食源性抗生素耐药菌的污染现状、传播扩散及健康风险研究进展
曹弘扬 ?汪庆?赵佳丽?梁海崟?郭绍月?万春云?骆慧晓

摘要:抗生素的过度使用加剧了环境耐药菌的产生,给人类健康造成潜在风险。在食品链多个环节过度使用或滥用抗生素,造成肉制品、水产品和水果蔬菜等多种食品中细菌耐药性逐年增强。携带耐药质粒的耐药菌通过“农场到餐桌”转移、定植在人体肠道,引起人体肠道菌群变化和免疫功能改变。食品的全球化贸易进一步加剧了细菌耐药性的全球性传播。本文综述了食源性耐药菌污染现状、传播途径和人体健康风险。同时,结合国内外研究现状,对食品环境中抗生素耐药菌的研究重点进行展望。
关键词:抗生素耐药菌;食源性;传播途径;健康风险
中图分类号:R978文献标志码:A
Progress on the contamination status, dissemination, and health risks of foodborne antibiotic resistant bacteria
Cao Hong-yang1, Wang Qing1, Zhao Jia-li2, Liang Hai-yin1, Guo Shao-yue1, Wan Chun-yun1, and Luo Hui-xiao1
(1 College of Energy and Environmental Engineering, Hebei Key Laboratory of Air Pollution Cause and Impact, Hebei University of Engineering, Handan 056038; 2 Medical College, Hebei University of Engineering, Handan 056038)
Abstract The abuse of antibiotics has aggravated the pollution of antibiotic resistant bacteria, posing a potential hazard to human health. Antibiotics have been widely used in many parts of the food chain, which resulted in the increase of antibiotic resistance in meat foods, aquatic foods, fruits, and vegetables. These antibiotic resistant bacteria with resistant plasmid are transferred and colonized in the human gut through 'farm-to-fork', causing changes in the human gut flora and immune functions. The global trade of food has further intensified the antibiotic resistance bacteria transfer and dissemination among the microorganisms. This paper introduced the current status of food-borne drug-resistant bacteria contamination, the transmission routes of antibiotic resistance, and human health risks. In addition, some countermeasures and prospects for the research on antibiotic resistant bacteria in food are reviewed.
Key words Antibiotic resistance bacteria; Food-borne; Transmission routes; Health risk
食品中抗生素耐藥性污染问题十分严重,已成为全球广泛关注的热点话题[1]。随着畜牧业、水产养殖和农业等迅速发展,人们在预防和治疗动植物疾病等方面过度使用抗生素,导致环境中细菌的耐药性逐年增强[1-3]。中国是抗生素生产和使用量最大的国家,仅在2013年抗生素使用量已到达1.62×105吨,占据全世界抗生素总使用量的23%[4]。并且抗生素的使用量依然会逐年增加,预估在2030年抗生素的使用量将是2010年的1.67倍[5]。证据表明,抗生素使用不当会加快抗生素耐药菌在大气[6]、海洋[7]和土壤[8]中的传播。美国疾病控制与预防中心表明,每年仅美国因感染抗生素耐药菌患病人数超200万人,死亡人数约2.3万人,如果不及时采取有效措施,抗生素耐药菌的污染将进一步扩大[9]。
目前,我国食源性耐药菌引发的疾病和耐药率呈现快速上升趋势,给人体健康造成潜在健康风险[10]。在鸡肉、猪肉、海产品、水果和蔬菜等多种食品中都已检测到抗生素耐药菌的存在,同时,至少已发现沙门菌、大肠埃希菌、空肠弯曲菌、单增李斯特菌和副溶血弧菌等致病菌表现出较强的耐药性[11-18]。因此,加强食品中耐药细菌的监测和管控迫在眉睫。本文综述了多种食品中抗生素耐药菌的研究现状,介绍了耐药菌在食物链中的来源、传播途径和人体健康风险,并对未来研究重点进行展望。
1 食源性耐药菌的污染现状
抗生素耐药菌广泛存在于多种食品,并且具有污染范围广和污染率高等特点[11]。近年来,不同国家和地区食品中检测出抗生素耐药菌种类和数量逐年上升[19]。在我国,食源性耐药菌的污染与传播同样不容忽视,其中肉类和水产食品被耐药菌污染最为严重,最高可达到59%[20]。
1.1 肉类食品
过度使用抗生素导致肉类食品中存在大量的抗生素耐药菌。研究人员从零售店的肉类食品中分离出了耐药菌[21-22],且种类多、丰度大[23-24]。根据大量数据调研表明(表1),发展中国家禽类食品中检测的沙门菌阳性菌株比例较高,南美地区阳性样本数量为13%~39%[25],非洲地区检测到阳性样品比例约为35%[26-27],亚洲地区检测到阳性样品比例为35%~60%[28-30],而在美国、英国等发达国家食品中检测到的抗生素耐药菌明显较低[22]。
肉类食品中分离出的耐药菌主要包括产志贺毒素大肠埃希菌[31-33]、金黄色葡萄球菌[33]、单核细胞增生李斯特菌[34-35]和鼠寒沙门菌[36]。由于耐药性具有可转移的特性,耐药菌中的抗性基因能够通过水平转移分子机制转移到肠道菌群内[37]。但根据最新的研究,停止给动物喂含有抗生素的饲料3~4周后,动物肠道、血液和肌肉中抗生素含量下降,其体内耐药菌丰度减少[38]。因此,可以在屠宰前停止喂含抗生素的饲料,以降低动物体内耐药菌的占比。
1.2 水产食品
水产养殖中大量使用的抗生素最终会进入到养殖水环境和水产食品中,导致产生越来越多耐药菌[39]。水产食品中耐药菌多数具有耐低温和耐高渗透压的特性,可以在极端环境中生存。人类在通过手拿、生吃等方式感染耐药菌,极易对人体健康造成潜在危害[40]。Tan等[41]对马来西亚的水产食品检测发现,副溶血弧菌污染了85.7%的水产食品,且对青霉素、氨苄西林和头孢唑林的耐药性较高,其中青霉素的耐药性为100%。Tran等[42]对越南水产食品中分离的菌株进行检测后同样发现,副溶血弧菌感染了86.2%的样品,且大多数菌株对氨苄西林、磺胺异恶唑和链霉素具有耐药性。Jiang等[43]通过检测发现,黄海和渤海水产食品中的副溶血弧菌污染较重,并对阿米卡星和头孢唑林等有多重耐药性。Ellis-Iversen等[44]收集丹麦零售店的水产食品进行耐药性检测,有89.7%的水产食品中检测到大肠埃希菌,其中45.6%的大肠埃希菌对至少一种抗生素具有耐药性。因此,水产食品中耐药菌污染已十分严重。
我国沿海沿河地区拥有丰富的水产食品,当地居民多以此为食。未被人体和动物利用的抗生素会随粪便或尿液进入环境,总量多达5.58×104吨,其中46%排放到水体环境[4],致使水产食品中抗生素耐药菌普遍增加[45],长期摄入这些耐药菌是否会对人体健康造成潜在风险值得深入探究。
1.3 乳类食品
生乳及乳制品中检出耐药菌对食品安全再次敲响了警钟[46]。生乳制品的微生物污染状况主要取决于动物的健康状况、养殖环境、挤奶环境和挤奶器的卫生状况[47]。生奶中由于水分含量高,pH中性,并且有丰富的营养物质,适合微生物繁殖。因此,乳品加工厂为了保证鲜奶的品质和延长鲜奶储存,不得已会在生乳的加工过程中添加微量的抗生素。
生乳[48]、巴氏灭菌奶[49]和奶酪[46]中均分离出了耐药菌。Sharma等[50]检测牛奶样品发现,生乳中有19.8%的样品被金黄色葡萄球菌污染,其具有凝固酶活性的金黄色葡萄球菌中,有90%的菌株对至少3种抗生素具有抗性。Ameen等[51]从患有乳腺炎的奶牛身上采集生乳样品,发现被金黄色葡萄球菌污染的样品有30%,且多数金黄色葡萄球菌对青霉素具有抗性。Aksomaitiene等[52]对来自牛奶的空肠弯曲杆菌菌株进行测试,结果表明分离的菌株均对至少一种抗生素具有抗性,其中对头孢曲松、环丙沙星和四环素耐药率分别为100%、90.2%和85.4%。Wang等[53]对乳制品中分离的乳酸菌进行检测,发现88.9%的分离株对至少一种抗生素具有耐药性。因此,不应仅检测乳制品中致病菌的耐药性,乳酸菌等益生菌的耐药性同样也需要进行监控。
1.4 水果蔬菜类食品
直食性的水果蔬菜中耐药菌可能对人体具有更大的潜在危害。Wang等[20]对超过一千种食品进行统计研究,发现超过六成直食性的水果蔬菜中检测出金黄色葡萄球菌,且与肉类食品相比,直食性的水果蔬菜中耐药菌比例更高,耐药性更强。Chaj?cka-Wierzchowska等[54]对270份植物源性食品进行鉴定发现,污染最严重的为肠球菌,其中对链霉素耐药性最高(55.6%),其次是利福平(51.4%)。与肉类食品相比,水果蔬菜中部分耐药菌耐药性更強,且这些耐药菌可直接进入人体肠道,加快了耐药菌在肠道中传播扩散,对人体健康有更大的潜在威胁[55]。
1.5 发酵、腌菜类食品
与其他食品相比,发酵、腌菜类食品中耐药菌丰度明显较高[56]。虽然发酵、腌制过程产生的酸、高盐和厌氧环境对耐药菌有较好的灭活作用,但是这些环境选择压力会进一步促进抗生素耐药菌的传播扩散[57-58]。蔡婷等[59]通过研究四川泡菜中乳酸菌的耐药性,发现经过发酵后耐药性普遍增强,食窦魏斯菌的抗生素耐药性甚至提高了3倍。许女等[56]对传统发酵食品中乳酸菌进行耐药性分析发现,泡菜、醋醅和发酵乳制品中分离出耐药表型≥6的多重耐药菌株明显增加。
腌肉和腊肠等传统肉类发酵食品中抗生素耐药菌污染同样严重[60]。Pisacane等[61]在对意大利传统香肠进行细菌培养,发现较高数量的葡萄球菌和肠球菌,且多数位于肠衣上。Chajecka-Wierzchowska等[3]对腌肉、腊肠和奶酪的研究显示,从146份样品中共分离出58株葡萄球菌,有41.3%的葡萄球菌对头孢西丁具有耐药性。Zeng等[62]调查中国在2011—2016年肉制品中的克罗诺杆菌,发现19.1%的腊肠中鉴定出克罗诺杆菌,在所有肉制品中污染最为严重。Wang等[63]通过对比中国腊肠和意大利腊肠,发现中国腊肠多为自发发酵,个别卫生条件较差,在细菌丰度和耐药性方面均超过意大利腊肠。发酵食品中存在的耐药菌种类较少、总量较低,但耐药性较强,直接食用含有抗生素耐药菌的发酵食品,对人体肠道健康的影响值得广泛关注。
2 食源性耐药菌在食物链的传播
食物链被认为是耐药菌从动植物传播到人体的重要途径[64]。农业和畜牧业的快速发展,使得产品的生产、加工、储存和分配方式产生了巨大变化,增加了抗生素耐药菌的传播风险[20,24,33]。
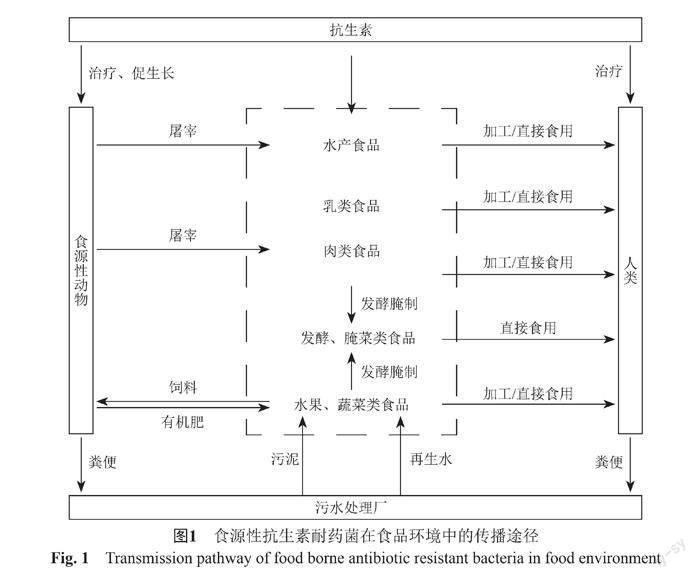
植物源性食品中耐药菌主要来源于农场环境[65]。进入动物体内的抗生素仅少量参与新陈代谢被生物利用,60%~90%的抗生素在动物体内诱导出抗生素耐药菌后,同耐药菌一起随粪便排出体外[66]。这些粪便直接或堆肥后施用于农田,造成土壤环境中耐药菌丰度显著增加。同时,粪便中残留的抗生素会对土壤中微生物产生选择压力,再次诱导产生抗生素耐药菌[67-68],且灌溉时使用再生水也可对农作物造成污染,这些耐药菌和抗生素通过食物链可以进入人体(图1)。Zhao等[69]研究农田土壤中施加动物粪便对农作物中细菌的影响,与未施肥相比,施加粪肥的土壤中细菌的耐药率明显增加,甚至高出5倍。Xiang等[70]对比有机肥种植与自然生长的蔬菜,发现有机肥种植的蔬菜不仅会增加耐药菌的丰度,也会增加耐药菌的多样性。常旭卉等[71]对施用粪肥的土壤进行检测,发现土壤中tetG、sulⅠ、qnrA、aadA2、aadD和intⅠ的绝对丰度明显增加。植物源性食品中耐药菌在肠道中的传播尤其值得关注。Zhu等[72]对灌溉再生水的农田进行研究,发现经过污水处理工艺和消毒工艺的再生水,仅含有少量的抗生素耐药菌,但对农田进行长期灌溉再生水后,依旧可导致农田中抗生素耐药菌的种类和丰度显著增加。Ma等[73]通过研究人体肠道和植物源性食品中的乳酸杆菌,发现人体肠道微生物可以从食物中获得抗生素耐药性。Losasso等[74]研究不同饮食习惯对肠道菌群耐药性的影响,发现素食主义者肠道内blaTEM耐药基因明显多于杂食者。植物源性食品中抗生素耐药菌转移到人体肠道仍是一个亟需解决的问题。
人类也可通过接触或食用动物源性食品而感染耐药菌。动物源性食品耐药菌增多主要有两种方式。首先,过量抗生素会导致动物肠道菌群等耐药菌增加[75]。Talley等[76]研究养牛场环境中的细菌,发现使用抗生素的养牛场中牛和果蝇均携带有耐药性的大肠埃希菌O157:H7。Liu等[77]研究发现了养殖场环境与产出猪肉中的抗生素耐药菌之间的关系,使用抗生素后,猪肉中抗生素耐药菌明显增加,且与养猪场环境中耐药菌种类大致相同,甚至和养猪场周围土壤也呈现出高度的相似性。Chen等[78]研究污水处理厂出水对水产食品的污染,发现污水处理厂出水中抗生素能够对10 km内的水环境造成不利影响,且可增加区域内水产食品中抗生素耐药菌丰度。其次,生产加工、运输储存和零售消费的各个阶段都可造成耐药菌的交叉污染[39,79-80]。在农场内,动物粪便中带有大量耐药菌,这些耐药菌可以通过粪口传播和接触传播再次污染动物[79,81]。养殖场内不完善的管理措施和较差的卫生条件,促使耐药菌在养殖动物之间传播[39]。Molechan等[80]对鸡的运输设备进行检测,发现运输设备中同样存在耐药菌,且可在运输的过程中通过接触污染鸡。另外,农贸市场、超市与餐厅中的砧板和刀具等也是传播耐药菌的重要途径[39]。肉类食品中鉴定出的耐药菌和耐药基因在人体中被找到,證明了被污染的食品可通过食物链感染人类[82]。Lu等[83]分析了中国上海在2006—2016年从腹泻患者中分离的沙门菌,从患者和当地猪肉中分离的mcr-1阳性菌株,大多来自同一社区,充分表明猪肉是主要传染源。Otto等[84]发现,在魁北克和安大略地区患者中分离的耐头孢噻呋肠道沙门菌主要来源于食用鸡肉。
人们通常会使用消毒、腌制、加热和冷冻等处理方式保证食品和食品加工环境的安全[85]。Yu等[86]研究经过消毒剂苯扎氯铵处理过的单核细胞增生李斯特菌,发现单核细胞增生李斯特菌增加了对头孢噻肟、头孢菌素和环丙沙星的耐药性。Govender等[20,33]对冷藏室内分离的金黄色葡萄球菌进行研究,在和国内未经过冷冻处理的金黄色葡萄球菌相比后,发现从冷藏库中分离到的金黄色葡萄球菌对青霉素有更强耐药性。
越来越多的证据表明,抗生素的不规范使用和滥用是导致食品中耐药菌增加的主要原因。因此,应减少抗生素的使用,并制定食源性食品中抗生素的使用指南。寻找合适的方法治疗食用动物,禁止抗生素作为生长促进剂,加强抗生素的管理,并采取合适的感染控制措施。
3 食品的全球贸易促进食源性耐药菌的全球传播
食品贸易全球化增加了食源性耐药菌的全球性传播扩散[87]。Cavaco等[88]在泰国患者、丹麦患者和进口食品中检测到有相同喹诺酮耐药基因qnr的科瓦利斯沙门菌,这些耐药致病菌多数来源于泰国,且最初在丹麦很少发现qnr耐药基因,随着丹麦从泰国进口越来越多的食品,耐药致病菌在丹麦正在快速传播。现今,在美国、中国、日本和丹麦检测到含有喹诺酮耐药基因的致病菌[88-89]。Vounba等[90]在加拿大、塞内加尔和越南的家禽中分离出的大肠埃希菌均携带mcr-1耐药基因。Van等[91]从尼日利亚、突尼斯和阿尔及利亚的家禽食品中分离出带mcr-1耐药基因的大肠埃希菌。
鸡肉已经成为食源性耐药菌在全球传播的主要载体[92]。中国、巴西和泰国等发展中国家已经成为动物源性食品的主要出口国[93],例如巴西生产的鸡肉会销往美国、中国和英国等142个国家[92]。有证据表明,鸡肉生产过程中常使用四环素类、磺胺类和氟喹诺酮类等多种抗生素,导致鸡肉中致病菌耐药性增强,食品贸易的全球化进一步把耐药性致病菌传播到世界的其他地区[91]。Roth等[92]利用数据库筛选出美国本土鸡肉和进口鸡肉的耐药菌信息,发现进口鸡肉中检测到大肠埃希菌对氟喹诺酮类药物的耐药率均高于40%,但在禁止使用氟喹诺酮类药物的美国本土鸡肉中耐药率则低于5%。Aarestrup等[94]使用数据库对丹麦食品中多重耐药链球菌进行分析,其中近一半的多重耐药链球菌来自泰国,且从泰国进口的鸡肉中分离的链球菌对萘啶酸、链霉素、庆大霉素、氨苄西林、氯霉素和环丙沙星的耐药率分别为88.6%、88.6%、63.6%、52.3%、36.4%和5%,均高于丹麦本土生产的鸡肉。食源性耐药菌在美国、加拿大、英国等发达国家也表现出迅速蔓延的趋势[95]。食品的贸易全球化能够把耐药性致病菌传播到世界各地,局部地区的管控并不能从整体上降低或消除人类感染耐药菌的风险。
不同的国家和地区,食源性耐药菌污染程度不同,但食品在世界各地消费量逐年增加,食品供应链全球化加剧了耐药菌的传播扩散。目前对食源性抗生素耐药菌在全球传播的研究多以数据库为基础进行,此种研究方法对解析耐药菌耐药机制和预防耐药菌传播等有不可比拟的优势,但庞大数据库的建立需要长时间大量的耐药菌与耐药基因信息,前期投入巨大。此外,抗生素耐药性已经成为一个全球性问题,它不仅限于某些地区或国家,各个国家都需要持续监测耐药性致病菌的迁移和进出口状况,采取多种方式来抑制抗生素耐药性的出现和传播[96]。
4 健康风险
人类肠道被认为是抗生素耐药基因的储存库[97]。有证据显示,农业和养殖业中的耐药菌是人体肠道微生物中主要的耐药菌,证明了“农场到餐桌”的假设,即食用被细菌污染的食品能明显改变人体肠道菌群[98]。肠道微生物在消化和代谢中发挥着至关重要的作用,长期不良的饮食习惯可能导致一些微生物丰度下降,另一些微生物丰度增加,易导致肥胖、炎症和过敏等不良反应。Takewaki等[99]利用宏基因组技术发现,多发性硬化症患者肠道菌群与健康人之间存在较大差异,并且证明改善饮食习惯可以改善病症。Haidar等[100]对一位急性胆囊炎患者进行病因分析,发现可能为食品中耐万古霉素肠球菌引发的疾病。食品中的耐药菌和耐药基因可以直接或间接影响肠道菌群结构,进而影响机体的消化系统[101]、免疫系统[102]和中枢神经系统[103]。
肉类食品引发的人体健康风险亟待解决[104]。Schoen等[105]使用定量微生物风险评估模型对猪肉中耐甲氧西林金黄色葡萄球菌进行评估,被感染的风险为3.20×10-3~1.30×10-2之间,存在人体被耐甲氧西林金黄色葡萄球菌污染的可能。Presi等[106]采用半定量模型对鸡肉、猪肉和牛肉中抗生素耐药菌造成的潜在风险进行评估,鸡肉、猪肉和牛肉的风险值分别为6.7、4.0和0.4,且冻肉的风险值最高。
人们普遍认为有机种植的水果、蔬菜更安全健康,然而研究发现有机肥种植的水果、蔬菜可能被抗生素耐药菌污染[107]。OFlaherty等[108]通过定量模型分析不同种植方式生菜中耐药大肠埃希菌,结果表明,人体对耐药大肠埃希菌的平均暴露水平为1.00×10-2~1.35×106 CFU/g之间,每食用100g被耐药致病性大肠埃希菌污染的生菜,患病的平均概率在1.46×10-9到1.88×10-2之间。Njage等[109]对莴苣中耐药大肠埃希菌进行模拟,3%的南非居民暴露水平在2.51×106到5.01×106 CFU/g之间。与欧盟食品微生物限量1441/2007的标准进行比较,生菜和莴苣中耐药大肠埃希菌暴露水平均已经超过1×103 CFU/g,因此,使用有机肥种植的水果蔬菜存在超过微生物限量标准的情况。
发酵食品和多数水果蔬菜都是直食性食品,这些食品中的耐药菌可以直接进入人体,增加病原菌感染人体的几率[20,80]。Lakhanpal等[110]研究游牧人直食性牛奶和肉類食品,发现食品中有6.7%被金黄色葡萄球菌所污染,其中对万古霉素耐药率为43.8%,游牧人患者万古霉素治疗的失败和较高的死亡率可能与直接食用携带耐万古霉素金黄色葡萄球菌的食品有关。
大量证据表明,抗生素耐药菌和抗生素耐药基因会破坏肠道菌群的稳定,改变菌群组成,进而对人体健康造成影响[11]。目前,食源性耐药菌污染在中国、印度、巴西和南非共和国等发展中国家较为严重,这些国家的人民需要承担更大健康风险,主要是因为缺乏适当的方法预防和控制食源性耐药菌的传播[89]。宏基因组测序技术可以获得耐药菌的全部遗传信息,可对环境中微生物菌群的多样性、功能活性等宏观特征进行研究。与传统技术相比,宏基因测序技术在发现新的耐药基因和耐药机制有着其他技术无可比拟的优势,但测序过程较为繁琐、价格昂贵。为了降低食源性耐药菌的感染,应尽可能从源头降低抗生素耐药菌的产生,减少抗生素的使用或使用抗生素替代物,同时完善监测体系,长期对食源性耐药菌进行监测。
5 研究展望
目前,国内外对食源性抗生素耐药菌做了大量研究,但研究结果多局限在食物中发现或检测到的耐药菌,而抗生素耐药菌在食品中的来源、传播和转移机制以及控制对策尚不清楚。因此,对未来食源性耐药菌的研究提供一些建议,主要包括:
(1)研究抗生素耐药菌在不同食品环境中的传播扩散机制,为有效遏制食源性耐药菌的水平转移提供新的思路。
(2)探究抗生素耐药菌通过食物链进入人体的传播机制,揭示耐药菌在人体肠道的污染水平及潜在健康风险,为控制耐药菌污染提供理论依据。
(3)加强畜牧业和水产养殖业等抗生素使用监管,探究食品生产、加工、储存和消费过程中耐药菌的传播或去除效果,并对过程中耐药菌的去除机制进行深入研究。
参 考 文 献
Duan M, Gu J, Wang X, et al. Factors that affect the occurrence and distribution of antibiotic resistance genes in soils from livestock and poultry farms[J]. Ecotoxicol Environ Saf, 2019, 180(5): 114-122.
Badi S, Cremonesi P, Abbassi M S, et al. Antibiotic resistance phenotypes and virulence-associated genes in Escherichia coli isolated from animals and animal food products in Tunisia[J]. FEMS Microbiol Lett, 2018. 365(10): 88.
Chajecka-Wierzchowska W, Zadernowska A, Nalepa B, et al. Coagulase-negative Staphylococci (CoNS) isolated from ready-to-eat food of animal origin-phenotypic and genotypic antibiotic resistance[J]. Food Microbiol., 2015, 46(4): 222-226.
Zhang Q Q, Ying G G, Pan C G, et al. Comprehensive evaluation of antibiotics emission and fate in the river basins of China: Source analysis, multimedia modeling, and linkage to bacterial resistance[J]. Environ Sci Technol, 2015, 49(11): 6772-6782.
Van Boeckel T P, Brower C, Gilbert M, et al. Global trends in antimicrobial use in food animals[J]. Proc Natl Acad Sci U S A, 2015, 112(18): 5649-5654.
付星宇, 汪庆, 毕聪聪, 等. 大气环境中抗生素耐药菌的来源与传播扩散研究进展[J/OL]. 中国抗生素杂志, https://doi.org/10.13461/j.cnki.cja.007088 1224, 2020.
赵小慧, 苏洁, 樊景凤, 等. 海洋环境中细菌耐药性研究进展[J]. 中国抗生素杂志, 2019, 44(4): 406-412.
Chen C, Pankow C A, Oh M, et al. Effect of antibiotic use and composting on antibiotic resistance gene abundance and resistome risks of soils receiving manure-derived amendments[J]. Environ Int, 2019, 128(4): 233-243.
Saharan V V, Verma P, Singh A P, et al. High prevalence of antimicrobial resistance in Escherichia coli, Salmonella spp. and Staphylococcus aureus isolated from fish samples in India[J]. Aquac Res, 2020, 51(3): 1200-1210.
Wu S, Huang J H, Zhang F, et al. Staphylococcus argenteus isolated from retail foods in China: Incidence, antibiotic resistance, biofilm formation and toxin gene profile[J]. Food Microbiol, 2020, 91(2): 103531.
Chen J, Ying G G, Deng W J. Antibiotic residues in food: Extraction, analysis, and human health concerns[J]. J Agr Food Chem, 2019, 67(27): 7569-7586.
楊承霖, 舒刚, 赵小玲, 等. 2010—2016年四川省食品动物源大肠杆菌的耐药性研究[J]. 西北农林科技大学学报(自然科学版), 2020, 48(9): 24-30.
陈伟冰, 李柏生, 卢向明, 等. 2012—2018年茂名市食品中食源性致病菌污染监测结果分析[J]. 应用预防医学, 2020, 26(2): 150-152.
邹志云, 严昕宇, 朱惠芳, 等. 2017年无锡地区食品从业人员及食源性疾病患者中分离沙门菌耐药情况[J]. 江苏预防医学, 2020, 31(5): 578-581.
李可维, 刘思洁, 赵薇, 等. 9274份肉及肉制品食源性致病菌监测结果分析[J]. 食品安全质量检测学报, 2020, 11(23): 9033-9038.
陈伟冰, 李柏生, 李振翠, 等. 广东省茂名市2017—2018年空肠弯曲菌食品分离株病原特征[J]. 中国热带医学, 2020. 20(5): 452-455.
陶勇, 刁保卫, 王利, 等. 马鞍山市不同来源副溶血弧菌生物学特征及分子流行病学研究[J]. 公共卫生与预防医学, 2013. 24(03): 18-22.
冯志宽, 殷文政, 杨荣杰, 等. 乳源性金黄色葡萄球菌的分离及耐药特性[J]. 食品工业, 2012, 33(6):102-104.
Yang S, Pei X, Wang G, et al. Prevalence of food-borne pathogens in ready-to-eat meat products in seven different Chinese regions[J]. Food Control, 2016, 65(18): 92-98.
Wang Y T, Lin Y T, Wan T W, et al. Distribution of antibiotic resistance genes among Staphylococcus species isolated from ready-to-eat foods[J]. J Food Drug Anal, 2019, 27(4): 841-848.
Osman K, Badr J, Al-Maary K S, et al. Prevalence of the antibiotic resistance genes in coagulase-Positive-and negative-Staphylococcus in chicken meat retailed to consumers[J]. Front Microbiol, 2016, 7(5): 1846.
de Jong A, Simjee S, Garch F E, et al. Antimicrobial susceptibility of enterococci recovered from healthy cattle, pigs and chickens in nine EU countries (EASSA Study) to critically important antibiotics[J]. Vet Microbiol, 2018, 216(2): 168-175.
Ta Y T, Nguyen T T, To P B, et al. Quantification, serovars, and antibiotic resistance of Salmonella isolated from retail raw chicken meat in Vietnam[J]. J Food Prot, 2014, 77(1): 57-66.
Chajecka-Wierzchowska W, Zadernowska A, Laniewska-Trokenheim L. Diversity of antibiotic resistance genes in Enterococcus strains isolated from ready-to-eat meat products[J]. J Food Sci, 2016, 81(11): 2799-2807.
Donado-Godoy P, Byrne B A, Leon M, et al. Prevalence, resistance patterns, and risk factors for antimicrobial resistance in bacteria from retail chicken meat in Colombia[J]. J Food Prot, 2015, 78(4): 751-759.
Adeyanju G T, Ishola O. Salmonella and Escherichia coli contamination of poultry meat from a processing plant and retail markets in Ibadan, Oyo State, Nigeria[J]. Springerplus, 2014, 3: 139.
Abd-Elghany S M, Sallam K I, Abd-Elkhalek A, et al. Occurrence, genetic characterization and antimicrobial resistance of Salmonella isolated from chicken meat and giblets[J]. Epidemiol Infect, 2015. 143(5): 997-1003.
Ta Y T, Nguyen T T, Phuong B T, et al. Quantification, serovars, and antibiotic resistance of Salmonella isolated from retail raw chicken meat in Vietnam[J]. J Food Protect, 2014, 77(1): 57-66.
Yang B, Cui Y, Shi C, et al. Counts, serotypes, and antimicrobial resistance of Salmonella isolates on retail raw poultry in the Peoples Republic of China[J]. J Food Protect, 2014, 77(6): 894-902.
Yoon R H, Cha S Y, Wei B, et al. Prevalence of Salmonella isolates and antimicrobial resistance in poultry meat from South Korea[J]. J Food Protect, 2014, 77(9): 1579-1582.
Grafskaia E N, Nadezhdin K D, Talyzina I A, et al. Medicinal leech antimicrobial peptides lacking toxicity represent a promising alternative strategy to combat antibiotic-resistant pathogens[J]. Eur J Med Chem, 2019, 180(6): 143-153.
Aguilar-Santelises M, Castillo-Vera J, Gonzalez-Molina R, et al. Clinical isolates of Escherichia coli are resistant both to antibiotics and organotin compounds[J]. Folia Microbiol, 2020, 65(1):87-94.
Govender V, Madoroba E, Magwedere K, et al. Prevalence and risk factors contributing to antibiotic-resistant Staphylococcus aureus isolates from poultry meat products in South Africa, 2015-2016[J]. J S Afr Vet Assoc, 2019, 90(2): 1-8.
Wu S, Wu Q, Zhang J, et al. Listeria monocytogenes prevalence and characteristics in retail raw foods in China[J]. PLoS One, 2015, 10(8): e0136682.
Oliveira T S, Varjao L M, da Silva L N N, et al. Listeria monocytogenes at chicken slaughterhouse: Occurrence, genetic relationship among isolates and evaluation of antimicrobial susceptibility[J]. Food Control, 2018, 88(6): 131-138.
Johnson T A, Stedtfeld R D, Wang Q, et al. Clusters of antibiotic resistance genes enriched together stay together in Swine agriculture[J]. Mbio, 2016, 7(2): e02214-15.
Bacanli M, Basaran N. Importance of antibiotic residues in animal food[J]. Food Chem Toxicol, 2019, 125(1): 462-466.
Wang Y, Hu Y, Cao J, et al. Antibiotic resistance gene reservoir in live poultry markets[J]. J Infect, 2019, 78(6): 445-453.
Monte D F, Lincopan N, Fedorka-Cray P J, et al. Current insights on high priority antibiotic-resistant Salmonella enterica in food and foodstuffs: A review[J]. Curr Opin Food Sci, 2019, 26(8): 35-46.
Jeong H W, Kim J A, Jeon S J, et al. Prevalence, antibiotic-resistance, and virulence characteristics of Vibrio parahaemolyticus in restaurant fish tanks in Seoul, South Korea[J]. Foodborne Pathog Dis, 2020, 17(3): 209-214.
Tan C W, Rukayadi Y, Hasan H, et al. Prevalence and antibiotic resistance patterns of Vibrio parahaemolyticus isolated from different types of seafood in Selangor, Malaysia[J]. Saudi J Biol Sci, 2020, 27(6): 1602-1608.
Tran T H T, Yanagawa H, Nguyen K T, et al. Prevalence of Vibrio parahaemolyticus in seafood and water environment in the Mekong Delta, Vietnam[J]. J Vet Med Sci, 2018, 80(11): 1737-1742.
Jiang Y, Chu Y, Xie G, et al. Antimicrobial resistance, virulence and genetic relationship of Vibrio parahaemolyticus in seafood from coasts of Bohai Sea and Yellow Sea, China[J]. Int J Food Microbiol, 2019, 290(19): 116-124.
Ellis-Iversen J, Seyfarth A M, Korsgaard H, et al. Antimicrobial resistant E. coli and Enterococci in pangasius fillets and prawns in Danish retail imported from Asia[J]. Food Control, 2020, 114(5): 106958.
Wu J J, Su Y L, Deng Y Q, et al. Prevalence and distribution of antibiotic resistance in marine fish farming areas in Hainan, China[J]. Sci Total Environ, 2019, 653(4): 605-611.
Tabaran A, Mihaiu M, Tabaran F, et al. First study on characterization of virulence and antibiotic resistance genes in verotoxigenic and enterotoxigenic E. coli isolated from raw milk and unpasteurized traditional cheeses in Romania[J]. Folia Microbiol, 2017, 62(2): 145-150.
Guo H, Pan L, Li L, et al. Characterization of antibiotic resistance genes from Lactobacillus isolated from traditional dairy products[J]. J Food Sci, 2017, 82(3): 724-730.
Pizauro L J L, de Almeida C C, Soltes G A, et al. Short communication: Detection of antibiotic resistance, mecA, and virulence genes in coagulase-negative Staphylococcus spp. from buffalo milk and the milking environment[J]. J Dairy Sci, 2019, 102(12): 11459-11464.
Yehia H M, Al-Masoud A H, Alarjani K M, et al. Prevalence of methicillin-resistant (mecA gene) and heat-resistant Staphylococcus aureus strains in pasteurized camel milk[J]. J Dairy Sci, 2020, 103(7): 5947-5963.
Sharma V, Sharma S, Dahiya D K, et al. Coagulase gene polymorphism, enterotoxigenecity, biofilm production, and antibiotic resistance in Staphylococcus aureus isolated from bovine raw milk in North West India[J]. Ann Clin Microbiol Antimicrob, 2017. 16(1): 65.
Ameen F, Reda S A, El-Shatoury S A, et al. Prevalence of antibiotic resistant mastitis pathogens in dairy cows in Egypt and potential biological control agents produced from plant endophytic actinobacteria[J]. Saudi J Biol Sci, 2019, 26(7): 1492-1498.
Aksomaitiene J, Ramonaite S, Tamuleviciene E, et al. Overlap of antibiotic resistant Campylobacter jejuni MLST genotypes isolated from humans, broiler products, dairy cattle and wild birds in Lithuania[J]. Front Microbiol, 2019. 10: 1377.
Wang K, Zhang H, Feng J, et al. Antibiotic resistance of lactic acid bacteria isolated from dairy products in Tianjin, China[J]. J Agr Food Res, 2019, 1(72): 100006.
Chaj?cka-Wierzchowska W, Zarzecka U, Zadernowska A. Enterococci isolated from plant-derived food-analysis of antibiotic resistance and the occurrence of resistance genes[J]. Lwt-food Sci Technol, 2020, 139(3): 110549.
Li Y, Cao W, Liang S, et al. Metagenomic characterization of bacterial community and antibiotic resistance genes in representative ready-to-eat food in southern China[J]. Sci Rep, 2020, 10(1): 15175.
許女, 李雅茹, 王超宇, 等. 传统发酵食品中乳酸菌的抗生素耐药性评估及耐药基因分析[J]. 中国食品学报, 2020, 20(7): 160-171.
向文良, 张庆, 卢倩文, 等. 获得性抗生素抗性基因:威胁发酵蔬菜食品安全的一种新型污染物[J]. 食品安全质量检测学报, 2015. 6(10): 3917-3922.
Liao X Y, Ma Y N, Daliri E B M, et al. Interplay of antibiotic resistance and food-associated stress tolerance in foodborne pathogens[J]. Trends Food Sci Technol, 2020, 95(1): 97-106.
蔡婷, 徐顾榕, 林凯, 等. 发酵用鲜辣椒中乳酸菌抗生素耐药性与耐药基因[J]. 食品与生物技术学报, 2016, 35(9): 941-949.
Lopez C M, Callegari M L, Patrone V, et al. Assessment of antibiotic resistance in Staphylococci involved in fermented meat product processing[J]. Curr Opin Food Sci, 2020, 31(4): 17-23.
Pisacane V, Callegari M L, Puglisi E, et al. Microbial analyses of traditional Italian salami reveal microorganisms transfer from the natural casing to the meat matrix[J]. Int J Food Microbiol, 2015, 207(16): 57-65.
Zeng H, Li C, Ling N, et al. Prevalence, genetic analysis and CRISPR typing of Cronobacter spp. isolated from meat and meat products in China[J]. Int J Food Microbiol, 2020, 321(3): 108549.
Wang X, Zhang Y, Ren H, et al. Comparison of bacterial diversity profiles and microbial safety assessment of salami, Chinese dry-cured sausage and Chinese smoked-cured sausage by high-throughput sequencing[J]. Lwt-food Sci Technol, 2018, 90(12): 108-115.
Gibson M K, Crofts T S, Dantas G. Antibiotics and the developing infant gut microbiota and resistome[J]. Curr Opin Microbiol, 2015, 27(20): 51-56.
Wei R, He T, Zhang S, et al. Occurrence of seventeen veterinary antibiotics and resistant bacterias in manure-fertilized vegetable farm soil in four provinces of China[J]. Chemosphere, 2019, 215(1): 234-240.
徐秋桐, 顾国平, 章明奎. 土壤中兽用抗生素污染对水稻生长的影响[J]. 农业资源与环境学报, 2016, 33(1): 60-65.
McKinney C W, Dungan R S, Moore A, et al. Occurrence and abundance of antibiotic resistance genes in agricultural soil receiving dairy manure[J]. Fems Microbiol Ecol, 2018, 94(3): 10.
Wang H, Chu Y, Fang C. Occurrence of Veterinary Antibiotics in Swine Manure from Large-scale Feedlots in Zhejiang Province, China[J]. Bull Environ Contam Toxicol, 2017, 98(4): 472-477.
Zhao X, Wang J, Zhu L, et al. Field-based evidence for enrichment of antibiotic resistance genes and mobile genetic elements in manure-amended vegetable soils[J]. Sci Total Environ, 2019, 654(5): 906-913.
Xiang W, Lu K, Zhang N, et al. Organic Houttuynia cordata Thunb harbors higher abundance and diversity of antibiotic resistance genes than non-organic origin, suggesting a potential food safe risk[J]. Food Res Int, 2019, 120(11): 733-739.
常旭卉, 賈书刚, 王淑平, 等. 粪源环丙沙星对潮土中抗生素抗性基因的影响[J]. 农业环境科学学报, 2018, 37(12): 2727-2737.
Zhu N, Jin H M, Ye X M, et al. Fate and driving factors of antibiotic resistance genes in an integrated swine wastewater treatment system: From wastewater to soil[J]. Sci Total Environ, 2020, 721(4): 137654.
Ma Q, Fu Y, Sun H, et al. Antimicrobial resistance of Lactobacillus spp. from fermented foods and human gut[J]. Lwt-food Sci Technol, 2017, 86(12): 201-208.
Losasso C, Di Cesare A, Mastrorilli E, et al. Assessing antimicrobial resistance gene load in vegan, vegetarian and omnivore human gut microbiota[J]. Int J Antimicrob Agents, 2018, 52(5): 702-705.
Onaran B, Goncuoglu M, Bilir Ormanci F S. Antibiotic resistance profiles of vancomycin resistant enterococci in chicken meat samples[J]. Ankara Univ Vet Fak, 2019, 66(4): 331-336.
Talley J L, Wayadande A C, Wasala L P, et al. Association of Escherichia coli O157:H7 with filth flies (Muscidae and Calliphoridae) captured in leafy greens fields and experimental transmission of E. coli O157:H7 to spinach leaves by house flies (Diptera: Muscidae)[J]. J Food Prot, 2009, 72(7): 1547-1552.
Liu Z, Klumper U, Shi L, et al. From pig breeding environment to subsequently produced pork: Comparative analysis of antibiotic resistance genes and bacterial community composition[J]. Front Microbiol, 2019, 10(10): 43.
Chen Y, Shen W T, Wang B, et al. Occurrence and fate of antibiotics, antimicrobial resistance determinants and potential human pathogens in a wastewater treatment plant and their effects on receiving waters in Nanjing, China[J]. Ecotoxicol Environ Saf, 2020, 206(12): 483-490.
Bai L, Lan R, Zhang X, et al. Prevalence of Salmonella isolates from chicken and pig Slaughterhouses and emergence of ciprofloxacin and cefotaxime co-resistant S. enterica serovar Indiana in Henan, China[J]. PLoS One, 2015, 10(12): e0144532.
Molechan C, Amoako D G, Abia A L K, et al. Molecular epidemiology of antibiotic-resistant Enterococcus spp. from the farm-to-fork continuum in intensive poultry production in KwaZulu-Natal, South Africa[J]. Sci Total Environ, 2019, 692(14): 868-878.
Mascaro V, Squillace L, Nobile C G, et al. Prevalence of methicillin-resistant Staphylococcus aureus (MRSA) carriage and pattern of antibiotic resistance among sheep farmers from Southern Italy[J]. Infect Drug Resist, 2019, 12: 2561-2571.
Smith K E, Besser J M, Hedberg C W, et al. Quinolone-resistant Campylobacter jejuni infections in Minnesota, 1992-1998. Investigation Team[J]. N Engl J Med, 1999, 340(20): 1525-1532.
Lu X, Zeng M, Xu J, et al. Epidemiologic and genomic insights on mcr-1-harbouring Salmonella from diarrhoeal outpatients in Shanghai, China, 2006-2016[J]. EBio Medicine, 2019, 42: 133-144.
Otto S J, Carson C A, Finley R L, et al. Estimating the number of human cases of ceftiofur-resistant Salmonella enterica serovar Heidelberg in Quebec and Ontario, Canada[J]. Clin Infect Dis, 2014, 59(9): 1281-1290.
Al-Nabulsi A A, Osaili T M, Shaker R R, et al. Effects of osmotic pressure, acid, or cold stresses on antibiotic susceptibility of Listeria monocytogenes[J]. Food Microbiol, 2015, 46(4): 154-160.
Yu T, Jiang X, Zhang Y, et al. Effect of benzalkonium chloride adaptation on sensitivity to antimicrobialagents and tolerance to environmental stresses in Listeria monocytogenes[J]. Front Microbiol, 2018, 9(11): 2906.
Thapa S P, Shrestha S, Anal A K. Addressing the antibiotic resistance and improving the food safety in food supply chain (farm-to-fork) in Southeast Asia[J]. Food Control, 2020, 108(2): 106809.
Cavaco L M, Hendriksen R S, Aarestrup F M. Plasmid-mediated quinolone resistance determinant qnrS1 detected in Salmonella enterica serovar Corvallis strains isolated in Denmark and Thailand[J]. J Antimicrob Chemother, 2007, 60(3): 704-706.
Ma Y, Xu X, Gao Y, et al. Antimicrobial resistance and molecular characterization of Salmonella enterica serovar Corvallis isolated from human patients and animal source foods in China[J]. Int J Food Microbiol, 2020, 335(24): 108859.
Vounba P, Rhouma M, Arsenault J, et al. Prevalence of colistin resistance and mcr-1/mcr-2 genes in extended-spectrum beta-lactamase/AmpC-producing Escherichia coli isolated from chickens in Canada, Senegal and Vietnam[J]. J Glob Antimicrob Resist, 2019, 19(4): 222-227.
Van T T H, Yidana Z, Smooker P M, et al. Antibiotic use in food animals worldwide, with a focus on Africa: Pluses and minuses[J]. J Glob Antimicrob Resist, 2020, 20(1): 170-177.
Roth N, Kasbohrer A, Mayrhofer S, et al. The application of antibiotics in broiler production and the resulting antibiotic resistance in Escherichia coli: A global overview[J]. Poult Sci, 2019, 98(4): 1791-1804.
Van T T, Nguyen H N, Smooker P M, et al. The antibiotic resistance characteristics of non-typhoidal Salmonella enterica isolated from food-producing animals, retail meat and humans in South East Asia[J]. Int J Food Microbiol, 2012, 154(3): 98-106.
Aarestrup F M, Hendriksen R S, Lockett J, et al. International spread of multidrug-resistant Salmonella schwarzengruna in food products[J]. Emerg Infect Dis, 2007, 13(5): 726-731.
Chen Y, Hammer E E, Richards V P. Phylogenetic signature of lateral exchange of genes for antibiotic production and resistance among bacteria highlights a pattern of global transmission of pathogens between humans and livestock[J]. Mol Phylogenet Evol, 2018, 125(8): 255-264.
Barrow P A, Jones M A, Smith A L, et al. The long view: Salmonella-the last forty years[J]. Avian Pathol, 2012, 41(5): 413-420.
Hu Y, Yang X, Qin J, et al. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota[J]. Nat Commun, 2013, 4(7): 2151.
Forslund K, Sunagawa S, Kultima J R, et al. Country-specific antibiotic use practices impact the human gut resistome[J]. Genome Res, 2013, 23(7): 1163-1169.
Takewaki D, Suda W, Sato W, et al. Alterations of the gut ecological and functional microenvironment in different stages of multiple sclerosis[J]. Proc Natl Acad Sci U S A, 2020, 117(36): 22402-22412.
Haidar G, Green M, American society of transplantation infectious diseases community of practice Intra-abdomina infections in solid organ transplant recipients: Guidelines from the American society of transplantation infectious diseases community of practice[J]. Clin Transplant, 2019, 33(9): e13595.
Requena T, Cotter P, Shahar D R, et al. Interactions between gut microbiota, food and the obese host[J]. Trends Food Sci Tech, 2013, 34(1): 44-53.
Dunn S J, Connor C, McNally A. The evolution and transmission of multi-drug resistant Escherichia coli and Klebsiella pneumoniae: The complexity of clones and plasmids[J]. Curr Opin Microbiol, 2019, 51(20): 51-56.
Esmaeil A M, Shomali N, Bakhshi A, et al. Gut microbiome and multiple sclerosis: New insights and perspective[J]. Int Immunopharmacol, 2020, 88(9): 107024.
Alban L, Nielsen E O, Dahl J. A human health risk assessment for macrolide-resistant Campylobacter associated with the use of macrolides in Danish pig production[J]. Prev Vet Med, 2008, 83(2): 115-129.
Schoen M E, Peckham T K, Shirai J H, et al. Risk of nasal colonization of methicillin-resistant Staphylococcus aureus during preparation of contaminated retail pork meat[J]. Microbial Risk Anal, 2020, 16(3): 100136.
Presi P, Stark K D, Stephan R, et al. Risk scoring for setting priorities in a monitoring of antimicrobial resistance in meat and meat products[J]. Int J Food Microbiol, 2009, 130(2): 94-100.
Zhu B, Chen Q, Chen S, et al. Does organically produced lettuce harbor higher abundance of antibiotic resistance genes than conventionally produced?[J]. Environ Int, 2017, 98(1): 152-159.
OFlaherty E, Solimini A G, Pantanella F, et al. Human exposure to antibiotic resistant-Escherichia coli through irrigated lettuce[J]. Environ Int, 2019, 122(1): 270-280.
Njage P M, Buys E M. Quantitative assessment of human exposure to extended spectrum and AmpC beta-lactamases bearing E. coli in lettuce attributable to irrigation water and subsequent horizontal gene transfer[J]. Int J Food Microbiol, 2017, 240(1): 141-151.
Lakhanpal P, Panda A K, Chahota R, et al. Incidence and antimicrobial susceptibility of Staphylococcus aureus isolated from ready-to-eat foods of animal origin from tourist destinations of North-western Himalayas, Himachal Pradesh, India[J]. J Food Sci Technol, 2019, 56(2): 1078-1083.
Kim J, Park H, Kim J, et al. Comparative analysis of aerotolerance, antibiotic resistance, and virulence gene prevalence in Campylobacter jejuni isolates from retail raw chicken and duckmeat in South Korea[J]. Microorganisms, 2019, 7(10): 433.
Maktabi S, Ghorbanpoor M, Hossaini M, et al. Detection of multi-antibiotic resistant Campylobacter coli and Campylobacter jejuni in beef, mutton, chicken and water buffalo meat in Ahvaz, Iran[J]. Vet Res Forum, 2019, 10(1): 37-42.
Abd-Elghany S M, Sallam K I, Abd-Elkhalek A, et al. Occurrence, genetic characterization and antimicrobial resistance of Salmonella isolated from chicken meat and giblets[J]. Epidemiol Infect, 2015, 143(5): 997-1003.
Narvaez-Bravo C, Taboada E N, Mutschall S K, et al. Epidemiology of antimicrobial resistant Campylobacter spp. isolated from retail meats in Canada[J]. Int J Food Microbiol, 2017, 253(8): 43-47.
Beier R C, Byrd J A, Andrews K, et al. Disinfectant and antimicrobial susceptibility studies of the foodborne pathogen Campylobacter jejuni isolated from the litter of broiler chicken houses[J]. Poultry Science, 2020, 100(2): 1024-1033.
收稿日期:2020-12-29
基金項目:国家自然科学基金(No. 42077393和No. 41703088);河北省重点研发计划项目(No.19273707D);
河北省自然科学基金青年基金(No.C2018402255)
作者简介:曹弘扬,男,生于1993年,在读硕士研究生,研究方向为环境微生物,E-mail: caohongyang086@163.com
*通讯作者, E-mail: wangqing@hebeu.edu.cn

