The role of Smad6 in immunity of the pearl oyster Pinctada fucata martensii*
Yu SHI , Xiaolan PAN , Meng XU , Huiru LIU , Hanzhi XU , Maoxian HE ,**
1 CAS Key Laboratory of Tropical Marine Bio-resources and Ecology, Guangdong Provincial Key Laboratory of Applied Marine Biology, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou 510301, China
2 Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou), Guangzhou 511458, China
Abstract Inhibitory Smads (I-Smads), which belong to the Smad family and inhibit bone morphogenic protein 2 (BMP2) signaling by a variety of mechanisms, can suppress innate immunity responses in vertebrates. However, there are no reports for the role of Smad6 in immunity in mollusks. In this study, we showed that Smad6 of the pearl oyster Pinctada fucata martensii was located in the Smad6 cluster of the phylogenetic tree; mRNA expression of Smad6 and Smad3 was up-regulated after lipopolysaccharide and polyinosinic: polycytidylic challenge; and transcript levels of Smad6 and Smad3 showed opposite patterns during wound healing. Under salinity stress, water inflow and outflow in the gills appear to be regulated by BMP2-Smads signals, and BMP2-Smads signaling may be closely related to the immune response. Our results indicate that Smad6 is involved in immunity, that it plays a positive role in the response to immune challenge and an inhibitory role during wound healing, and that Smad6 and Smad3 may work against each other.
Keyword: Smad6; BMP2-Smads signal pathway; expression; immunity; Pinctada fucata martensii
1 INTRODUCTION
The transforming growth factor β (TGF-β) family plays crucial roles in embryonic development,differentiation of cells, formation of organs, and adult tissue homeostasis. The family includes TGF-βs,activins, bone morphogenetic proteins (BMPs), and growth and differentiation factors (GDFs). TGF-βs and activins are preferentially phosphorylated by Smad2 and Smad3, whereas BMP signals are preferentially transmitted by Smad1/5/8 (Zhang et al.,1996).
The Smad protein family is an important medium for transducing BMP-Smads signals and includes R-Smads (receptor-regulated Smads, Smad1, 5, 8, 9),Co-Smads (common partner Smads, Smad4), and I-Smads (inhibitory Smads, Smad6, 7). BMPs trigger signaling through binding the transmembrane receptor BMPR-II. The formed ligand-receptor complex phosphorylates and activates BMPR-I, which has intrinsic serine-threonine kinase activities. The activated BMPR-I phosphorylates R-Smads, which then form heterotrimeric complexes with Co-Smads.The formed complexes then translocate to the nucleus to bind to co-activating factors or inhibitory factors to activate or repress gene expression (von Bubnoffand Cho, 2001).
Inhibitory Smads (I-Smads) contain a conserved carboxy-terminal MH2 domain (Goto et al., 2007).Smad6 inhibits BMP2 signaling by a variety of mechanisms, including interaction with activated type I receptors or with activated phosphorylation of Smad1/5 (Ishida et al., 2000; Itoh et al., 2001), and the MH2 domain is required for these interactions.Smad6 can also interact with Smad4 to inhibit BMP2 signaling (Hata et al., 1998). In the nucleus, Smad6 inhibits the BMP signaling pathway by interacting with homeobox C8 (Hoxc8) and Hoxc9 or by binding to the target DNA (Bai et al., 2000; Bai and Cao,2002). In vertebrates, Smad3 preferentially transfers TGF-β signals, and TGF-β signaling is inhibited by Smad7 (Hanyu et al., 2001). However, Smad7 was not found in mollusc, there was only one inhibitory Smad (Smad6) in mollusc. Therefore, if Smad3 can transfer BMP2 signals and if Smad6 can inhibit BMP2 signaling by interacting with Smad3 inPinctadafucatamartensiineeds to be studied. To date, most studies of BMP-Smads signaling pathwayrelated genes focused on characterization of ligands,receptors, and Smad family members and their involvement in development and shell formation in invertebrates, such asCrassostreagigas(Herpin et al., 2005; Lelong et al., 2007; Le Quéré et al., 2009;Liu et al., 2014),Lymnaeastagnalis(Iijima et al.,2008; Shimizu et al., 2011),P.f.martensii(Miyashita et al., 2008; Shi et al., 2020),Patellavulgate(Nederbragt et al., 2002),Drosophila(Raftery and Sutherland, 1999),Hydra(Reinhardt et al., 1996),Caenorhabditiselegans(Savage et al., 1996), and various corals (Zoccola et al., 2009). However, little is known about the roles of these genes in molluscan immunity.
Members of the TGF-β superfamily are involved in innate immunity in both vertebrates and invertebrates(Lieber and Luckhart, 2004; Nicholas and Hodgkin,2004). In vertebrates, I-Smads can mediate the effect of TGF-β on anti-inflammatory activity. Smad6 suppresses innate immunity responses by recruiting Smurfs to MyD88 and triggers its polyubiquitylation and proteasomal degradation to inhibit the MyD88-dependent pathway (Lee et al., 2011). Smad6 also can interfere with the toll-like receptor 4 signaling pathway (Choi et al., 2006). Smad6 methylation represses nuclear factor κB (NF-κB) activation and periodontal inflammation (Zhang et al., 2018). In mollusks, a TGF-b/activin homologue fromC.gigas(Lelong et al., 2007), Smad4 ofBiomphalariaglabrata(Adema et al., 2010), Smad3 and Smad5 ofHyriopsiscumingii(Hu et al., 2017; Li et al., 2020),and Smads1/5 ofP.f.martensii(Shi et al., 2021) were reported to be involved in immunity against bacterial challenge and in wound healing. However, whether Smad6 plays a role in immunity in mollusks is unknown.
The pearl oysterP.f.martensiiis mainly cultivated in China and Japan for pearl production. This marine bivalve also is an important model for studying biomineralization, genetic variation, growth, and resistance to disease. The process of artificially inserting an extraneous piece of mantle into the recipient oyster to form a pearl sac can injure the animal and lead to bacterial and viral diseases, which in turn can cause large-scale mortality and enormous economic losses (Miyazaki et al., 1999; Kitamura et al., 2000; Potasman et al., 2002; De Zoysa and Lee,2009). Thus, it is important to understand the innate immunity of this species to improve culture conditions and pearl production.
In our previous studies, we characterized the BMP2-Smads signals inP.f.martensiiand explored the signaling pathway by which Smad1/5 transduces the BMP2 signal to regulate shell formation (Zhao et al., 2016; Shi et al., 2020). We identified Smad6,which regulates BMP2-Smads signaling with a negative feedback in vertebrates. However, whether Smad6 is involved in immunity and whether it plays a similar negative role inP.f.martensiiwas not known.Therefore, the goals of this study were to phylogenetically analyze the protein sequences of Smad6, examine the mRNA expression ofSmad6in different tissues and developmental stages,characterize the role of Smad6 and Smad3 after immune challenge, and investigate the expression of Smad6 and Smad3 during wound healing. We also assessed the expression of genes involved in BMP2-Smads signaling [BMP2(AB176952),Smad3(ABX57736),Smad4(AGY49100),Smad1/5(AGI96394),Smad6(AGI96395), and muscle segment homeobox (MSX) (KJ028208)] and immunity [NF-κB(JQ061255), matrix metalloproteinase1 (MMP, KC881251), interleukin 17 (IL-17, JX971444), tissue inhibitor of metalloproteinase (TIMP, KP852352), copper-zincsuperoxide dismutase (CuZn-SOD, PB 4757.2 from the genome ofP.f.martensii), and lipopolysaccharide(LPS)-induced tumor necrosis factor α (LITAF, PB 1747.11 from the genome ofP.f.martensii)] after oysters were exposed to salinity stress.

Table 1 Gene-specific primers1 used for qRT-PCR analysis
2 MATERIAL AND METHOD
2.1 Animal
Pinctadafucatamartensii(2 years old) were obtained from the Marine Biology Research Station at Daya Bay of the Chinese Academy of Sciences(Shenzhen, Guangdong, China). These oysters were cultivated in the sea and acclimated in floating net cages under natural conditions. All animal experiments were conducted in accordance with the guidelines and approval of the respective Animal Research and Ethics Committees of the Chinese Academy of Sciences (Shi et al., 2020).
2.2 Phylogenetic tree
Multiple alignments of the Smad6 amino acid sequences were performed using ClustalW 1.81.Protein phylogenetic analysis was conducted with MEGA (Molecular Evolutionary Genetics Analysis)6.0 using the neighbor-joining method.
2.3 Expression of Smad6 in tissues of P. f. martensii
The expression pattern ofSmad6mRNA in the tissues ofP.f.martensiiwas analyzed using quantitative real-time PCR (qRT-PCR). Total RNA was isolated from eight tissues (intestines, adductor muscle, foot, mantle, heart, testis, digestive gland,and gill) from three individuals. qRT-PCR was performed on the samples following Shi et al. (2012).Smad6transcript levels were normalized against 18s transcripts levels, as 18s was expressed equally in all tested tissues. The level of expression was calculated using the 2-ΔΔCtmethod. Table 1 lists the primers used for qRT-PCR analysis.
2.4 Expression of Smad6 in different developmental stages of P. f. martensii
To analyze the developmental expression patterns ofSmad6inP.f.martensii, 10 developmental stages(polar body stage, four cell, eight cell, blastula stage,gastrula stage, trochophores, D-shaped larvae, umbo larvae, eyespot larvae, and attached larvae) were collected and stored at -80 °C. The total RNA extraction and qRT-PCR analyses were performed following Shi et al. (2012) for three replicates of each developmental stage. The mRNA expression of 18s was stable in all tested developmental stages, so it was used to calculate the relative level ofSmad6expression.
2.5 LPS and polyinosinic:polycytidylic (poly (I∶C))challenge
Pinctadafucatamartensiiwere randomly divided into three groups for intramuscular injection with i)100 μL of LPS (Sigma-Aldrich, St. Louis, MO, USA)dissolved in sterile phosphate-buffered saline (PBS)at a final concentration of 1 μg/μL, ⅱ) 100 μL of poly(I∶C) (Invivogen, San Diego, CA, USA) dissolved in sterile PBS at a final concentration of 1 μg/μL, or ⅲ)100 μL of sterile PBS as the control. After treatment,nine individuals form each group were randomly sampled at 0, 12, 24, 36, 48, and 72 h post injection.Mantle tissues were quickly dissected, frozen in liquid nitrogen, and stored at -80 °C for subsequent RNA extraction. Expression ofSmad6andSmad3mRNA was examined using qRT-PCR as described previously (Shi et al., 2020).
2.6 Wound healing assay
The V-shaped notch experiment was performed according to a previously described method (Shi et al., 2020). The primers used to measure the expression levels ofSmad6andSmad3are listed in Table 1.
2.7 Salinity stress experiment
For the salinity stress experiment, we soaked five plastic buckets in 10 mg/L of potassium permanganate solution for 10 min, rinsed them, and then filled with 70 L of filtered seawater. Fresh water and salt crystals were mixed to create a salinity gradient of 16, 20, 29,32, and 36. Because the salinity of seawater in the aquaculture area was 29, 29 was used as the control group. Two hundredP.f.martensiiwere randomly divided into five groups and placed in the buckets.Five individuals from each group were randomly sampled at 4, 24, 72, 120, and 168 h after treatment.Gill tissues were quickly dissected, frozen in liquid nitrogen, and stored at -80 °C for subsequent RNA extraction.BMP2,Smad3,Smad4,Smad1/5,Smad6,MMP,MSX,NF-κB,IL-17,TIMP,CuZn-SOD, andLITAFmRNA expression levels were measured using qRT-PCR, which was performed as described previously (Shi et al., 2020).
2.8 Statistical analysis
Quantitative data are shown as the mean ±standard deviation (SD). Figures were constructed using GraphPad Prism 8 (La Jolla, CA, USA). All values were compared by one-way analysis of variance (ANOVA) followed by Duncan’s multiple range tests, two-way ANOVA followed by posttests,or at-test, all performed using SPSS 13.0 (SPSS Inc., Chicago, IL, USA). Differences were considered to be statistically significant atP<0.05 orP<0.01.
3 RESULT
3.1 Phylogenetic analysis of the Smad family of P. f. martensii
Phylogenetic analysis showed that the Smad family genes are clustered into five separate clades inP.f.martensii: the Smad1/5/8/9 cluster and Smad2/3 cluster are aggregated into R-Smads, the Smad6 cluster and Smad7 cluster are aggregated into I-Smads, and the Smad4 cluster is aggregated into Co-Smad. Smad5 ofP.f.martensiiis located in the Smad1/5/8/9 cluster and is most closely related to that ofCrassostreavirginica; Smad3 is located in the Smad2/3 cluster and is most closely related to that ofC.gigas; Smad4 is located in the Smad4 cluster and is most closely related to that ofC.virginica; and Smad6 is located in the Smad6 cluster and is most closely related to that ofC.gigasandC.virginica(Fig.1).
3.2 Tissue distribution of Smad6 in P. f. martensii
TheSmad6mRNA was expressed in all tissues examined, with the highest level in the gill, a moderate level in adductor muscle, mantle, testis, intestines,and foot, and a low level in the digestive gland and heart (Fig.2).
3.3 Expression of Smad6 in different developmental stages of P. f. martensii
TheSmad6mRNAs were expressed at all developmental stages examined. The mRNA levels were moderate in the polar body stage, peaked at the eight-cell stage, decreased to a moderate level from the blastula to the gastrula stage, and then decreased significantly in the trochophore stage. The expression level was low from the D-shaped larva stage to the attached larval stage (Fig.3).
3.4 Responses of Smad6 and Smad3 in P. f. martensii challenged with LPS and poly (I∶C)
To determine whetherSmad6andSmad3in the mantle ofP.f.martensiiare involved in innate immune defense, their temporal expression patterns were monitored after the oysters were stimulated by LPS and poly (I:C). After LPS stimulation, the mRNA expression ofSmad3significantly increased at 36 h but returned to the original level at 48 and remained there until the end of the experiment;expression was significantly up-regulated at 12 h,peaked at 48 h, and remained high thereafter in the poly (I:C) stimulation group. The mRNA expression ofSmad6was significantly increased at 24 h,remained high until 48 h, and peaked at 72 h after LPS stimulation; expression was significantly upregulated at 48 h and 72 h after poly (I:C) stimulation(Fig.4).
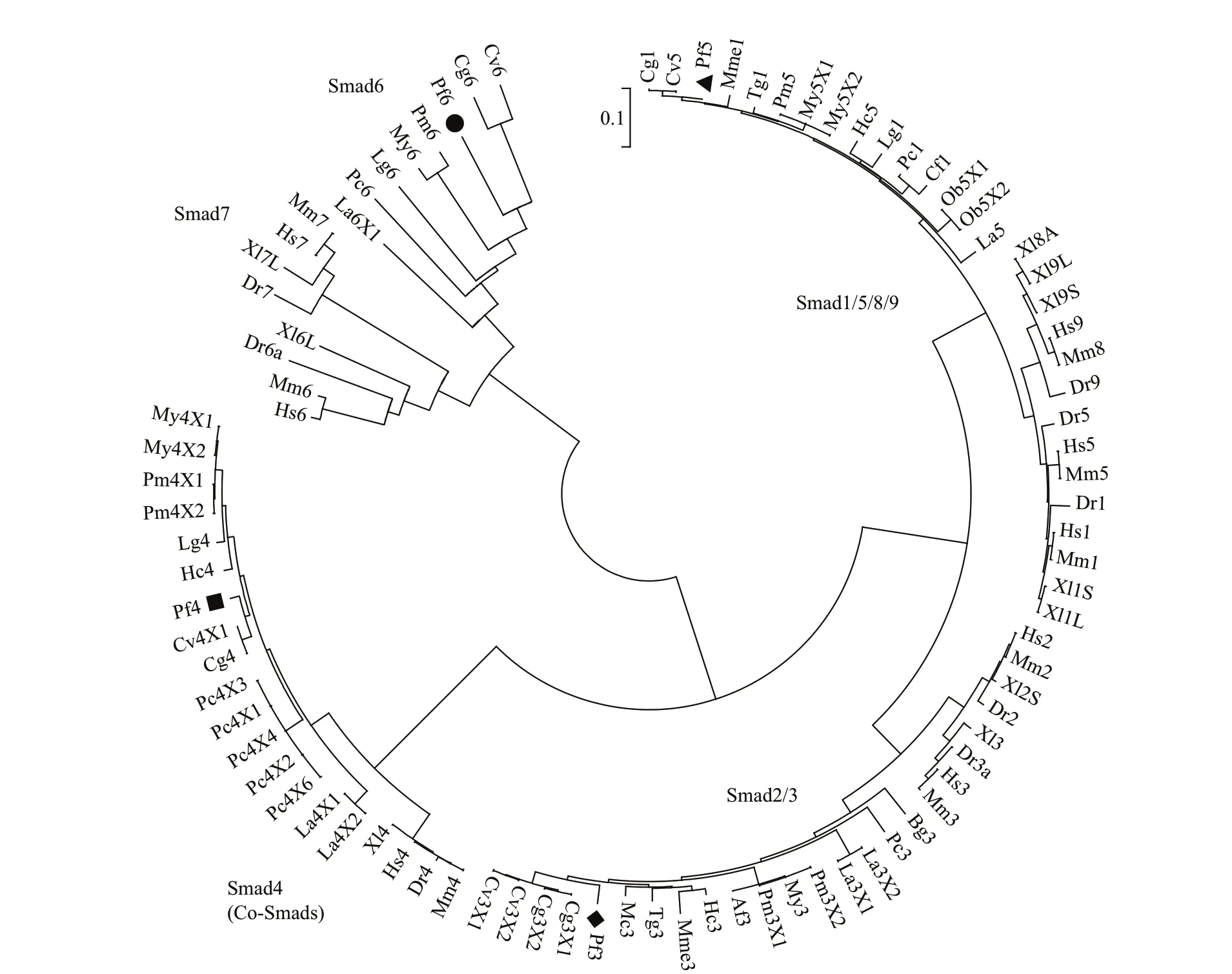
Fig.1 Phylogenetic analysis of the Smads family
3.5 Smad6 and Smad3 mRNA expression during wound healing
In the wound healing experiment, the mRNA expression ofSmad3was significantly down-regulated from 2 to 24 h compared with 0 h, then it dramatically increased at 120 h post wounding. The transcript levels ofSmad6were significantly higher at 24 h and 120 h post wounding compared with 0 h (Fig.5).
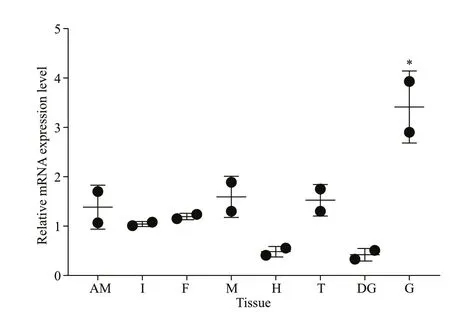
Fig.2 Expression pattern of Smad6 mRNA in various tissues of P. f. martensii
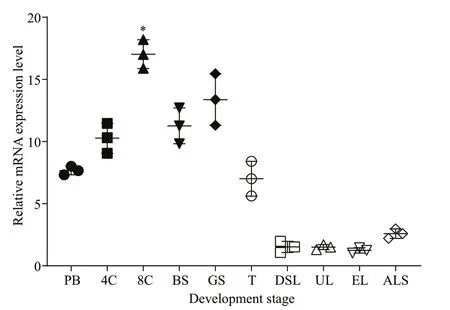
Fig.3 Expression pattern of Smad6 mRNA at the developmental stages of P. f. martensii
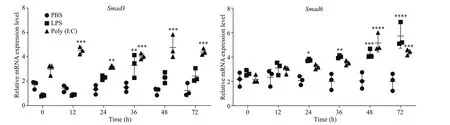
Fig.4 mRNA expression levels of Smad6 and Smad3 in mantle tissues of P. f. martensii after challenge with LPS and poly(I:C)
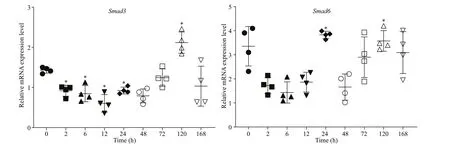
Fig.5 mRNA expression levels of Smad6 and Smad3 in the mantle of P. f. martensii after wound healing
3.6 Expression of genes involved in BMP2-Smads signaling and immunity after exposure to salinity stress
The temporal expression of genes involved in BMP2-Smads signaling and immunity in the gills ofP.f.martensiiafter exposure to salinity stress was evaluated to better understand their role mantle immunity function. The mRNA expression ofBMP2,Smad4,Smad1/5,Smad6,MSX,IL-17,TIMP,CuZn-SOD, andLITAFwas significantly up-regulated compared to the control (29) during low salinity (16 or/and 20) treatment, whereas the levels ofNF-κBandMMPtranscripts were significantly decreased.Smad3expression increased in the 20 salinity treatment at 24 h but then decreased from 72 to 120 h. High salinity treatment (32 or/and 36) resulted in significantly increased expression ofBMP2,Smad4,Smad1/5,Smad6,MSX,IL-17,TIMP,CuZn-SOD, andLITAFand decreased expression ofSmad3andNF-κBcompared to the control (29) from 4 to 168 h.MMPexpression was significantly down-regulated at 4 h and from 72 to 168 h after 32 salinity treatment, but it was higher than the control at 24 h. At 36, expression ofMMPdecreased at 4 h, peaked at 24 h, remained high until 120 h, and then decreased at 168 h (Fig.6).
4 DISCUSSION
We conducted multiple sequence alignment to study the phylogenetic relationships between Smad6 and other members of the Smads family inP.f.martensiiand found that five independent branches (Smad1/5/8/9, Smad2/3, Smad4, Smad6,Smad7) are present in the phylogenetic tree. Smad6 is located in the Smad6 cluster and is most closely related to that ofC.gigasandC.virginica.
Smad6was expressed in all tissues examined,showing its wide distribution in the adult body. This pattern indicates that Smad6 has diverse functions in physiological processes ofP.f.martensii. Moreover,the highest expression level in the gill and moderate level in the mantle suggest that Smad6 may be involved in immunity ofP.f.martensii.
Smad6mRNA also was expressed in all 10 development stages tested, with high levels in embryonic stages and low levels in the larval stages.This result suggests that Smad6 may play an important role in early development ofP.f.martensii. The period from the trochophore stage to D-shaped larvae is very important for metamorphosis in bivalves, and key activities such as cell proliferation and shell formation takes place during this period (Liu et al.,2014). In our study, the mRNA expression ofSmad6decreased beginning at the D-shaped larval stage,which suggests that the inhibitory effect of Smad6 was down-regulated.
The mRNA expression levels ofSmad3andSmad6were up-regulated in the mantle ofP.f.martensiiafter LPS and poly (I:C) stimulation. These data suggest the potential role of Smad3 and Smad6 in innate immunity to defend against bacterial and viral infections. The mRNA expression ofSmad3was significantly up-regulated 12 h after Poly (I:C)treatment and remained high at 72 h. It also increased 24 h after LPS treatment and remained high until 36 h. These results indicate that Smad3 was more sensitive and more persistent to Poly (I:C) than to LPS stimulation. Hu et al. (2017) reported that HcSmad3 expression inH.cumingiiwas up-regulated in hemocytes and the hepatopancreas afterAeromonashydrophilaand peptidoglycan stimulation, suggesting that Smad3 may play a role in innate immunity in bivalves. However, Smad6 seemed be more sensitive and more persistent to LPS than to Poly (I:C),suggesting that it may be involved in immunity in a different way from that of Smad3. Additionally,Smad6 seemed to play a positive role after LPS and poly (I:C) challenge inP.f.martensii.
Wound healing is an essential process in which the separated or destroyed tissue attempts to restore itself to its normal state (Mokoena et al., 2018). TGF-β is one of the essential growth factors involved in wound healing, and it is dependent on the Smads pathway.TGF-β activates downstream mediators Smad2 and Smad3, which results in the differentiation of fibroblasts into alpha smooth muscle-expressing myofibroblasts(Xu et al., 2016). In our study,Smad3expression significantly decreased from 2 to 24 h after wounding,but then increased at 120 h. In contrast,Smad6expression was significantly up-regulated at 24 and 120 h after wounding. This opposite expression pattern suggests that Smad3 may play a positive role and Smad6 may play a negative role during wound healing.However, whether up-regulated Smad6 expression suppresses Smad3 expression within 24 h of wounding requires further investigation. InH.cumingii, Smad3 expression did not change significantly after wounding,whereas expression of Smad4 and HcSmad5 was significantly up-regulated post wounding (Hu et al.,2017; Li et al., 2020).BiomphalariaglabrataSmad4 and Cgi-SMAD1/5/8 levels were also increased post wounding (Adema et al., 2010; Liu et al., 2014). These results indicate that Smads family members may play a complex role in mollusk immunity.
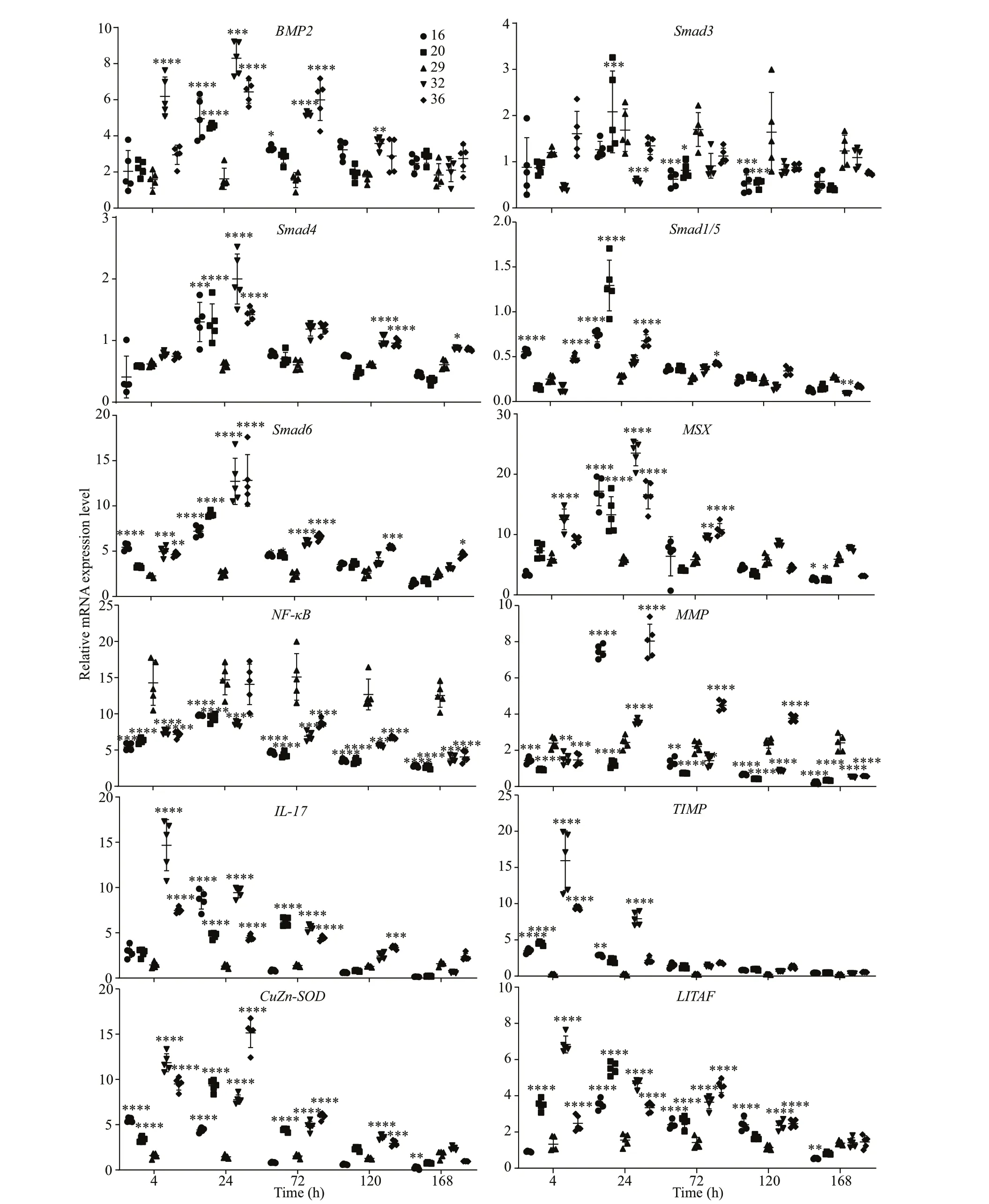
Fig.6 mRNA expression levels of BMP2, Smad3, Smad4, Smad1/ 5, Smad6, MSX, NF- κB, MMP, IL- 17, LITAF, CuZn- SOD,and TIMP in the gills of P. f. martensii cultured under different salinity concentrations (29 was the control salinity concentration)
Most marine bivalves can vary their osmotic pressure. When encountering salinity change, they first respond by closing the shell and sealing the mantle cavity, and then they regulate the expression of genes related to ion exchange and water transport in the body (Navarro, 1988). Under static osmotic pressure, the main osmotic effectors in shellfish hemolymph are balanced by the organic and inorganic ions in the external water environment, and cells will not expand or shrink and the organism can carry out normal life processes (Pasantes-Morales and Schousboe, 1997; Wilder et al., 1998). Although marine shellfish do not have a specific osmotic regulation organ, the gill tissue performs most of this function. Gill tissue and the hepatopancreas of the scallopArgopectenirradianswere substantially damaged under extreme salinity stress. Previous studies demonstrated that increased dietary sodium chloride (salt) intake stimulated endothelial cells to produce TGF-β (Ying and Sanders, 1998, 1999, 2002,2003; Ying et al., 2008, 2012), which led to increased levels of phosphorylated Smad2 (S465/467) (Ying et al., 2013). However, the role of Smads in regulation of salt concentration by the gills in mollusks is poorly understood. Therefore, we investigated the temporal expression of genes involved in BMP2-Smads signaling and immunity in the gills ofP.f.martensiiafter stress treatment with different salinity concentrations.
Both low salinity (16 or/and 20) and high salinity(32 or/and 36) stress significantly up-regulated the mRNA expression ofBMP2,Smad4,Smad1/5,Smad6, andMSX, indicating that the gills may control water flow in and out of the organism under salinity stress by increasing the expression level of BMP2-Smads signals. The effect oflow salinity on BMP2-Smad signaling occurred mainly within 24 h, whereas the effect of high salinity lasted for more than 24 h.This result suggests thatP.f.martensiimay be better able to adapt to a low salinity environment than to a high salinity environment, which is consistent with our previous studies of osmoregulation in this species(Pan et al., 2020). The pattern of Smad3 expression was opposite that of Smad6, which was similar to the results for wound healing, and it suggests that Smad3 and Smad6 may have opposing function.
Many studies have reported that salinity stress is closely related to immunity and that salinity stress can cause an immune response. Therefore, we evaluated the expression of important immune genes inP.f.martensiiafter salinity stress. The patterns of mRNA expression ofIL-17,TIMP,CuZn-SOD, andLITAFwere similar to those observed for the BMP2-Smads signaling genes, which suggests that BMP2-Smads signaling may be closely related to the immune response ofP.f.martensiiin addition to participating in osmotic pressure regulation. The duration of upregulation of these genes during high salinity stress was longer than that during low salinity stress, which also supports the premise thatP.f.martensiimay more easily to adapt to a low salinity environment.
Previous researchers found that the TGF-β/Smad pathway can regulate IL-17 as part of immunity in vertebrates (Xie et al., 2016; Chen et al., 2018), and several studies reported that CuZn-SOD is involved in the immune response in mollusks (Kim et al., 2007;Ni et al., 2007; Lin et al., 2008; Bao et al., 2009; Li et al., 2010). Additionally, LITAF homologs were identified and characterized fromC.gigas, the scallopChlamysfarreri, andP.f.martensiithat were exposed to LPS or virus challenge (Yu et al., 2007; Park et al.,2008; Zhang et al., 2009).
The Smad DNA-binding motif was previously found in the promoter regions of MMP and TIMP,which suggests that they may be the direct target genes of Smads (Verrecchia et al., 2001; Qureshi et al., 2008). Kubota et al. (2017) showed that TIMP and MMP were involved in shell wound repair, and Montagnani et al. (2001) reported similar functions were for TIMP inC.gigas. We found that the mRNA expression ofMMPinP.f.martensiiwas up-regulated or down-regulated by high salinity or low salinity stress, respectively, which suggests that MMP may be involved in the immune response to salinity stress via a complex signaling pathway. NF-κB is a classic,evolutionarily conserved, mediator of immune responses in vertebrates (Ghosh et al., 1998), and it also plays an important role in innate immune responses inP.f.martensii(Huang et al., 2012). The down-regulated expression of NF-κB after both high and low salinity stress suggests that it was involved in the immune response to salinity stress.
5 CONCLUSION
Phylogenetic analysis showed that Smad6 ofP.f.martensiiis located in the Smad6 cluster, which illustrates conservation of the Smads family. The tissue distribution of Smad6 suggests its possible role in immunity, and the expression pattern of Smad6 in different developmental stages indicates that it likely plays a crucial role in development at embryonic stages, but not larval stages. The increased expression ofSmad3andSmad6mRNAs in mantle tissue after LPS and Poly (I∶C) challenge show that they may be involved in innate immunity against bacterial and viral infections. Their expression patterns during wound healing suggest that Smad3 may play a positive role while Smad6 may play a negative role in this process. The temporal expression patterns of genes involved in BMP2-Smads signaling and immunity in the gills after stress treatment with different salinities suggest that the gills control water flow during salinity stress by increasing the expression level of BMP2-Smads signals and that BMP2-Smads signaling may also be closely related to the immune response ofP.f.martensii. Finally, Smad6 and Smad3 may counteract each other during wound healing and salinity stress.
6 DATA AVAILABILITY STATEMENT
All data generated or analyzed during this study are included in this article.
7 AUTHOR CONTRIBUTION
Yu SHI planned and designed the research.Maoxian HE supervised the experiments. Yu SHI performed most of the experiments, analyzed data,and wrote the paper. Meng XU performed the challenge assay, Xiaolan PAN and Huiru LIU performed the wound healing experiment; and Hanzhi XU helped perform the salinity stress assay. All authors reviewed, edited, and revised the manuscript.
 Journal of Oceanology and Limnology2022年3期
Journal of Oceanology and Limnology2022年3期
- Journal of Oceanology and Limnology的其它文章
- Erratum to: Impact of typhoon Lekima (2019) on material transport in Laizhou Bay using Lagrangian coherent structures*
- Erratum to: Chinia gen. nov.—the second diatom genus simonsenioid raphe from mangroves in Fujian, China*
- Characterization of Amphidinium (Amphidiniales,Dinophyceae) species from the China Sea based on morphological, molecular, and pigment data*
- Cytological and transcriptional analysis reveal phosphatidylinositol signaling pathway plays key role in mitotic division of Pyropia yezoensis*
- Carnosine concentration and expression profiles of carnosine related genes in Mytilus after beta-alanine injection*
- Microsatellite marker development and population genetic analysis revealed high connectivity between populations of a periwinkle Littoraria sinensis (Philippi, 1847)*
