The potential of gene therapies for spinal cord injury repair: a systematic review and meta-analysis of pre-clinical studies
Catriona J. Cunningham , Mindaugas Viskontas Krzysztof Janowicz Yasmin Sani Malin E. Håkansson Anastasia Heidari Wenlong Huang , Xuenong Bo
Abstract Currently, there is no cure for traumatic spinal cord injury but one therapeutic approach showing promise is gene therapy. In this systematic review and meta-analysis, we aim to assess the efficacy of gene therapies in pre-clinical models of spinal cord injury and the risk of bias. In this metaanalysis, registered at PROSPERO (Registration ID: CRD42020185008), we identified relevant controlled in vivo studies published in English by searching the PubMed, Web of Science, and Embase databases. No restrictions of the year of publication were applied and the last literature search was conducted on August 3, 2020. We then conducted a random-effects meta-analysis using the restricted maximum likelihood estimator. A total of 71 studies met our inclusion criteria and were included in the systematic review. Our results showed that overall, gene therapies were associated with improvements in locomotor score (standardized mean difference [SMD]: 2.07, 95% confidence interval [CI]: 1.68–2.47, Tau2 = 2.13, I2 = 83.6%) and axonal regrowth (SMD: 2.78, 95% CI: 1.92–3.65, Tau2 = 4.13, I2 = 85.5%). There was significant asymmetry in the funnel plots of both outcome measures indicating the presence of publication bias. We used a modified CAMARADES (Collaborative Approach to Meta-Analysis and Review of Animal Data in Experimental Studies) checklist to assess the risk of bias, finding that the median score was 4 (IQR: 3–5). In particular, reports of allocation concealment and sample size calculations were lacking. In conclusion, gene therapies are showing promise as therapies for spinal cord injury repair, but there is no consensus on which gene or genes should be targeted.
Key Words: animal models; gene delivery; meta-analysis; regenerative medicine; spinal cord injury; systematic review; viral vectors
Introduction 299 Methods 299 Results 300 Discussion 301
Introduction
Traumatic spinal cord injury (SCI) is a devastating life event leaving around 90% of patients with significant long-term disability including sensorimotor impairment, loss of independence, and decreased quality of life (Ahuja et al., 2017). There are an estimated 27 million people worldwide living with the consequences of SCI (GBD 2016 Traumatic Brain Injury and Spinal Cord Injury Collaborators, 2019). Central nervous system (CNS) regeneration is often referred to as the holy grail of neuroscience. The obstacles to overcome are vast including the physical barrier of cystic cavities, the presence of numerous inhibitory molecules such as chondroitin sulphate proteoglycans which create a non-permissive environment and the limited intrinsic regenerative capacity of adult CNS neurons (Fawcett, 2019).
Gene therapy is the application of genetic material to correct diseasecausing mutations, downregulate genes contributing to disease or deliver genes encoding molecules with therapeutic potential. The most widely used vectors for gene therapy delivery are viral vectors including adenoviruses, adeno-associated viruses (AAV), and lentiviruses (LV), the latter of which is a genus of retroviruses (Cring and Sheffield, 2022). Non-viral vectors including liposomes and nanoparticles might be used in favor of viral vectors due to their capacity to carry longer genetic sequences (Uchida et al., 2014). Another approach is genome editing, revolutionized by the development of CRISPRCas9 technology in 2012 (Dunbar et al., 2018). Gene therapy approaches being investigated in preclinical models of SCI includein vivo
delivery of genes to the spinal cord to increase the expression of promoters of axonal regrowth or silence inhibitors with short hairpin RNA (shRNA) or small interfering RNA (siRNA) (Bo et al., 2011; Zavvarian et al., 2020). This has the potential to overcome some of the limitations of direct administration of drugs and recombinant proteins. For example, it has long since been established that Chondroitinase ABC (ChABC) can degrade chondroitin sulphate proteoglycan chains and promote functional recovery after SCI in animal models (Bradbury et al., 2002) but due to the short half-life of the enzyme, repeated invasive dosing would be required. In more recent years, gene therapy approaches have therefore been explored to deliver ChABC (Burnside et al., 2018).Several narrative reviews have discussed the potential of gene therapy for spinal cord repair (Bo et al., 2011; Uchida et al., 2014; Zavvarian et al., 2020), but to the best of our knowledge, there is no existing systematic review and meta-analysis. Our review, therefore, aims to assess the efficacy of gene therapies in preclinical models of SCI and the risk of bias.
Methods
This systematic review followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses statement (PRISMA) guidelines (Moher et al., 2009) and the protocol of this meta-analysis has been published in the PROSPERO database under registration No. CRD42020185008.
Search strategy
We conducted searches in PubMed, Web of Science, and Embase (OVID) for articles published in English using the following keywords search strategy:(gene therapies OR gene therapy OR gene delivery OR CRISPR OR CRISPR/cas9 OR CRISPR cas9 OR CRISPR-cas9 OR viral vector$ OR viral construct$ OR lentivirOR lentiviral construct$ OR lentiviral vector$ OR adeno-associated viral vector$ OR adeno-associated viral construct$ OR adenoassociated viral vector$ OR adenoassociated viral construct$ OR adeno associated viral vector$ OR adeno associated viral construct$ OR adenoviral OR adenoviral vector$ OR adenoviral viral construct$ OR AAV$ OR AAV-induced) AND (spinal cord injury OR spinal cord injuries OR SCI OR injured spinal cord OR spinal cord injured OR damaged spinal cord OR spinal cord damage OR spinal cord contusion OR contused spinal cord OR lacerated spinal cord OR spinal cord laceration OR spinal cord compression OR compressed spinal cord OR spinal cord trauma OR transected spinal cord OR spinal cord transection OR spinal cord hemisection OR axon regeneration OR axon growth OR axon regrowth).To limit the searches to animal studies, we used previously published filters (Hooijmans et al., 2010; de Vries et al., 2014). We also screened relevant review articles for additional studies. We applied no limits to the year of publication and the last search was performed on August 3, 2020.
Study screening
Study screening was conducted by two independent reviewers (MH, AH) and disagreements were resolved through discussion with a third reviewer (CC). After removing duplicates, we first screened titles and abstracts to exclude clearly irrelevant studies (e.g. reviews, irrelevant disease models). In the second phase, we then assessed the full texts of identified studies against the complete inclusion and exclusion criteria. Controlled studies which delivered a gene therapy (in vivo
vector delivery, administration ofex vivo
transduced cells, CRISPR/cas9) to a preclinical model of traumatic SCI and assessed functional recovery using a locomotor score were included. The criteria for the comparator group were SCI + one of the following: vector expressing a reporter protein only; cells transduced with a vector expressing a reporter protein only or a CRISPR non-targeting control. Studies with naïve, sham surgery, and SCI only controls were excluded.Data extraction
Qualitative data for the systematic review were extracted by two independent reviewers (MV,KJ) and any disagreements were resolved through discussion with a third reviewer (CC). The study design information extracted included were: (1) SCI model; (2) species; (3) age; (4) sex; (5) total animal number; (6) number of groups; (7) gene therapy; (8) timing of administration; (9) route of administration; (10) dose and (11) combination therapy. We defined the primary outcome measure as locomotor score (any scale) and the secondary outcome measures as axon regrowth and evoked hind paw mechanical hypersensitivity. We extracted means and SEM or SD values from the main text where possible. When data was only presented graphically, we used the online graphical tool WebPlotDigitizer (https://automeris.io/WebPlotDigitizer/) to extract values from the graphs. In instances when outcome measures were assessed at multiple time points, only data from the last time point was extracted. When one control group was shared by more than one treatment group, we corrected for this by dividing the number of animals in the control group by the number of treatment groups served. Locomotor data were extracted by two independent reviewers (MV, KJ) and then the secondary outcome data were extracted by a second team (CC, YS). We cross-checked the estimates and where these varied by < 10%, means were taken. Any differences > 10% were resolved through discussion. Where exact n numbers were not reported, it was unclear if SD or SEM was used and if not possible to extract data using WebPlotDigitizer, we emailed the authors for clarification. If after two attempts the data were not made available, we excluded those studies from the meta-analysis.
Risk of bias assessment
We assessed the risk of bias using an adapted 7-point CAMARADES (Collaborative Approach to Meta-Analysis and Review of Animal Data in Experimental Studies) Risk of Bias Checklist (Macleod et al., 2004) including the following: 1) peer-reviewed publication; 2) random allocation to group; 3) allocation concealment; 4) blinded assessment of outcome; 5) sample size calculation/power calculation; 6) compliance with animal welfare regulations; 7) statement of potential conflict of interest.
Statistical analysis
We conducted all statistical analysis and graphing using the metafor package (Viechtbauer, 2010) in RStudio V1.3.959 (RStudio, Boston, MA, USA), R version 4.0.1. Effect sizes were calculated using Hedge’s g with a positive standardized mean difference (SMD) favoring treatment for all outcome measures. We then conducted random effects meta-analyses using the restricted maximum likelihood estimator. We assessed heterogeneity usingI
(percentage of between-study variance due to heterogeneity rather than sampling error) and Tau(between-study variance) (Vesterinen et al., 2014). To visualize publication bias, we used funnel plots and confirmed asymmetry with Egger’s regression test. We used trim-and-fill analysis to estimate the number of “missing” unpublished studies and calculate an adjusted effect size accounting for publication bias (Duval & Tweedie, 2000). Lastly, we conducted subgroup analyses to explore the following study characteristics as sources of heterogeneity: gene therapy platform; SCI model; timepoint of administration; route of administration; randomization and blinding. Independent random effects models were fitted to subgroups and then estimates were compared using a Wald-type test. We only conducted analysis when there were at least four comparisons in a subgroup. Significance was defined asP
< 0.05.Results
Study characteristics
We identified 2487 records from our literature search and following screening, 71 studies were included in the systematic review (Figure 1
). Study characteristics including genes delivered, SCI models and total animal numbers are presented inAdditional Table 1
. As expected, the majority of studies were conducted in rats (n
= 55) and mice (n
= 12) with the remaining studies using dogs (n
= 3) and rabbits (n
= 1) models. The models used were as follows: contusion (n
= 35); compression (n
= 17); complete transection (n
= 14); hemisection (n
= 4) and electrolytic lesion (n
= 1). The vast majority of studies induced a thoracic SCI (n
= 66) with almost half (n
= 35) using T10 as the injury level. The remaining studies induced injury in the lumbar (n
= 3) and cervical (n
= 1) spinal cord and one final study did not specify the level.
Figure 1|PRISMA flow diagram. Summary of the number of studies identified, screened, and ultimately included in the systematic review and meta-analysis. PRISMA: Preferred Reporting Items for Systematic Reviews and Meta-Analyses statement.
All but one study used viral vectors as the gene therapy platform, employing eitherin vivo
delivery (n
= 44) orex vivo
transduction of cells (n
= 26). Ito et al. (2009) instead administered liposomes containingE. coli
plasmid vectors expressing interferon β. The most commonly used viral vector was lentiviruses (n
= 32) followed by AAV (n
= 20), retroviruses (n
= 7), adenoviruses (n
= 6) and herpes simplex viruses (n
= 5). A wide range of cell types were used including the following: stem cells, namely mesenchymal stem cells (n
= 15) and neural stem cells (n
= 5); adult somatic cells such as Schwann cells (n
= 2) and microglia (n
= 1); and a T-REx-293 cell line (n
= 1). A substantial range of genes encoding for potentially therapeutic molecules were delivered including growth factors glial cell-derived neutrotrophic factor (n
= 7), brainderived neurotrophic factor (BDNF;n
= 4), and neutrophin-3 (NT-3;n
= 3). A smaller number of studies (n
= 11) administered siRNAs or shRNAs to silence genes including tumour necrosis factor-α (Zhang et al., 2015) and Nogo (Liu et al., 2016).The vast majority of studies administered gene therapies locally by intralesional (n
= 41), intraspinal (n
= 20), intrathecal (n
= 5), or a combination of intralesional and intraspinal injections (n
= 1). There was great variability in the timing of administration ranging from 1 week before (n
= 1) to 3 weeks (n
= 1) after injury with the most common timepoint being 0 hours (n
= 34). Of note, there were five studies that did not clearly specify the time point of administration.A total of seven studies administered gene therapies in combination with biomaterials (n
= 4) such as fibrin gels (Tsai et al., 2017; Shi et al., 2019), stem cells (n
= 1) and ChABC (n
= 2). Blits et al. (2003) implanted a Schwann cell bridge into the lesion at the same time as AAV vectors containing brainderived neurotrophic factor and NT-3. Only three studies co-administered cyclosporin as an immunosuppressant (Hwang et al., 2009; Lee et al., 2009, 2011).Meta-analysis results
A total of 56 studies (68 comparisons) were assessed in the meta-analysis of our primary outcome measure, locomotor score. Two studies could not be included as the data were expressed as median ± interquartile range (IQR) and 13 studies were excluded after the authors did not respond to our requests for clarification of their data. As summarized inFigure 2
, our results suggested that gene therapies improved locomotor scores compared with control (SMD: 2.07, 95% confidence interval [CI]: 1.68–2.47,P
< 0.0001, Tau= 2.13,I
= 83.6%).We then proceeded to analyze our secondary outcome measures. We included 24 studies (28 comparisons) in the meta-analysis of the axonal regrowth data (Figure 3
). Overall, gene therapies led to an improvement in axonal regrowth compared with control (SMD: 2.78, 95% CI: 1.92–3.65,P
< 0.0001, Tau= 4.13,I
= 85.5%). Only 3 studies included data on evoked hind paw mechanical hypersensitivity and as we defined 10 as the minimum in our protocol, we were not able to analyze this secondary outcome measure.Subgroup analysis
Lastly, we conducted a subgroup analysis to explore sources of heterogeneity. As shown inAdditional Tables 3
and4
, we did not observe any significant effects of randomization, blinding, SCI model, or gene therapy platform in either the locomotor score or axonal regrowth datasets.Study quality and risk of bias
As shown inTable 1
, the median score was 4 (IQR: 3–5). Although most studies reported blinding (71.8%), less than half (45.1%) reported using randomization. Only two studies (Li et al., 2017; Chen et al., 2020) specified that sample size calculations (Additional Table 2
) were conducted and it was only possible to determine that one study used allocation concealment (Cen et al., 2013).
Table 1 |The risk of bias assessed using a 7 item modified CAMARADES checklist
Furthermore, our results suggested there was a publication bias in both outcome measures. There was asymmetry in the funnel plot of the locomotor score data (Figure 4A
) with most studies reporting positive effect sizes favoring gene therapies. This asymmetry was confirmed by Egger’s regression (P
< 0.0001). Trim-and-fill analysis estimated that there were 2 “missing” unpublished studies with negative effect sizes (Figure 4B
). When included in the meta-analysis, this led to a small reduction in SMD from 2.07 to 1.99. In similarity, there was also significant (P
< 0.0001, Egger’s regression) funnel plot asymmetry in the axonal regrowth dataset (Figure 4C
). Trim-and-fill analysis again estimated that there were 2 “missing” studies (Figure 4D
) and when included, reduced the SMD from 2.78 to 2.59.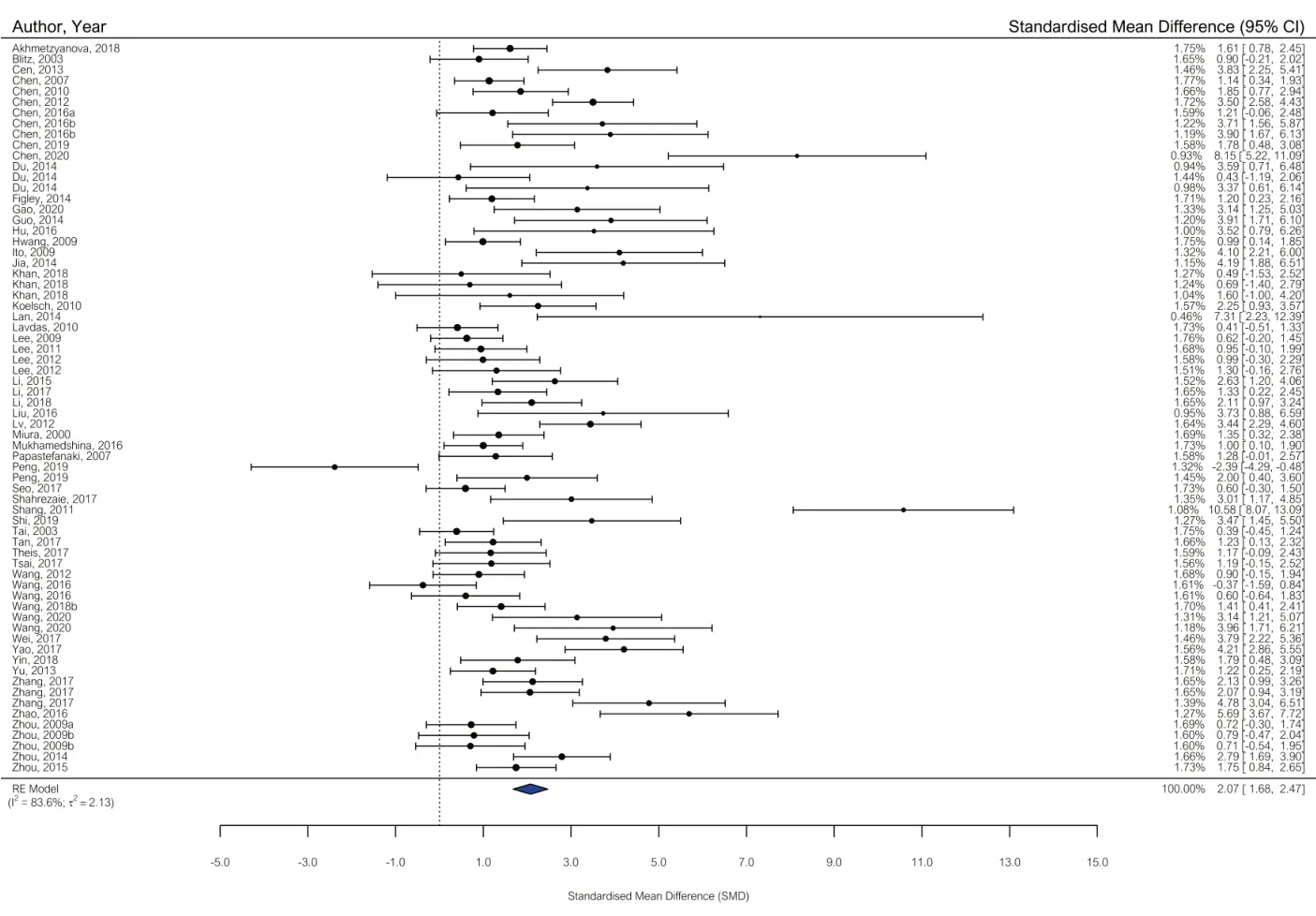
Figure 2|Forest plot of locomotor score data. Summary of the effect sizes of each comparison included in the meta-analysis expressed as standardized mean difference (SMD) and 95% confidence intervals (CIs). A positive SMD favors treatment and represents an improvement in locomotor score. The overall estimated effect size, as represented by the blue diamond, was 2.07 (95% CI: 1.68–2.47).
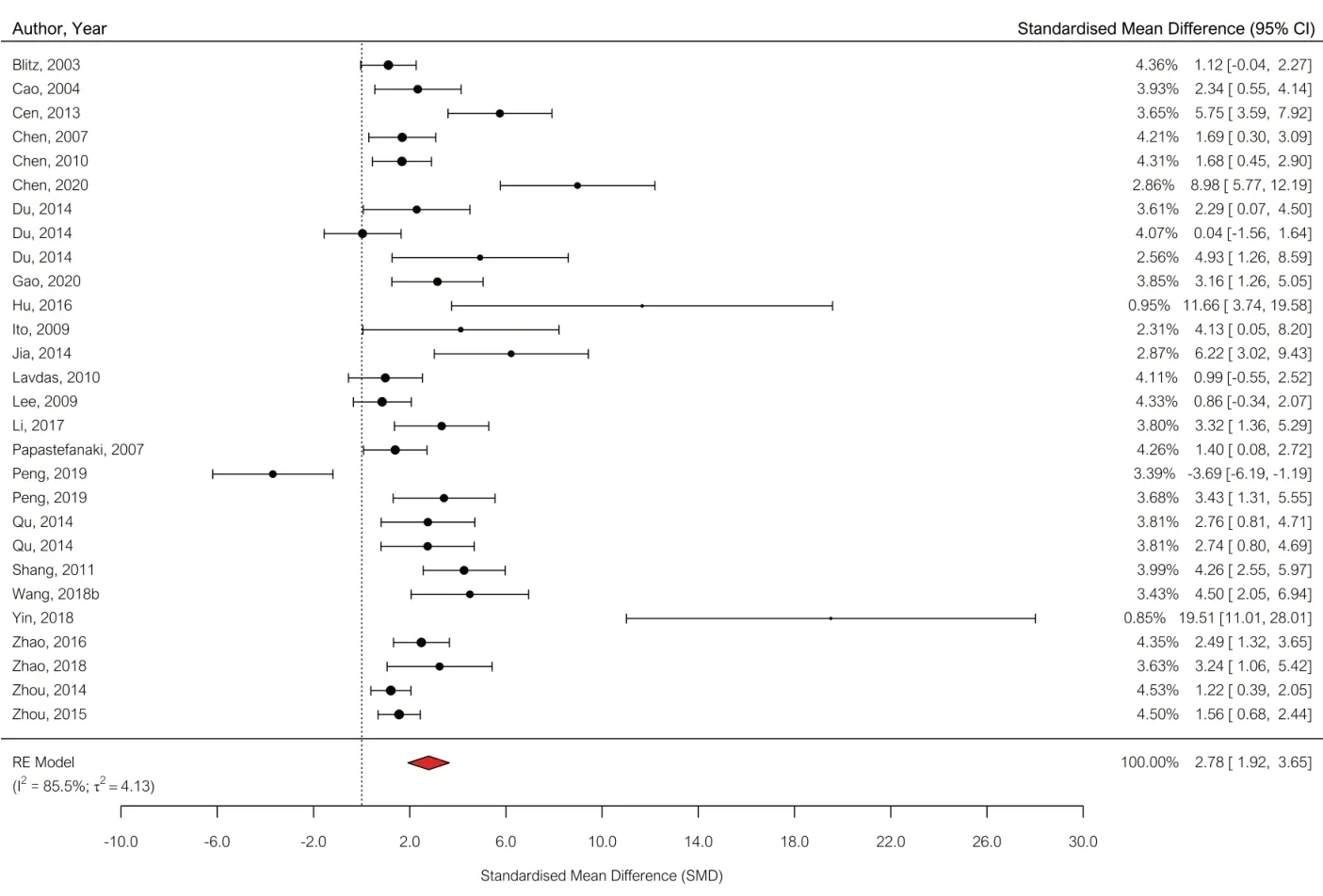
Figure 3|Forest plot of axonal regrowth data. Effect sizes are expressed as standardized mean difference (SMD) and 95% confidence intervals (CIs). A positive SMD represents increased axonal regrowth. The overall estimated effect size was 2.78 (95% CI: 1.92–3.65).
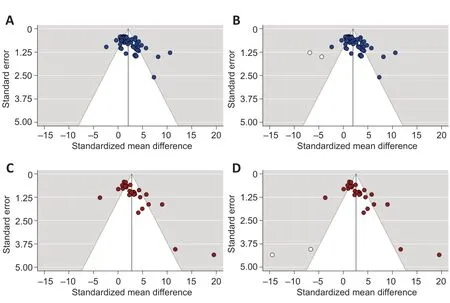
Figure 4|Funnel plots for assessing the risk of publication bias. (A, B) The asymmetric funnel plot in the locomotor score dataset is suggestive of publication bias (A) and the trim-and-fill analysis (B) estimated there were 2 “missing” unpublished studies (unfilled circles) on the left-hand side of the plot with negative effect sizes favoring control. (C, D) Similarly, there was also pronounced asymmetry in the funnel plot of the axonal regrowth outcome (C) with trim-and-fill analysis again estimating 2 “missing” studies (D). The dotted lines represent 95% confidence intervals.
Discussion
We identified 71 studies which met the inclusion criteria for our systematic review and meta-analysis of gene therapies in preclinical models of SCI. Overall, our meta-analysis favored treatment with gene therapies leading to significant improvements in the locomotor score and enhanced axonal regrowth compared to controls. We observed significant asymmetry in the funnel plots of both outcome measures suggesting the presence of publication bias. However, the trim-and-fill analysis only estimated a modest number of “missing” studies with negative or neutral effect sizes in both datasets. We next reported that the median risk of bias score was 4 (IQR: 3–5) as assessed using a modified CAMARADES checklist. Although the vast majority of studies used blinding, reporting of allocation concealment and power calculations was lacking.
While gene therapies were associated with improvements in both outcome measures, there was great heterogeneity in the therapies administered. A total of 58 different genes were targeted including the delivery of promoters of axonal regrowth brain-derived neurotrophic factor, NT-3, and NGF. As an alternative approach, some studies silenced inhibitors of axonal regeneration such as Nogo (Liu et al., 2016) and its receptor-negative growth regulatory protein 1 (Lv et al., 2012; Zhao et al., 2018) using siRNAs or shRNAs. Given the complex pathophysiology of SCI, it seems unlikely that gene therapy alone will be able to induce substantial recovery in patients (Griffin and Bradke, 2020). However, only 7 out of 71 included studies that administered gene therapies as part of the combinatorial approach.
A potential limitation of our meta-analysis was the focus on motor recovery. By requiring studies to have assessed locomotor scores to meet our inclusion criteria, we may have excluded studies focussing on the potential of gene therapies for sensory nerve regeneration and treatment of neuropathic pain. This could explain why just three studies assessed evoked hind paw mechanical hypersensitivity and it was therefore not feasible to conduct a meta-analysis on this outcome. We chose locomotor scores (including Basso, Beattie and Bresnahan locomotor rating scale and Basso mouse scale) as our primary outcome measure as these are the most widely used behavioral tests for assessing functional recovery in preclinical models of SCI (Watzlawick et al., 2019). While simple to conduct and inexpensive, we do acknowledge that a disadvantage of locomotor scores is subjectivity (Silva et al., 2014). Another possible limitation in our meta-analysis was that all the comparisons in our subgroup analysis were insignificant. This could be explained by a lack of power. In our analysis, we reported high heterogeneity and unbalanced subgroups which can increase the number of studies required to reach sufficient power in the subgroup analysis (Cuijpers et al., 2021).
An important finding of the studies included in our meta-analysis was that only one study used a cervical SCI model. This is in stark contrast to the clinical population in which ~60% of traumatic injuries are at the cervical level (Ahuja et al., 2017). Around 32% of patients have a thoracic SCI but 93% of our included studies used a thoracic model. We, therefore, recommend that future studies focus more on assessing the efficacy of gene therapies in cervical models to increase the translational potential. However, we do acknowledge the challenges this can introduce including respiratory dysfunction, higher mortality rates, and increased post-operative care needs (Sharif-Alhoseini et al., 2017). Another limitation that may impact clinical translation was the lack of studies which explored subacute and chronic timepoints of administration for gene therapies. While this will increase research costs, these timepoints also need to be explored because the pathophysiology is substantially different from acute SCI.
An ongoing issue in preclinical research is inadequate study reporting and in particular, measures to reduce bias such as blinding and randomization. To address this, the National Centre for the Replacement, Refinement and Reduction of Animals (NC3Rs) worked with a group of experts to develop the Animal Research: Reporting ofin vivo
Experiments (ARRIVE) Guidelines (Percie du Sert et al., 2020). While many journals endorse these guidelines and began requiring authors to submit a completed ARRIVE checklist, a randomized controlled trial of manuscripts submitted toPLoS One
showed that compliance with the guidelines was still lacking (Hair et al., 2019). The Nature Publishing Group implemented its checklist to reduce the risk of bias in preclinical studies. While this did lead to some improvements compared with manuscript submissions in journals without this policy, there were still improvements needed including reporting of power calculations (NPQIP Collaborative Group, 2019). In similarity with this, we found that very few studies included power calculations and reported allocation concealment.After decades of clinical trials, several gene therapies have been approved in recent years including AAV9-based Zolgensma for the treatment of spinal muscular atrophy (European Medicines Agency: EMA/173982/2020, 2020). The safety of several types of viral vectors, particularly AAV (Nathwani et al., 2014; Colella et al., 2018), has been widely reported. While clinical trials of cell therapies for SCI have been ongoing for several years, gene therapy for SCI is yet to reach clinical trial. A consortium called CHASE-IT, supported by International Spinal Research Trust, plans to conduct a clinical trial on lentiviruses mediated expression of mammalinized ChABC (International Spinal Research Trust, 2021). However, the degradation of the inhibitory chondroitin sulphate proteoglycans at the injury site alone may not be enough to lead to successful axonal regeneration and functional recovery. As previously mentioned, combinatorial approaches likely hold the key to successful SCI repair. Gene therapies may need to be combined with cell therapies, biomaterials, and other strategies such as rehabilitation and epidural stimulation (Bo et al., 2011).
In conclusion, our systematic review and meta-analysis demonstrate that gene therapies are showing promise in preclinical models of SCI. Although a wide range of genes have been studied, there is no consensus on which gene or genes are most effective in promoting axonal regrowth. It is highly likely that multiple genes in different cell types such as injured neurons in the CNS or primary sensory neurons and glial cells at the injury site may need to be targeted to achieve successful axonal regrowth.
Acknowledgments:
We would like to thank Dr Ying Jin (Drexel University) and Dr Alexandros Lavdas (Eurac Research) for replying to our requests for clarification of the exact n numbers.
Author contributions:
Concept and design of the study: CJC, WH, XB; acquisition of data: MV, KJ, YS, MEH, AH; analysis and interpretation of data: CC; drafting manuscript: CJC; revising manuscript: CJC, WH, XB; acquisition of funding: WH. MEH and AH contributed equally to this work. WH and XB also contributed equally to this work. All authors read and approved the final manuscript.
Conflicts of interest:
None declared.
Availability of data and materials:
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Open access statement:
This is an open access journal, and articles are distributed under the terms of the Creative Commons AttributionNonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
Additional files:
Summary of studies included in the systematic review.
Extended risk of bias checklist data.
Subgroup analysis of the locomotor score data.
Subgroup analysis of the axonal regrowth outcome.
- 中国神经再生研究(英文版)的其它文章
- c-Abl kinase at the crossroads of healthy synaptic remodeling and synaptic dysfunction in neurodegenerative diseases
- The mechanism and relevant mediators associated with neuronal apoptosis and potential therapeutic targets in subarachnoid hemorrhage
- Microglia depletion as a therapeutic strategy: friend or foe in multiple sclerosis models?
- Brain and spinal cord trauma: what we know about the therapeutic potential of insulin growth factor 1 gene therapy
- Functions and mechanisms of cytosolic phospholipase A2 in central nervous system trauma
- Cre-recombinase systems for induction of neuronspecific knockout models: a guide for biomedical researchers

