PbPH5,an H+ P-ATPase on the tonoplast,is related to malic acid accumulation in pear fruit
SONG Jun-xing,CHEN Ying-can,LU Zhao-hui,ZHAO Guang-ping,WANG Xiao-li,ZHAl Rui,WANG Zhi-gang,YANG Cheng-quan,XU Ling-fei
College of Horticulture,Northwest A&F University,Yangling 712100,P.R.China
Abstract Organic acid content is one of the most important factors influencing fruit flavors. The predominant organic acid in most pear cultivars is malic acid,but the mechanism controlling its accumulation remains unclear. In this study,by comparing gene expression levels and organic acid contents,we found that the expression of PbPH5,which encodes a P3A-ATPase,is highly correlated with malic acid accumulation in four different pear species,with correlation coefficients of 0.932**,0.656*,0.900**,and 0.518* (*,P<0.05;**,P<0.01) for Pyrus bretschneideri Rehd.,P.communis Linn.,P.pyrifolia Nakai.,and P.ussuriensis Maxim.,respectively. Moreover,the overexpression of PbPH5 in pear significantly increased the malic acid content. In contrast,silencing PbPH5 via RNA interference significantly decreased both its transcript level and the pear fruit malic acid content. A subcellular localization analysis indicated that PbPH5 is located in the tonoplast.Additionally,a phylogenetic analysis indicated that PbPH5 is a PH5 homolog gene that is clustered with the Petunia hybrida,Malus domestica,and Citrus reticulata genes. Considered together,these findings suggest that PbPH5 is a functionally conserved gene. Furthermore,the accumulation of malic acid in pear fruit is at least partly related to changes in the PbPH5 transcription levels.
Keywords:pear,P3A-ATPase,PH5 homolog,malic acid accumulation,proton pump
1.lntroduction
Pear fruits are very popular among consumers because of their unique flavors,which can be evaluated based on several characteristics,including taste,aroma,and mouthfeel (Mayuoni-Kirshinbaum and Porat 2014;Manzooret al.2020). Organic acids are important compounds influencing the taste of various fruits (Visseret al.1968;Harkeret al.2002),and different kinds of organic acids are associated with diverse tastes (Hulme 1958). For example,people sense both malic acid and citric acid in foods as either tartness or sourness,although the sensation is more intense with citric acid (Ramoset al.2007). Similar to other fruits,the main organic acid in the fruit of most pear cultivars is malic acid,although citric acid has been detected in a few cultivars (Hudina and Śtampar 2000;Liuet al.2016). The citric acid biosynthesis pathway was first proposed by Haffaker and Wallace (1959),whereas the mechanism controlling malic acid biosynthesis was not fully explored until 1993(Notton and Blanke 1993). Both acids are synthesized from oxaloacetic acid,which is produced from carbon dioxide and phosphoenolpyruvate in a reaction catalyzed by phosphoenolpyruvate carboxylase. However,malate dehydrogenase and citrate synthetase are the enzymes responsible for the final synthesis of malic acid and citric acid,respectively.
In most fruit cells,organic acids are mainly stored in the vacuole,which is a large organelle that occupies a considerable portion of the plant cell volume and affects multiple physiological activities (Moskowitz and Hrazdina 1981;Shohei 1984). Many soluble substances,such as ions,pigments,and proteins,are transported to the vacuolesviavarious tonoplast transporters,enabling vacuoles to regulate the cellular osmotic balance and pH homeostasis,while also enhancing plant stress resistance(Rayle and Cleland 1977;Szczypkaet al.1997;Martinoiaet al.2007;Quintanaet al.2007;Ahmadet al.2016;Sunet al.2021). In fruit cells,malic acid and citric acid are almost exclusively stored in vacuoles in their protonated or monoanionic forms because of the low pH environment within the vacuoles. Under neutral or alkaline conditions(in the cytosol),these acids dissociate into their dianionic and trianionic forms,which are immediately protonated in the vacuole (Oleskiet al.1987;Rentsch and Martinoia 1991;Etienneet al.2013). The transport of malic acid from the cytoplasm to the vacuole is mediated by many proteins in the vacuolar membrane. For example,previous studies revealed that aluminum-activated malate transporter family members help in transporting malate into the vacuole of fruit cells (Baiet al.2012),whereas the tonoplast dicarboxylate transporter is required for transporting malate inArabidopsisand grape cells(Terrieret al.1998;Emmerlichet al.2003). Additionally,the H+-ATPase (V-ATPase) and H+-PPiase (V-PPase)detected in some pulp cells may be functional at different developmental stages of the fruit (Maeshima 2000;Suzukiet al.2000;Terrieret al.2001). Notably,the efficiency of organic acid accumulation is influenced by the degree of vacuolar acidification and the electric potential gradient across the tonoplast (Δψ) (Etienneet al.2013).
Proton pumps are important for the accumulation of organic acids because they transport protons into the vacuoles to maintain an acidic environment and generate a positive Δψ. Recently,there has been increasing interest in P-ATPases,with the “P-type”indicating a phosphorylated intermediate,which have diverse functions related to anthocyanin accumulation(Walteret al.2008),biotic or abiotic stress responses(Gévaudantet al.2007),and organic acid accumulation(Shiet al.2015). The P-ATPases,which are encoded by a large gene family,are involved in various ion transport processes. On the basis of ion types and sequence identities,this gene family has been divided into five major subfamilies with several branches (Axelsen and Palmgren 1998,2001;Maet al.2020). Three subfamilies (P1,P2,and P3) are responsible for the transport of metals or hydrogen ions. The first subfamily (P1) has two branches,which are related to the transport of K+(P1A) and heavy metals (P1B). The second subfamily (P2),which is divided into four branches,transports various ions,including Ca2+(P2Aand P2B) and Na+(H+)/K+(P2Cand P2D) (Palmgren and Nissen 2011). Of the two branches of P3 (P3Aand P3B),P3Aincludes P-ATPases localized to the plasma membrane,where they mediate the transport of H+(Walteret al.2008),whereas the enzymes in the P3Bbranch are associated with the transport of Mg2+(Faracoet al.2014). The P4 subfamily mainly comprises putative lipid flippases. The P5 subfamily remains to be functionally characterized (Baxteret al.2003).
In the last decade,the functions of a series of P-ATPases have been studied in many species (Sichulet al.2007;Wanget al.2013;Chenet al.2018). InPetunia,the lack of a P-ATPase proton pump (PH5)belonging to the P3Asubfamily prevents the accumulation of anthocyanins in the seed coat and causes the flowers to turn blue (Walteret al.2008). Another study revealed that thePH5-likegene is expressed at very low levels in an acid-freeCitruscultivar (Shiet al.2015). Additionally,CsPH8andMdMa10,which are P-type proton pump genes,are involved in the accumulation of organic acids inCitrusand apple fruits (Maet al.2019;Shiet al.2019). Moreover,a P-ATPase gene associated with malate accumulation in pear has been identified (Zhanget al.2020). Earlier research confirmed the importance of proton pumps for facilitating the accumulation of organic acids related to fruit flavors. Because fruit flavors influence consumer demand,exploring the utility of the P-ATPase gene family for developing new pear cultivars that produce fruits with desirable flavors is warranted. In this study,we identified the P3Asubfamily genes in pear.By comparing the changes of the P3Asubfamily gene expression levels with the malic acid content in Chinese white pear (PyrusbretschneideriRehd.),we identified a candidate gene that may affect the accumulation of malic acid during various fruit developmental stages. The cultivars of several different pear species and transiently transformed samples were used to verify the function of the candidate gene. The results of this study will be useful for characterizing the P3Asubfamily genes more thoroughly,and may be relevant for future related studies.
2.Materials and methods
2.1.Plant materials
This study was completed with the following five pear cultivars:‘Dangshansuli’ (DS;P.bretschneideriRehd.),‘Zaosu’ (ZS;P.bretschneideriRehd.),‘Abate Fetel’ (AF;P.communisLinn.),‘Huangjinli’ (HJL;P.pyrifoliaNakai.),and ‘Jinxiangshui’ (JXS;P.ussuriensisMaxim.). The DS,ZS,and AF fruits were harvested from the same orchard(Yangling,Shaanxi Province,China) and immediately sent to the laboratory. The HJL and JXS fruits were respectively harvested from orchards in Qingdao,Shandong Province,China and Mudanjiang,Heilongjiang Province,China. Fruit with a normal shape and uniform size were randomly harvested at each fruit developmental stage that was analyzed (Hussainet al.2017),with three biological replicates comprising four to six fruit per stage. For DS,flower,root,stem,leaf,and pericarp samples were collected. Additionally,ZS fruit collected at 130 days after full bloom (DAFB) were used for transient transformations. Fruit samples were peeled and cored,after which the pulp was frozen and ground into a power in liquid nitrogen,then transferred to a marked tinfoil-lined paper bag and stored at -80°C until use.
2.2.ldentification of P3A-type ATPase genes
To identify the pear P3A-type ATPase genes,the P3A-type ATPase subfamily genes ofArabidopsisserved as a query for screening the NCBI database (https://www.ncbi.nlm.nih.gov/) using the Basic Local Alignment Search Tool(BLAST) Software. The obtained full-length sequences of the pear (P.bretschneideriRehd.) P3A-type ATPase genes were examined,and repeated sequences were eliminated. In addition,the sequence authenticity was confirmed by PCR amplification and sequencing results,and the specific primers are listed in Appendix A. The P3A-type ATPase genes previously identified in various species (Walteret al.2008;Liet al.2016;Maet al.2019;Shiet al.2019) are listed in Appendix B.
2.3.Phylogenetic analysis
The protein sequences encoded by theP.bretschneideriRehd. P3Asubfamily genes,along with theArabidopsisprotein sequences,were used to construct a phylogenetic tree using the MEGA 7.0 Program. Specifically,the neighbor-joining method and the JTT+G Model were used,with 1 000 bootstrap replicates. The accession numbers of the sequences included in this tree are listed in Appendix C. The phylogenetic trees for different species were built using the same method.
2.4.RNA isolation and quantitative real-time PCR(qRT-PCR)
The Polysaccharides &Polyphenolics-rich RNAprep Pure Plant Kit (Tiangen,Beijing,China) was used to isolate total RNA from the samples collected at each fruit developmental stage. The isolated RNA content was quantified using a UV-1700 spectrophotometer (Shimadzu,Kyoto,Japan),whereas RNA quality was assessed by agarose gel electrophoresis. The high-quality total RNA (2 µg) served as the template for synthesizing firststrand cDNA using the PrimeScript RT Reagent Kit with gDNA Eraser (TaKaRa,Dalian,China). The cDNA was analyzed in qRT-PCR assays,which were performed using the iCycler iQ5 System (Bio-Rad,Berkeley,CA,USA). The reaction mixtures included 5 µL SYBR PremixEx TaqII (TaKaRa) and the specific primers (Appendix D).Analyses were completed with three biological replicates,each comprising three technical replicates. The relative expression levels were calculated using the 2−ΔΔCtmethod(Livak and Schmittgen 2001).
2.5.Measurement of malic acid content
The malic acid content was determined by ultraperformance liquid chromatography (UPLC). To prepare samples,about 1 g powered material and 6 mL ddH2O were added to a centrifuge tube,after which the samples were sonicated for 30 min and then centrifuged for 15 min at 13 000×g. The supernatants were collected and analyzed using the Nexera LC-30A UPLC System(Shimadzu,Kyoto,Japan),which included a C18 column(4.6 mm×150 mm,5 µm). For each sample,a 10.0-µL aliquot of the supernatant flowed at 0.8 mL min-1through the column,which was maintained at 40°C,with the help of KH2PO4buffer (pH 2.4). An SPD detector was used to detect the absorption peak of each sample (210 nm wavelength). Malic acid standards (Yuanye,Shanghai,China) were used to prepare a standard curve,which was then used to quantify the malic acid contents of samples based on the peak areas.
2.6.Subcellular localization assay
The full-lengthPbPH5coding sequence was amplified from the first-strand cDNA for the DS fruit.The following primer pair was used to construct the pHBT-PbPH5-GFP-NOS recombinant plasmid(5´-GCTTCGAATTCTGCAGTCGACAGAATCGATAGGCAGATGGATGA-3´/5´-GCCCTTGCTCACTACGGATCCGACAGTGTATGATTGCTGGATTGTGT-3´). Fourweek-oldArabidopsisseedlings were used for analyzing the fluorescence of the PbPH5-GFP fusion protein.Arabidopsismesophyll protoplasts were isolated and cotransformed with the recombinant plasmid and a vacuolar membrane marker (vac-rk CD3-975) using a commercial kit designed for preparing and converting the protoplasm ofArabidopsisleaves (Protein Interaction,Wuhan,China).The co-transformedArabidopsismesophyll protoplasts were exposed to a weak-light environment for 12-18 h.A TCS SP8 confocal laser scanning microscope (Leica,Weztlar,Germany) was used to detect the fusion protein,marker,and chlorophyll autofluorescence at wavelengths of 488,651,and 750 nm,respectively (Maet al.2019).
2.7.Transient transformation of pear fruit
The transient transformation was performed as previously described (Zhaiet al.2016;Songet al.2020). To overexpressPbPH5in pear,the full coding sequence was amplified using specific primers (5´-tagaactagtggat ccAGAATCGATAGGCAGAGATGGATGA-3´/5´-cggtatcg ataagcttCGAGACTCGGAGACTCAGACA-3´) and then inserted into the pGreen II 0029-62SK (OE) vector to construct the OE-PbPH5recombinant plasmid. The OE and OE-PbPH5plasmids were inserted into separateAgrobacteriumtumefaciensstrain EHA105 cells,and OE was used as the control. The resulting transformants containing the correct plasmid were cultured in liquid medium supplemented with 25 mg L-1rifampicin and 50 mg L-1kanamycin. When the medium turned golden yellow,the cells were centrifuged for 5 min at 13 000×g and then resuspended in a buffer comprising 10 mmol L-1MES,10 mmol L-1MgCl2,pH 5.6,and 100 mmol L-1acetosyringone. The optical density (at 600 nm) of the resuspension was adjusted to 0.6,after which the cells were cultured in darkness for 2 h prior to injection into healthy fruits. To silencePbPH5in pear,an approximately 500-bp gene fragment was amplified using the following primer pair (5´-gcctccatggggatcCGCTGTAGTTTACCTGC AAGT-3´/5´-atgcccgggcctcgaATGATTGCTGGATTGTGT CTATGTC-3´). The amplified sequence was inserted into the TRV2 vector. The TRV1,TRV2,and TRV2-PbPH5plasmids were inserted into separateA.tumefaciensstrain EHA105 cells using the transformation procedure described above,TRV2 was used as the control. TheA.tumefacienscells were injected into healthy fruit with a uniform shape. Pulp was collected from the infected fruit after three days for gene expression analysis using qRTPCR assays. Additionally,the malic acid content of pulp collected after seven days was measured by UPLC. To verify the transformation of fruit with theA.tumefacienscells,an OE-GUSvector was inserted intoA.tumefaciensstrain EHA105 cells,which were then injected into fruits for a GUS staining analysis as previously described (Zhaiet al.2019).
2.8.Statistical analyses
Data were collected,arranged,and analyzed using the SPSS 17.0 Software (SPSS Inc,Chicago,IL,USA). The significance of the differences among correlated data was determined by a one-way ANOVA and Pearson’s correlation analysis. The threshold for significance was set at either*,P<0.05 or**,P<0.01.
3.Results
3.1.ldentification and phylogenetic analysis of P3A-ATPase genes in pear
TheAtAHA10gene encodes a P3A-ATPase proton pump that facilitates the accumulation of proanthocyanidin precursors in vacuoles by generating the required driving force. Therefore,using the subfamily of P3A-type ATPase genes ofArabidopsisas the query for a BLAST search,we identified all of the pear P3A-ATPase genes. Moreover,the PCR amplification and sequencing results of these genes were used to confirm the sequence authenticity in the genome database (Appendix E). Taken together,a total of 12 genes were used to construct a neighborjoining phylogenetic tree (Fig.1),which indicated the pear P3A-ATPase genes were accurately identified.

Fig.1 Neighbor-joining phylogenetic tree of the P3A-ATPases in pear and Arabidopsis.
3.2.The candidate gene was identified by correlation analysis between the P3A-ATPase gene expression levels and malic acid contents in different Pyrus species
To identify the candidate gene affecting pear fruit malic acid content,we analyzed the gene expression levels in different DS tissues,including the root,stem,leaf,flower,pericarp,and pulp at various developmental stages (Fig.2-A). For all tissues examined,we detected several highly expressed genes;however,none of the genes were highly expressed in all tissues. Thus,these genes may have different functions in the various pear tissues. Most of the genes were highly expressed in the root,leaf,and flower. The following six genes were highly expressed in three or more tissues:PbATPase1-Like(b),PbATPase1-Like(c),PbATPase4-Like(a),PbATPase 4-Like(b),PbATPase5-LikeandPbATPase8. Three genes,PbATPase1-Like(a),PbATPase4,andPbATPase9,were highly expressed in two tissues. Of the remaining three genes,PbATPase11-Like(c)was highly expressed only in the leaf,whereasPbATPase11andPbATPase11-Like(b)were highly expressed exclusively in the root. In the pulp,several highly expressed genes were identified.
The pulp of DS fruit at different developmental stages was analyzed by UPLC to determine the organic acid content. The UPLC data revealed that malic acid was the most abundant organic acid. Accordingly,the malic acid content was used to represent the total organic acid content in DS fruit. Changes in the level of malic acid accumulation are presented in Fig.2-B. The malic acid content increased gradually during the early fruit developmental stage,peaked at 60 DAFB,and then subsequently decreased until the fruits matured. Only a few genes showed expression-level changes that were similar to the malic acid content changes. A correlation analysis revealed a significant positive correlation between the expression of these genes and the malic acid content (Fig.2-C).
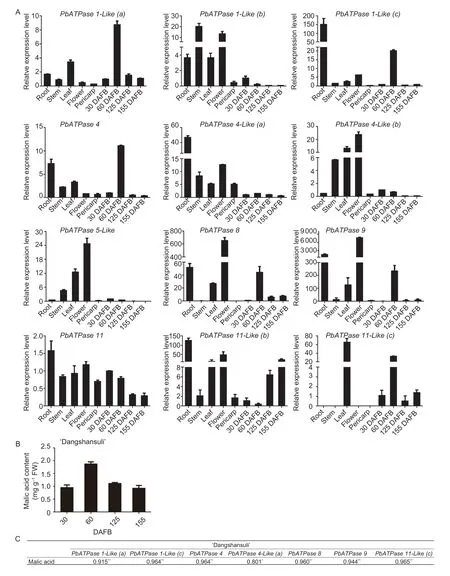
Fig.2 Correlation analysis of P3A-ATPase gene expression levels and malic acid contents in ‘Dangshansuli’ (DS). A,expression patterns of P3A-ATPase genes in various tissues and at different developmental stages. B,fruit malic acid contents at different developmental stages. DAFB,days after full bloom. C,significant positive correlations between the malic acid contents and the relative expression levels of several P3A-ATPase genes. The gene expression data are presented as the averages of three replicates,with the standard deviations (SD) indicated by error bars. *,P<0.05;**,P<0.01.
Moreover,the correlation of these genes was further analyzed in anotherP.bretschneideriRehd.cultivar.The organic acid content in ZS was also analyzed by UPLC,and the data indicated the organic acid contents were similar in ZS and DS. The malic acid level in ZS fruits increased during the cell expansion stage,peaked(at approximately 2.4 mg g-1fresh weight) at 60 DAFB,and then gradually decreased as the fruit continued to develop. An analysis was performed between the expression levels of these genes which had a significant positive correlation and the malic acid content,and the results of the correlation are listed in Fig.3-B. However,the expression level of one gene is not listed because it was too low. On the contrary,one gene,PbATPase1-Like(a),had a correlation coefficient of 0.932**(**,P<0.01). These results imply that this gene,which was designated asPbPH5,affects the accumulation of malic acid during the pear fruit developmental stages.

Fig.3 Further correlation analysis of malic acid contents and expression levels of genes with significant positive correlations in another Pyrus bretschneideri Rehd.cultivar. A,comparison of the changes to the malic acid content and gene expression level in‘Zaosu’ (ZS). DAFB,days after full bloom. The gene expression data are presented as the averages of three replicates,with the standard deviations (SD) indicated by error bars. B,results of the correlation analysis performed with the SPSS 17.0 Software.**,P<0.01.
To further confirm the correlation betweenPbPH5expression and the malic acid content in developing pear fruit,we also analyzedPbPH5transcript levels and malic acid accumulation in the fruits of a few other pear species. In AF,there were no major changes in the malic acid content,which peaked (at approximately 1.3 mg g-1fresh weight) at 125 DAFB (Fig.4-A). ThePbPH5gene was more highly expressed during the fruit cell expansion stage than in the cell division and maturation stages,which was similar to the changes in the malic acid level. In HJL,the malic acid content and thePbPH5expression level were the highest at 25 DAFB,after which they both decreased as the fruit developed (Fig.4-A). In contrast with the other pear cultivars analyzed,the malic acid content in JXS was maintained at a relatively high level in the developing fruit (Fig.4-A). The malic acid content rapidly increased,peaking at 60 DAFB,with no subsequent decreases until the fruits matured. Although the malic acid level decreased at maturity,it was still higher than those of the other cultivars. Regarding thePbPH5expression level,it initially increased and then decreased,similar to the malic acid content.
A correlation analysis of these cultivars revealed a significant positive correlation between the malic acid content and thePbPH5expression level (Fig.4-B). These results suggest that malic acid accumulation is related toPbPH5expression during pear fruit developmental stages.
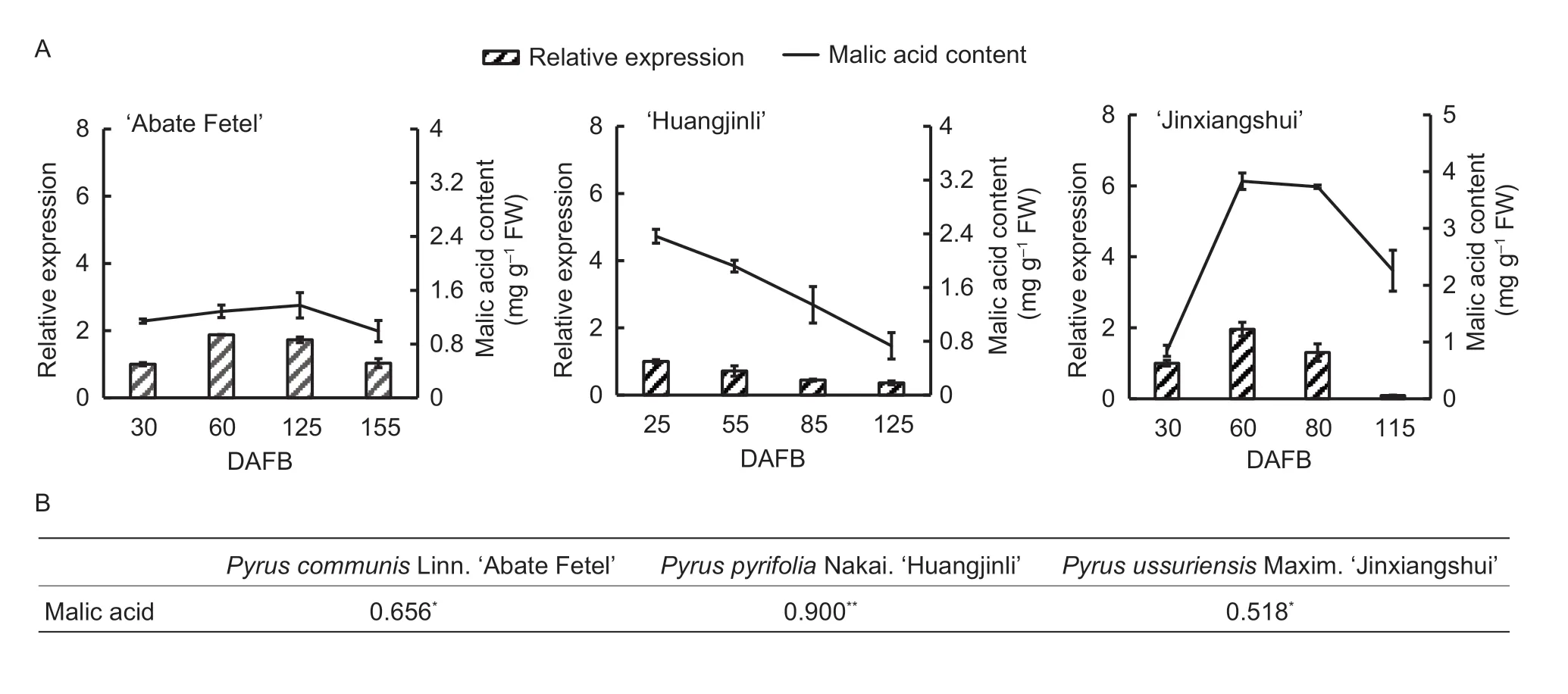
Fig.4 Further confirmation of the correlation between malic acid contents and relative expression levels of the putative gene in different pear species. A,comparison of the changes to gene expression levels and malic acid contents in ‘Abate Fetel’ (AF),‘Huangjinli’ (HJL),and ‘Jinxiangshui’ (JXS). DAFB,days after full bloom. The gene expression data are presented as the averages of three replicates,with the standard deviations (SD) indicated by error bars. B,significant positive correlations were revealed for all three cultivars. *,P<0.05;**,P<0.01.
3.3.Subcellular localization of PbPH5
To determine the subcellular localization of PbPH5,Arabidopsismesophyll protoplasts transformed with the pHBT-PbPH5-GFP-NOS recombinant plasmid were examined. The PbPH5-GFP fusion protein was detected in the tonoplast (Fig.5-A),and a tonoplast marker,vac-rk CD3-975,was used to verify the subcellular localization of PbPH5-GFP. Specifically,the red fluorescence of the tonoplast marker (Fig.5-B) and the green fluorescence of the PbPH5-GFP fusion protein were detected at the same cellular location (Fig.5-E).

Fig.5 Subcellular localization of PbPH5 in Arabidopsis mesophyll protoplast. A,the fluorescence of PbPH5-GFP fusion protein.B,vac-rk CD3-975 marks the tonoplast (red signal). C,chloroplasts show auto fluorescence. D,optical photomicrographs (bright field). E,an overlay of the bright field and fluorescence illumination.
3.4.Functional analysis of PbPH5 in pear
To functionally characterizePbPH5,OE-PbPH5and TRV2-PbPH5recombinant plasmids were prepared and used for pear fruit transient transformations. To assess the utility of our fruit transformation procedure,A.tumefaciensEHA105 cells carrying the OE-GUSvector were injected into fruit. Three days later,the fruit was harvested and subjected to a GUS staining analysis. Only the injection site turned blue,indicating the recombinant plasmid was expressed in the pear fruit (Appendix F). Similar to other fruits,pear fruit contain many kinds of organic acids,but malic acid is the predominant organic acid in ZS (Shaet al.2011;Zhanget al.2020). Therefore,the malic acid content was used to clarify the function ofPbPH5. Compared with the control,the malic acid content at the injection site was significantly higher in the samples in whichPbPH5was overexpressed (Fig.6-B). Additionally,RNA interference ofPbPH5in pear significantly decreased both thePbPH5transcript level and the malic acid content (Fig.6-D).These findings imply that thePbPH5expression level is related to the accumulation of malic acid because of the associated changes in the transport of H+into the vacuole of pear fruit cells.

Fig.6 Transient transformation for the overexpression and silencing of PbPH5 in pear fruit. A and B,gene expression levels (A) and malic acid contents (B) in control (CK) and PbPH5-overexpressing pear fruit,respectively. C and D,gene expression levels (C) and malic acid contents (D) in CK and PbPH5-silenced pear fruit. The gene expression data are presented as the averages of three replicates,with the standard deviations (SD) indicated by error bars. *,P<0.05;**,P<0.01.
3.5.Evolution of the tonoplast P-ATPase proton pump
The results of this study indicate thatPbPH5is related to the pear fruit malic acid content. To determine whether this function is conserved,we analyzed thePbPH5homologs in other species. First,thePbPH5homolog in apple (MdMa10) was identified in a BLAST search. A previous study proved thatMdMa10,which encodes a P-type ATPase influencing fruit organic acid accumulation,is aPH5homolog (Maet al.2019). Therefore,thePH5homologs in various species were included in a phylogenetic analysis (Walteret al.2008;Liet al.2016;Shiet al.2019). On the basis of their phylogenetic relationships,PbPH5is also aPH5homolog and is clustered with the genes fromPetuniahybrida,Malus domestica,andCitrusreticulata(Fig.7).

Fig.7 Phylogenetic analysis of the PH5 homologs in different species. The pear PbPH5 gene is indicated with an asterisk.
4.Discussion
Fruit acidity is a quantitative trait controlled by multiple genes and its genetic mechanism is complex (Maet al.2019). The main organic acids contained in different kinds of fruit are different,and they are associated with diverse taste sensations (Hulme 1958;Hudina and Śtampar 2000;Ramoset al.2007;Liuet al.2016). Therefore,the organic acid content in mature pears will partly affect flavor. In recent years,some problems are restricting the development of the pear industry,such as low yields of high-quality fruit,low contents of soluble solids,poor resistance to stress,etc. Hence,clarifying the mechanism regulating the accumulation of organic acids in pear fruit will likely help breeders to generate new pear cultivars that produce fruit with improved flavor characteristics.
The developing fruits of many pear cultivars primarily accumulate malic acid (Hudina and Śtampar 2000;Shaet al.2011;Liuet al.2016;Zhanget al.2020). In fruit cells,malic acid is synthesized in the cytoplasm and then stored in the vacuole (Notton and Blanke 1993;Etienneet al.2013). Additionally,the main factors limiting malic acid accumulation are vacuolar pH and a positive internal Δψ,which are related to the accumulation of protons in the vacuole (Etienneet al.2013). Two distinct types of proton pumps (V-ATPase and V-PPase) have been identified in several fruit crops (Maeshima 2000;Suzukiet al.2000;Terrieret al.2001). Although both types can pump protons into the vacuole,the associated energy sources differ (Etienneet al.2013). Recent research has confirmed that P-type ATPases are crucial regulators of organic acid accumulation in some fruits,including apple (Maet al.2019),Citrus(Shiet al.2019),and pear(Zhanget al.2020). The P-type ATPases form a large plasma membrane protein family (Axelsen and Palmgren 1998). Although P-ATPases also use ATP as an energy source,they are structurally different from the V-ATPases.More specifically,the V-ATPases consist of two subunits,whereas P-ATPases contain only one catalytic subunit(Maet al.2020),and different members in this superfamily transport various ions,such as Na+,Ca2+,Cu2+,and H+,which are involved in a variety of metabolic pathways. The P-ATPase genes are distributed on different chromosomes,and many of them are specifically expressed in more than one tissue (Maet al.2020;Manzooret al.2020). Their expression patterns also vary among tissues and are significantly associated with several metabolic pathways.For example,previous studies found that the transcription ofMD15G1108400in apple roots is negatively correlated with the hydroponic solution pH (Maet al.2020),and thatAtAHA1andAtAHA2are needed to regulate apoplastic pH(Elenaet al.2018). Similarly,in pear,30 P-ATPase genes are located on different chromosomes. The expression patterns of these genes differ among developmental stages and are modulated by hormones,implying that the encoded P-ATPases affect certain physiological activities related to pear fruit development (Manzooret al.2020). In the current study,the expression patterns of P3A-ATPase subfamily genes varied in the root,stem,leaf,flower,pericarp,and pulp among the developmental stages.Additionally,only a few genes were significantly associated with the pear fruit malic acid content.
The P3A-ATPase genes encode proton pumps,which are primarily responsible for transporting H+,making them important for the accumulation of malic acid in vacuoles (Moskowitz and Hrazdina 1981;Shohei 1984).One of the limiting factors of malic acid accumulation is vacuolar acidification (Etienneet al.2013). Recently,the relationship between several P3A-ATPase genes and organic acid accumulation was confirmed. The expression patterns of these genes,includingCsPH8,MdMa10,andPbr029767.1,are significantly related to the organic acid contents (Maet al.2019;Shiet al.2019;Zhanget al.2020). In the present study,we identified a P3A-ATPase gene (PbPH5) that is substantially associated with the pear fruit malic acid content. ThePbPH5transcript level was significantly (*,P<0.05;**,P<0.01) correlated with the malic acid contents in four different pear species (with correlation coefficients of 0.915**,0.932**,0.656*,0.900**,and 0.518*). The results of a transient transformation experiment indicated that the pulp malic acid content was significantly influenced by thePbPH5expression level.Taken together,these results suggest thatPbPH5may be partly related to the accumulation of malic acid in pear.There are many proteins involved in the accumulation of organic acids (Etienneet al.2013),and the correlation coefficients ofPbPH5are also variable among different pear species,which means the accumulation of organic acid in pear is controlled by multiple genes. In addition,it is not clear whether the regulatory networks based on this gene or other transcription factors are involved in organic acid accumulation (Zhanget al.2020). Therefore,our study will not only help to elucidate the molecular mechanism regulating fruit acidity,but it will also lay a foundation for revealing the related genetic mechanism.
Although most P3A-ATPases are located in the plasma membrane,one was initially discovered in the vacuolar membrane,where it has critical functions related to vacuolar acidification. The function of this enzyme (PH5)was verified inPetunia(Walteret al.2008). To clarify the differences in the subcellular localizations of PH5 and other P3A-ATPases,their N-and C-terminal cytoplasmic domains were compared (Arangoet al.2003). Analyses of the sequence differences and subcellular localizations indicated that PH5 is localized to the vacuolar membrane rather than the plasma membrane because of an alteration in the N-terminal sequence,which may have occurred in aPH5ancestor that evolved from other P3A-ATPases (Liet al.2016). TheMdMa10gene also encodes a P-ATPase located in the vacuolar membrane.Our subcellular localization assay proved that PbPH5 is located in the tonoplast. Thus,PbPH5may be aPH5homolog that is involved in vacuolar acidification.
To clarify the relationship betweenPbPH5andPH5homologs,a phylogenetic analysis of various species was conducted on the basis of earlier investigations(Walteret al.2008;Liet al.2016;Shiet al.2019). ThePH5homologs evolved from the P3A-ATPase genes that existed in the early stages of the evolution of green plants(Hedgeset al.2004;Kühlbrandt 2004). Over time,the P-ATPase genes evolved into diverse types,and the P3Asubfamily existed before angiosperms first appeared(Palmeret al.2004). During evolution,PH5homologs evolved from P3Asubfamily genes before the existence of angiosperms and colored flowers,suggesting that they are functionally conserved genes. In our phylogenetic analysis,PbPH5was classified in the same branch as the relatedP.hybrida,M.domestica,andC.reticulatagenes,meaning that it is aPH5homolog encoding an enzyme with a conserved function.
5.Conclusion
PbPH5is significantly associated with pear fruit malic acid accumulation. This gene,which is aPH5homolog,encodes a functionally conserved enzyme. Additionally,the variations in the malic acid contents among pear species is at least partly due to the differences inPbPH5transcription. Furthermore,there are many factors that affect the accumulation of organic acids in developing pear fruit. The data presented herein may be useful for characterizing the P3Asubfamily genes more thoroughly,with potential implications for the breeding of new pear cultivars.
Acknowledgements
This research was funded by the National Key Research and Development Program of China (2019YFD1001400)and the National Natural Science Foundation of China(31601715).
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendicesassociated with this paper are available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
 Journal of Integrative Agriculture2022年6期
Journal of Integrative Agriculture2022年6期
- Journal of Integrative Agriculture的其它文章
- Genetic diversity analysis and GWAS reveal the adaptive loci of milling and appearance quality of japonica rice (Oryza sativa L.) in Northeast China
- A major and stable QTL for wheat spikelet number per spike validated in different genetic backgrounds
- The GhMAX2 gene regulates plant growth and fiber development in cotton
- Optimization of nitrogen fertilization improves rice quality by affecting the structure and physicochemical properties of starch at high yield levels
- Source-sink relations and responses to sink-source manipulations during grain filling in wheat
- lmage-based root phenotyping for field-grown crops:An example under maize/soybean intercropping
