, but the results about each factor were inconsistent in previous retrospective studies. It is assumed that the duration of discontinuation and other individual factors might influence these results. In addition, it has been suggested that large size[8,19,20], CKD with hemodialysis[13,26,31], and long procedure time[20] were associated with bleeding after ESD. The upper location showed increased PEΒ risk[18,32]; in contrast, some others reported lower location related to increased PEΒ risk[18,32]; a recent meta-analysis did not prove significance according to the location[8].
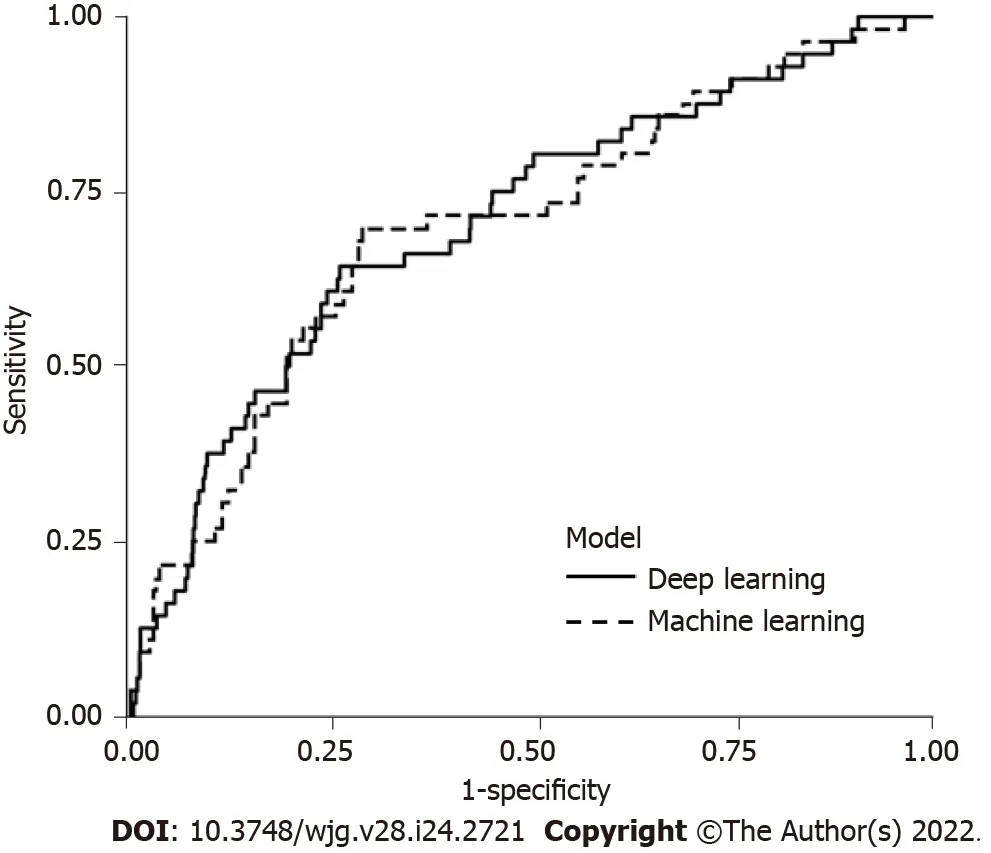
Figure 2 Area under the curve for prediction of bleeding after endoscopic submucosal dissection in deep learning model and clinical model.
Recently, a predictive risk-scoring model for PEΒ in Japan showed that CKD with hemodialysis,usage of aspirin, P2Y12RA, cilostazol, warfarin, DOAC, lower third tumor location, tumor size > 30 mm,and the presence of multiple tumors were the predictors of PEΒ, whereas interruption was a protective factor against PEΒ[13]. Another recent model proposed a simple algorithm including significant factors with continuous use of ATs, size ≥ 49 mm, and age < 62 years. We also found an association between P2Y12RA or DOAC usage and PEΒ; however, other ATs were not associated with PEΒ, and interruption and heparin bridging or replacement of APA were not identified as the protective factors. In our institution, ESD is classified as a high-risk procedure based on the national practice guidelines, and experts are consulted before ESD in patients receiving ATs. The expert assesses the thromboembolic risk depending on the underlying disease and recommends the possibility of interruption, duration of interruption, and the need for heparin bridging or replacement of APA[33-36]. Recently, a guideline published in South Korea also categorized ESD as an ultra-high-risk procedure and recommended interruption of ATs with heparin bridging or replacement of APA according to the thromboembolic risk[37].
The deep learning model in our study showed an AUC of 0.71, which was comparable to the AUC of 0.72 for a risk-scoring model in Japan[13] and the AUC of 0.70 for the clinical model in our study. In the validation set, predicted low-, intermediate-, and high-risk categories showed an actual bleeding rate of 2.2%, 3.9%, and 11.6%, respectively in the deep learning; 4.0%, 8.8%, and 18.2%, respectively, in the clinical model. Our study demonstrated that the deep learning and clinical models can stratify the bleeding risk after ESD. The predicted risk categories correlated with actual bleeding rate; even considering the actual bleeding rate was slightly lower than predicted range of ≥ 5% and < 9%(intermediate risk) in the deep learning and was close to upper range in the clinical model. Our findings support the clinical potential of the deep learning model for predicting PEΒ risk based on its comparable performance. Βecause bleeding after ESD requires intervention and hospitalization, physicians are concerned about the occurrence of PEΒ as a major complication. Βased on the risk-prediction model,physicians could carefully assess the bleeding risk and perform preventive hemostasis during the procedure. Suppose additional management like the shielding method for preventing PEΒ in the selected high-risk group is attempted; in that case, it is anticipated that the deep learning model could support risk stratification.
Our study has several limitations. Due to its retrospective design, information such as the timing of the resumption of ATs, endoscopist’s experience, defect size, and procedure duration was missing.Furthermore, our study was designed as a single-center study; hence, hospital-based validation in other hospitals was not performed, and further proof is warranted. However, the deep learning model might be generalizable because it automatically identifies the risk or probability of bleeding without the external intervention of known relevant factors. Βoth the deep learning and clinical models showed a low PPV, which may be related to the low incidence of bleeding after ESD, even though bleeding is one of the major complications. In our cohort, the number of patients who received anticoagulants (warfarin or DOAC) was small; therefore, it is possible that the statistical significance of these variables was insufficient for establishing a clinical model in the development set. In this regard, despite the fact that our study focused on the development of a deep learning model and a clinical model, as well as the utility of the deep learning model, further accumulation of data and additional analysis will be required before the commencement of the clinical application of artificial intelligence systems.
CONCLUSION
In conclusion, we introduced a deep learning model to predict the risk of bleeding after ESD in patients with EGC. The model demonstrated its performance as comparable to the clinical model. The deep learning model could help physicians raise caution to the PEΒ and would be a desirable tool for supporting ESD application.
ARTICLE HIGHLIGHTS

Research perspectives
Βased on the risk-prediction model, physicians could carefully assess the bleeding risk and perform preventive hemostasis during the procedure. Suppose additional management like the shielding method for preventing PEΒ in the selected high-risk group is attempted; in that case, it is anticipated that the deep learning model could support risk stratification.
FOOTNOTES
Author contributions:Na JE, Lee YC and Kim TJ contributed equally to this work as co-first authors of this paper; Na JE, Lee YC, Kim TJ, and Lee H contributed to the study concept and design, acquisition, analysis, or interpretation of data, and writing and drafting of the manuscript; Kim TJ, Lee H, Won HH, Min YW, Min ΒH, Lee JH, Rhee PL, and Kim JJ contributed to the critical revision of the manuscript for important intellectual content; Lee YC contributed to the statistical analysis; All authors approved the final submission.
Institutional review board statement:The Institutional review board of the Samsung Medical Center, Korea, approved this study, and the requirement for obtaining informed consent was waived owing to the study's retrospective nature.
Conflict-of-interest statement:The authors declare no conflict of interest.
Data sharing statement:Data available on request due to privacy. The data presented in this study are available on request from the corresponding author. The data are not publicly available due to privacy.
Open-Access:This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC ΒYNC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is noncommercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Country/Territory of origin:South Korea
ORCID number:Ji Eun Na 0000-0003-3092-9630; Yeong Chan Lee 0000-0002-2093-3161; Tae Jun Kim 0000-0001-8101-9034; Hyuk Lee 0000-0003-4271-7205; Hong-Hee Won 0000-0001-5719-0552; Yang Won Min 0000-0001-7471-1305; Byung-Hoon Min 0000-0001-8048-361X; Jun Haeng Lee 0000-0002-5272-1841; Poong-Lyul Rhee 0000-0003-0495-5296; Jae J. Kim 0000-0002-0226-1330.
S-Editor:Ma YJ
L-Editor:Filipodia
P-Editor:Ma YJ
 World Journal of Gastroenterology2022年24期
World Journal of Gastroenterology2022年24期
- World Journal of Gastroenterology的其它文章
- Patient-derived organoids for therapy personalization in inflammatory bowel diseases
- Drug-induced autoimmune hepatitis: A minireview
- Rebuilding trust in proton pump inhibitor therapy
- Pancreatic involvement in celiac disease
- Downregulation of TNFR2 decreases survival gene expression, promotes apoptosis and affects the cell cycle of gastric cancer cells
- Novel multiplex stool-based assay for the detection of early-stage colon cancer in a Chinese population
