人参bZIP基因家族生物信息学分析
王思嘉,孙嘉莹,刘美琦,任伟超,于欣欣,刘秀波,马 伟, 3*
人参基因家族生物信息学分析
王思嘉1,孙嘉莹1,刘美琦1,任伟超1,于欣欣1,刘秀波2*,马 伟1, 3*
1.黑龙江中医药大学药学院,黑龙江哈尔滨 150040 2.黑龙江中医药大学佳木斯学院,黑龙江佳木斯 154007 3.教育部北药基础与应用研究重点实验室,黑龙江哈尔滨 150040
通过人参基因家族生物信息学分析,为人参功能基因开发利用提供理论依据。运用PlantTFDB数据库预测人参基因组中基因;通过ExPASy网站得到人参基因家族信息和特征;使用MEME网站对基因家族保守基因序列进行分析;运用MEGA软件建立基因家族系统进化关系;利用TBtools软件进行表达分析。人参基因组中含有157个基因家族成员,其中152个定位在细胞核,其余5个分别定位在叶绿体和内质网,相对分子质量在14 009.93~83 440.38,等电点在4.53~10.05,氨基酸数目在120~760 aa,除以外,均为亲水蛋白。在干旱胁迫下的表达量为83.47;在根和盐胁迫条件下表达量均最高,分别为119.82和117.86;在叶中表达量为48.54;花中表达量最高为79.55;成熟果实中表达量最高是,为119.32;在未成熟果实中的表达量最高,且为65.32。推测与同源基因功能相似,可以调控根的分生组织活性;在干旱胁迫条件下表达量最高,推测对干旱胁迫有重要调控作用;A亚族成员/()、()、/()参与干旱胁迫和盐胁迫调控,推测与A亚族成员具有同源关系的、、、在干旱胁迫和盐胁迫条件下具有相似调控作用。
人参;;生物信息学;基因家族;盐胁迫;干旱胁迫
作为成员众多的转录因子家族,在植物领域研究较为透彻[1]。该基因家族对植物的生长发育有重要调节作用,参与植物次生代谢和非生物胁迫的调控[2-3]。通过前人研究成果发现,在拟南芥、陆地棉、花生、小麦、番茄等植物中分别含有75、151、112、156、70个基因成员[4-8]。前期研究将基因家族分为10个亚族[9],分为A、B、C、D、E、F、G、H、I和S。其中,A亚族的和参与应激响应脱落酸(ABA)信号[10],可激活多个晚期胚胎发育[11];C亚族中的、与共同表达可调控种子特异性[12];在D亚族中,确定花器官数目,并参与茎部和花分生组织的表达[13];E亚族中和通过其他基因家族成员相互作用参与植株发育[14]。除此之外,在芹菜中发现基因在干旱、低温、盐处理条件下的表达量不同,并随时间不同发生变化[15];蒺藜苜蓿中基因参与盐胁迫,和均参与干旱胁迫[16]。
人参C.A.Meyer是五加科、人参属多年生草本植物[17],在明代《理虚元鉴》中提到其具有大补元气、复脉固脱之效[18],所以对人体机能恢复有一定作用。人参用于治疗神经衰弱和抑郁型精神病,也可用于治疗冠心病[19],且人参皂苷Rg1有保护糖尿病大鼠肾脏的作用[20],人参皂苷Rg3与紫杉醇结合可抑制肿瘤细胞增长[21],如Luminal型乳腺癌[22]。同时,人参的一些活性成分在美容领域有一定研究[23]。近期研究表明影响人参皂苷的合成[24],可见,人参在临床上具有较高的药用价值且应用广泛。
本研究利用了生物信息学技术,对人参基因家族成员进行鉴定,分析基因家族信息、系统进化、保守基序以及基因表达等,为深入研究人参转录因子在干旱、盐、低温等协迫条件下的调控作用提供理论基础。
1 材料
人参基因组序列文件下载自人参基因组数据库(ginseng genome database,http://ginsengdb.snu.ac.kr/data.php),拟南芥基因家族来自TAIR数据库(http://www.arabidopsis.org/index.jsp)。
2 方法
2.1 人参bZIP基因家族的鉴定
使用生信软件TBtools(v1.086)提取人参全部基因序列,将得到的蛋白序列文件提交plantTFDB数据库(http://planttfdb.cbi.pku.edu.cn/)进行预测,得到人参基因家族成员,用TBtools软件去除重复转录本,并利用Expasy网站(https://web.expasy.org/ protparam/)获取人参的等点量、相对分子质量、蛋白质疏水性、不稳定系数、脂肪族系数等信息,利用WoLFPSORT (https://wolfpsort.hgc.jp/)对人参基因家族成员进行亚细胞定位。
2.2 人参bZIP保守基序分析
使用MEME网站(http://meme-suite.org/tools/ meme),将人参bZIP家族成员蛋白序列的文件进行保守基序预测,最大保守基序数目设为10,其余条件默认,并使用TBtools软件进行可视化。
2.3 人参bZIP基因家族的系统进化
把人参bZIP蛋白序列与拟南芥bZIP蛋白序列整合,将所得文件使用MUSCLE程序进行多序列比对,利用Mega-X软件采用邻接法(neighbor-joining,NJ),Bootstrap值设置为1000次重复,模型选择Poisson model,构建系统发育树。
2.4 人参bZIP基因表达分析
根据人参的等基因数据,在TBtools中绘制热图,进一步对人参基因家族进行表达分析。
3 结果与分析
3.1 人参bZIP基因家族信息和特征
根据人参基因家族的信息(表1)可知,氨基酸数目在120 aa(PgbZIP19)~760 aa(PgbZIP22),相对分子质量的变化范围为14 009.93(PgbZIP19)~83 440.38(PgbZIP22),等电点的范围介于4.53~10.05(PgbZIP76~PgbZIP4),亚细胞定位显示除PgbZIP5、PgbZIP11、PgbZIP79、PgbZIP116、PgbZIP130以外均定位在细胞核,PgbZIP5、PgbZIP116定位在叶绿体,PgbZIP11、PgbZIP79、PgbZIP130定位在内质网;蛋白质疏水性(GRAVY)除PgbZIP157外均为负值;PgbZIP5、PgbZIP115、PgbZIP126的不稳定系数小于40,为稳定蛋白,其余为不稳定蛋白。
表1 人参bZIP基因家族信息和特征
Table 1 Information and characteristics of ginseng bZIP gene family
基因名称基因序列不稳定系数氨基酸数量相对分子质量等电点蛋白质疏水性亚细胞定位脂肪族系数 PgbZIP1Pg_S0015.258.5142047 358.916.49−0.838细胞核67.62 PgbZIP2Pg_S0054.554.5340746 070.726.10−0.814细胞核71.43 PgbZIP3Pg_S0157.158.8332636 432.058.65−0.489细胞核86.01 PgbZIP4Pg_S0161.2961.5320421 865.7510.05−0.750细胞核61.23 PgbZIP5Pg_S0185.134.0616718 520.479.85−0.278叶绿体87.07 PgbZIP6Pg_S0227.3247.5136340 718.146.94−0.886细胞核57.36 PgbZIP7Pg_S0266.1766.6237040 533.445.57−0.791细胞核61.03 PgbZIP8Pg_S0270.157.8445549 459.836.20−0.753细胞核64.15 PgbZIP9Pg_S0292.4145.3833737 695.697.75−0.736细胞核74.07 PgbZIP10Pg_S0431.2552.7333036 872.618.34−0.814细胞核69.73 PgbZIP11Pg_S0431.545.9475682 233.396.04−0.632内质网63.43 PgbZIP12Pg_S0447.753.3338341 604.956.24−0.840细胞核58.36 PgbZIP13Pg_S0500.3441.0020123 151.389.52−0.040细胞核97.01 PgbZIP14Pg_S0602.2541.5736140 739.216.09−0.419细胞核83.88 PgbZIP15Pg_S0624.1961.2415517 986.097.91−1.008细胞核64.84 PgbZIP16Pg_S0659.1055.9946051 724.917.06−0.482细胞核75.57 PgbZIP17Pg_S0662.3860.8150455 852.426.88−0.586细胞核74.42 PgbZIP18Pg_S0662.4051.2933137 878.636.37−0.888细胞核64.20 PgbZIP19Pg_S0662.4355.7712014 009.939.64−0.828细胞核70.75 PgbZIP20Pg_S0684.2852.3331635 490.074.63−0.456细胞核88.83 PgbZIP21Pg_S0687.150.7016918 223.945.94−0.691细胞核64.67 PgbZIP22Pg_S0741.1645.6376383 440.386.28−0.487细胞核75.48 PgbZIP23Pg_S0741.3858.4643047 802.186.85−0.658细胞核69.70 PgbZIP24Pg_S0745.2167.2937642 193.557.82−0.891细胞核63.40 PgbZIP25Pg_S0762.1964.2937640 738.815.72−0.649细胞核65.48 PgbZIP26Pg_S0788.342.2213315 153.859.57−0.377细胞核87.29 PgbZIP27Pg_S0872.672.6418320 453.586.59−0.997细胞核51.86 PgbZIP28Pg_S0884.151.7628731 741.486.67−0.206细胞核79.16 PgbZIP29Pg_S0889.6139.6525527 585.928.95−0.634细胞核76.94 PgbZIP30Pg_S0905.134.6127529 893.275.86−0.683细胞核64.18 PgbZIP31Pg_S0938.951.1029533 135.474.72−0.467细胞核86.88 PgbZIP32Pg_S0967.2773.7025228 878.759.91−0.616细胞核73.45 PgbZIP33Pg_S0981.1549.6640142 806.986.04−0.818细胞核51.17 PgbZIP34Pg_S0992.2254.1859765 001.448.66−0.727细胞核61.68 PgbZIP35Pg_S1022.244.7219921 669.626.15−0.778细胞核65.68 PgbZIP36Pg_S1055.2752.2729033 092.188.72−0.890细胞核63.55 PgbZIP37Pg_S1084.852.5445449 502.749.53−0.649细胞核72.80 PgbZIP38Pg_S1097.3269.3914316 053.836.96−0.694细胞核55.24 PgbZIP39Pg_S1144.256.8543546 958.299.11−0.694细胞核65.49 PgbZIP40Pg_S1164.663.1117220 021.326.16−0.836细胞核77.15 PgbZIP41Pg_S1176.1459.0241243 666.805.86−0.942细胞核46.04 PgbZIP42Pg_S1222.150.0333738 200.436.03−0.433细胞核83.18 PgbZIP43Pg_S1241.249.2813515 270.995.91−0.696细胞核57.78 PgbZIP44Pg_S1242.2368.0833837 650.935.89−0.715细胞核67.87 PgbZIP45Pg_S1290.3562.9144148 732.357.15−0.655细胞核68.82 PgbZIP46Pg_S1338.2056.0458965 153.926.35−0.527细胞核76.53 PgbZIP47Pg_S1348.1064.9336339 935.995.82−0.796细胞核64.10 PgbZIP48Pg_S1349.160.0013215 826.019.46−0.883细胞核74.55 PgbZIP49Pg_S1389.3351.3040843 739.046.19−0.884细胞核49.80
续表1
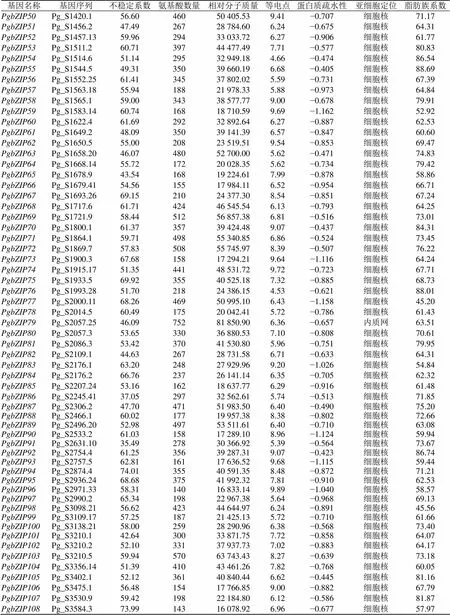
基因名称基因序列不稳定系数氨基酸数量相对分子质量等电点蛋白质疏水性亚细胞定位脂肪族系数 PgbZIP50Pg_S1420.156.6046050 405.539.41−0.707细胞核71.17 PgbZIP51Pg_S1456.247.4926728 784.606.24−0.675细胞核64.31 PgbZIP52Pg_S1457.1359.9629433 033.726.27−0.906细胞核61.77 PgbZIP53Pg_S1511.260.7139744 477.497.71−0.577细胞核80.83 PgbZIP54Pg_S1514.651.1429532 949.184.66−0.474细胞核86.54 PgbZIP55Pg_S1544.549.3135039 660.196.68−0.405细胞核88.69 PgbZIP56Pg_S1552.2561.4134537 802.025.59−0.731细胞核67.39 PgbZIP57Pg_S1563.1855.9418821 978.335.88−0.973细胞核64.84 PgbZIP58Pg_S1565.159.0034338 577.779.00−0.678细胞核79.91 PgbZIP59Pg_S1583.1460.7416818 710.599.69−1.162细胞核52.92 PgbZIP60Pg_S1622.461.6929232 892.646.27−0.887细胞核62.53 PgbZIP61Pg_S1649.248.0935039 141.396.57−0.847细胞核60.60 PgbZIP62Pg_S1650.555.0020823 519.519.54−0.853细胞核69.47 PgbZIP63Pg_S1658.2046.0748052 700.005.62−0.471细胞核74.83 PgbZIP64Pg_S1668.1455.7217220 028.355.62−0.734细胞核79.42 PgbZIP65Pg_S1678.943.5416819 224.617.99−0.878细胞核58.86 PgbZIP66Pg_S1679.4154.5615517 984.116.52−0.954细胞核66.71 PgbZIP67Pg_S1693.2669.1521024 377.308.54−0.851细胞核67.24 PgbZIP68Pg_S1717.661.7142446 545.546.13−0.793细胞核64.25 PgbZIP69Pg_S1721.958.4451256 857.386.81−0.516细胞核73.01 PgbZIP70Pg_S1800.161.3735739 424.489.07−0.437细胞核84.31 PgbZIP71Pg_S1864.159.7149855 340.856.86−0.524细胞核73.45 PgbZIP72Pg_S1869.757.8350855 745.978.39−0.507细胞核76.22 PgbZIP73Pg_S1900.367.6815817 294.219.64−1.116细胞核64.24 PgbZIP74Pg_S1915.1751.3544148 531.729.72−0.723细胞核67.71 PgbZIP75Pg_S1933.569.9235540 525.187.32−0.885细胞核68.73 PgbZIP76Pg_S1993.2851.7021824 386.154.53−0.621细胞核88.01 PgbZIP77Pg_S2000.1168.2646950 995.106.43−1.158细胞核45.20 PgbZIP78Pg_S2014.560.4917520 042.415.72−0.786细胞核61.43 PgbZIP79Pg_S2057.2546.0975281 850.906.36−0.657内质网63.51 PgbZIP80Pg_S2057.353.6533036 880.537.10−0.808细胞核70.61 PgbZIP81Pg_S2086.353.4237041 530.805.96−0.751细胞核79.95 PgbZIP82Pg_S2109.144.6326728 731.586.71−0.633细胞核64.31 PgbZIP83Pg_S2176.163.2024827 929.969.20−1.026细胞核54.84 PgbZIP84Pg_S2176.266.7623726 141.146.35−0.705细胞核62.32 PgbZIP85Pg_S2207.2453.1616218 637.776.29−0.916细胞核61.48 PgbZIP86Pg_S2245.4137.0529732 562.615.74−0.513细胞核71.85 PgbZIP87Pg_S2306.247.7047151 983.506.40−0.490细胞核75.20 PgbZIP88Pg_S2466.160.0217719 957.388.38−0.802细胞核72.66 PgbZIP89Pg_S2496.2052.9849753 511.616.40−0.710细胞核63.08 PgbZIP90Pg_S2533.261.0315817 289.108.96−1.124细胞核59.94 PgbZIP91Pg_S2631.1035.4927830 366.925.39−0.564细胞核73.67 PgbZIP92Pg_S2754.461.2535639 287.319.07−0.423细胞核86.74 PgbZIP93Pg_S2757.562.8116117 636.529.68−1.115细胞核59.44 PgbZIP94Pg_S2874.474.0135540 591.358.48−0.872细胞核71.21 PgbZIP95Pg_S2936.2468.6837541 992.327.81−0.910细胞核62.53 PgbZIP96Pg_S2971.3358.3114016 833.149.89−1.040细胞核58.57 PgbZIP97Pg_S2990.265.3419822 967.385.64−0.968细胞核69.13 PgbZIP98Pg_S3098.2156.6242344 644.976.24−0.891细胞核45.56 PgbZIP99Pg_S3109.1757.2518721 425.135.72−0.710细胞核61.66 PgbZIP100Pg_S3138.2158.0025928 290.966.38−0.568细胞核73.40 PgbZIP101Pg_S3210.142.6430033 871.757.72−0.858细胞核64.07 PgbZIP102Pg_S3210.252.1033137 937.737.02−0.883细胞核64.17 PgbZIP103Pg_S3210.559.9457063 743.438.27−0.639细胞核73.18 PgbZIP104Pg_S3356.1451.3941043 461.267.82−0.768细胞核60.05 PgbZIP105Pg_S3402.152.1236140 840.446.62−0.445细胞核81.16 PgbZIP106Pg_S3475.156.4815417 766.859.00−0.882细胞核67.79 PgbZIP107Pg_S3530.959.4219822 184.806.12−0.586细胞核81.87 PgbZIP108Pg_S3584.373.9914316 078.926.96−0.677细胞核57.97
续表1
3.2 人参bZIP家族保守基序分析
使用MEME网站查找人参bZIP蛋白保守基序,可知PgbZIP的保守基序个数为1~6个。Motif9只在A亚族中存在一小部分;D亚族中普遍含有6个保守基序;Motif5只存在于I、E家族中;PgbZIP中均含有Motif1,且少数PgbZIP成员中只含有Motif1,见图1。
3.3 人参bZIP基因家族的系统进化关系
将人参的157个bZIP蛋白和拟南芥中74个蛋白进行系统进化树的构建(图2),bZIP家族分为10个亚家族,分别为A、B、C、D、E、F、G、H、I和S,其中A亚族中包括33个人参bZIP成员;B亚族中只含有2个成员PgbZIP53、PgbZIP144;C亚族中包括12个PgbZIP成员;D亚族中含有22个PgbZIP成员;E亚族中含有15个PgbZIP成员;F亚族中含有4个PgbZIP成员:PgbZIP151、PgbZIP91、PgbZIP86、PgbZIP30;G亚族中含有8个PgbZIP成员;H亚族中含有10个PgbZIP成员;I亚族中含有16个成员;S亚族含有35个成员,是含有人参bZIP成员数最多的亚族。
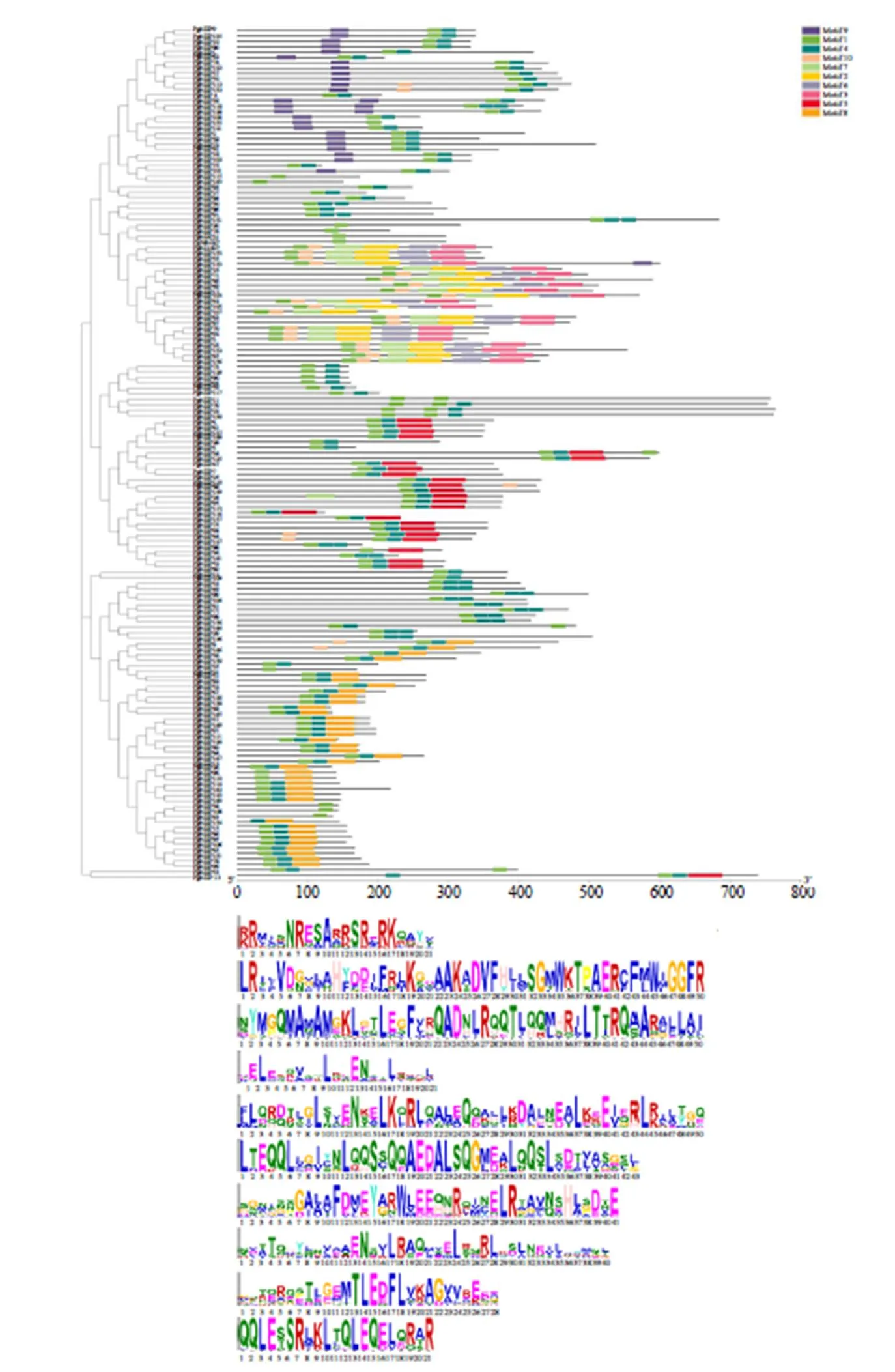
图1 人参bZIP蛋白基序以及Motif分析
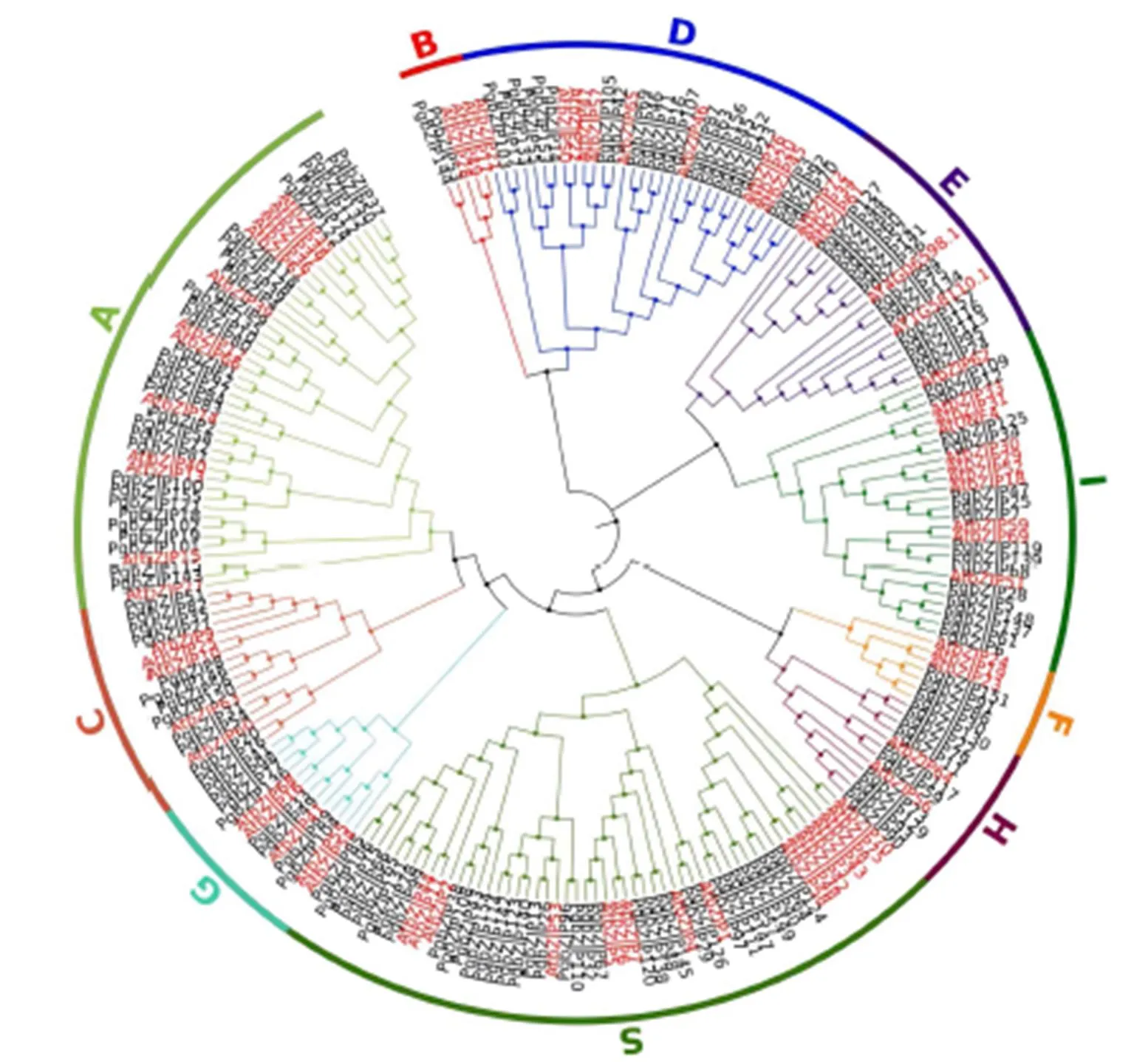
图2 人参和拟南芥bZIP转录因子的系统发育树
3.4 人参bZIP基因在植物不同器官中的表达
通过TBtools软件制作热图,对基因家族的表达进行分析。如图3,表明、在成熟果实中表达量较高,表达最明显,在未成熟果实中表达量最高;在花中表达最明显;、在根中表达量较高;在根和盐胁迫条件下中的表达量最高,在盐胁迫下表达量较高;在干旱胁迫下表达量最高。
4 讨论
bZIP是一类参与植物生长、生物与非生物胁迫的转录因子,通过图2和图3分析,人参中在根中的表达量最高,在茎和花中的表达量逐渐降低,与其同源的拟南芥基因具有抑制分生组织活性与根生长的功能[25],推测其在也具有负调控作用。()、()、()、()、()、()为拟南芥中TGA中的成员,相互作用参与植物疾病调控[26-27],推测其同源基因、、、也有相似的功能。
非生物胁迫包括干旱胁迫、盐胁迫、低温胁迫等[28],Yoshida等[29]发现bZIP转录因子对抗逆性有重要作用。在干旱胁迫条件下表达量最高,属于S亚族,该亚族对非生物胁迫有较强响应,其中AtbZIP44在干旱胁迫中发挥重要作用[30],因与为同源基因,推测在人参中起到相似作用。
ABA在植物的生长发育中对环境胁迫发挥重要作用[31],其中拟南芥成员/()、()、/()主要参与ABA、盐胁迫、干旱胁迫、热胁迫等[32],成员、、与其为同源基因,且在盐胁迫条件下表达较高,推测、、可能参与盐胁迫调控。对盐胁迫有较强反应[31],、在盐胁迫条件下的表达量较高,推测其在人参盐胁迫条件下发挥重要作用。Fujita等[32]发现拟南芥中的ABRE结合因子家族成员依赖ABA信号,增强植物营养组织的抗旱性。烟草研究在ABA的处理下使A亚族中的成员表达量上升[33],结合图3发现成员、、、、、表达量在叶、花、干旱胁迫下或盐胁迫有明显增加,推测在ABA处理下也有相似功能,参与人参的生长发育。
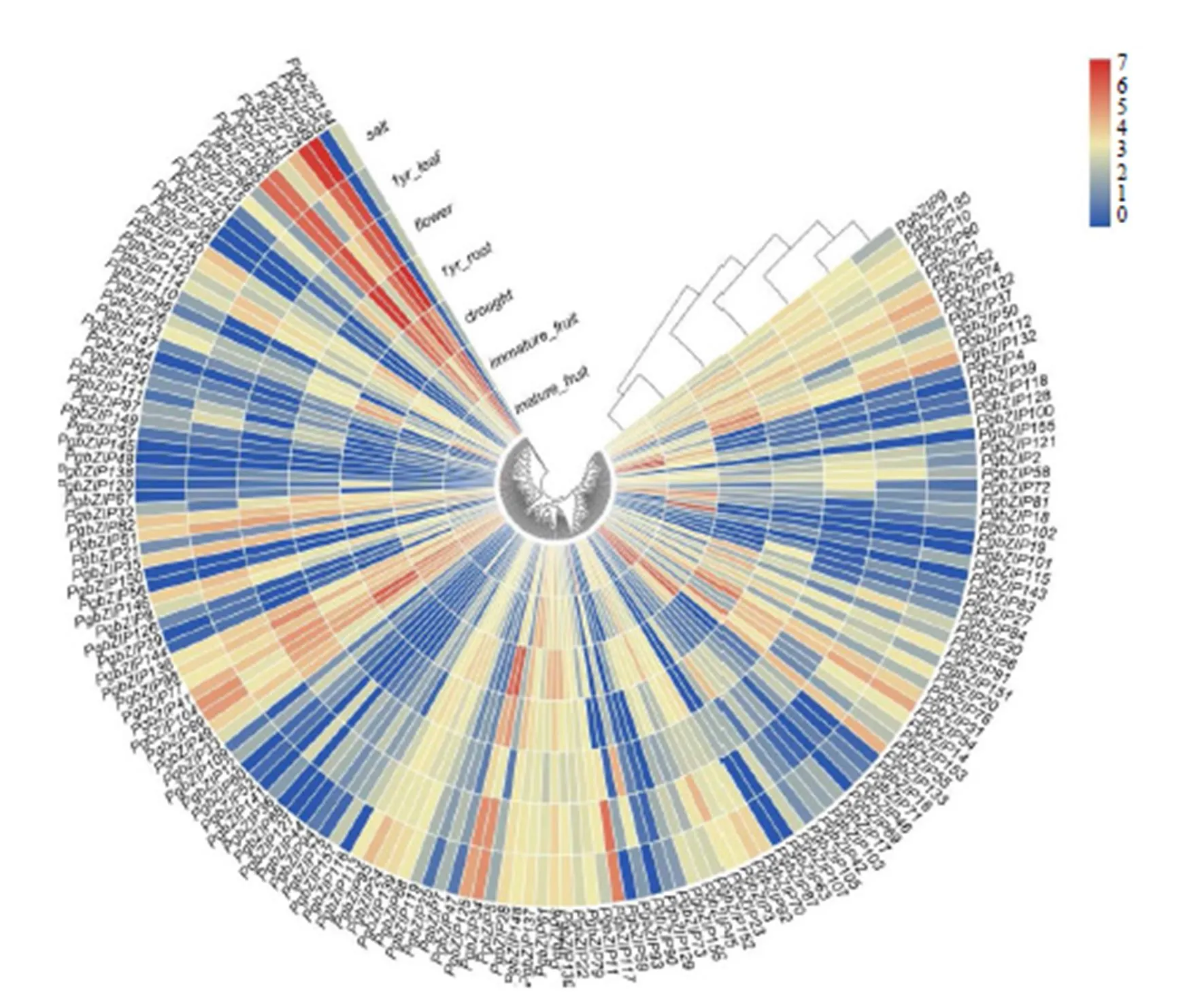
图3 人参bZIP基因在植物不同器官和非胁迫条件下的表达分析
通过人参基因家族的生物信息学分析,对可能参与人参非生物胁迫的基因进行功能预测,为深入研究基因提供分子基础,为进一步探索人参次生代谢调控以及药用活性成分临床应用提供理论依据。
利益冲突 所有作者均声明不存在利益冲突
[1] 刘莉.DREBs、转录因子与植物抗旱性研究进展 [J].浙江农业科学, 2013, 54(1): 98-102.
[2] 沈迪, 陈龙正, 陶建平, 等.芹菜bZIP转录因子基因AgbZIP16的逆境响应分析 [J].植物生理学报, 2019, 55(12): 1817-1826.
[3] 曹红利.茶树家族基因的非生物胁迫响应及C亚家族CsbZIP6和CsbZIP4的功能初步分析 [D].北京: 中国农业科学院, 2016.
[4] Jakoby M, Weisshaar B, Dröge-Laser W,.bZIP transcription factors in[J]., 2002, 7(3): 106-111.
[5] 邢宇鹏.棉花基因家族全基因组鉴定及分析 [D].泰安: 山东农业大学, 2020.
[6] 高斌, 陈娟娟, 崔顺立, 等.花生基因家族全基因组鉴定及抗旱表达分析 [J].植物遗传资源学报, 2020, 21(1): 174-191.
[7] 李雪垠.小麦转录因子家族中花药发育及抗逆相关基因的功能研究 [D].杨凌: 西北农林科技大学, 2016.
[8] 朱芸晔, 薛冰, 王安全, 等.番茄转录因子家族的生物信息学分析 [J].应用与环境生物学报, 2014, 20(5): 767-774.
[9] Dröge-Laser W, Snoek B L, Snel B,.Thetranscription factor family-an update [J]., 2018, 45(Pt A): 36-49.
[10] Kang J Y, Choi H I, Im M Y,.basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling [J]., 2002, 14(2): 343-357.
[11] Bensmihen S, Rippa S, Lambert G,.The homologousandtranscription factors function antagonistically to fine-tune gene expression during late embryogenesis [J]., 2002, 14(6): 1391-1403.
[12] Lara P, Oñate-Sánchez L, Abraham Z,.Synergistic activation of seed storage protein gene expression inby ABI3and two bZIPs related to OPAQUE2 [J]., 2003, 278(23): 21003-21011.
[13] Fletcher J C.The ULTRAPETALA gene controls shoot and floral meristem size in[J]., 2001, 128(8): 1323-1333.
[14] Shen H S, Cao K M, Wang X P.A conserved proline residue in the leucine zipper region of AtbZIP34 and AtbZIP61 ininterferes with the formation of homodimer [J]., 2007, 362(2): 425-430.
[15] 沈迪, 陈龙正, 陶建平, 等.芹菜转录因子基因的逆境响应分析 [J].植物生理学报, 2019, 55(12): 1817-1826.
[16] 齐晓, 张正社, 闵学阳, 等.紫花苜蓿基因家族的鉴定、进化及表达分析 [J].草业科学, 2017, 34(8): 1635-1648.
[17] 杨秀伟, 富力.人参中三萜类化学成分的生物学活性和药理学作用 [J].中国现代中药, 2016, 18(1): 36-55.
[18] 龙玉婷, 孙志会, 王志文, 等.人参大补元气的功效内涵浅析 [J].中国中医急症, 2019, 28(9): 1679-1682.
[19] 柳良燕.人参西洋参功效比较及用法研究 [J].实用医技杂志, 2008, 15(34): 41-43.
[20] 马小芬, 谢席胜, 左川, 等.人参皂甙Rg1对糖尿病肾病大鼠肾脏保护作用的机制研究 [J].生物医学工程学杂志, 2010, 27(2): 342-347.
[21] 张奉海, 张瑞荣, 陈淑娟, 等.人参皂苷Rg3联合紫杉醇抑制人肝癌细胞HepG2增殖和对裸鼠移植瘤模型的作用机制研究 [J].中医药学报, 2020, 48(10): 16-20.
[22] 郑颖娟, 王辉, 刘晓静, 等.人参皂苷对Luminal型乳腺癌中Fas、FasL表达水平的影响 [J].辽宁中医杂志, 2020, 47(10): 185-189.
[23] 熊晨阳, 许明良, 易帆, 等.人参不同部位主要活性成分及其在美容护肤方面的研究进展 [J].日用化学工业, 2019, 49(3): 193-198.
[24] 李宏杰.吉林人参转录因子基因家族系统分析及PgbZIP48-3基因功能的初步验证 [D].长春: 吉林农业大学, 2020.
[25] Weiste C, Pedrotti L, Selvanayagam J,.Thetranscription factor links low-energy signalling to auxin-mediated control of primary root growth [J]., 2017, 13(2): e1006607.
[26] Kesarwani M, Yoo J, Dong X N.Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in[J]., 2007, 144(1): 336-346.
[27] 田义, 张彩霞, 康国栋, 等.植物TGA转录因子研究进展 [J].中国农业科学, 2016, 49(4): 632-642.
[28] 王冰, 程宪国.干旱、高盐及低温胁迫下植物生理及转录因子的应答调控 [J].植物营养与肥料学报, 2017, 23(6): 1565-1574.
[29] Yoshida T, Fujita Y, Sayama H,.AREB1, AREB2, and ABF3are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation [J]., 2010, 61(4): 672-685.
[30] Weltmeier F, Rahmani F, Ehlert A,.Expression patterns within theC/S1transcription factor network: Availability of heterodimerization partners controls gene expression during stress response and development [J]., 2009, 69(1/2): 107-119.
[31] Sun X L, Li Y, Cai H,.Thetranscription factor is a positive regulator of plant tolerance to salt, osmotic and drought stresses [J]., 2012, 125(3): 429-438.
[32] Fujita Y, Fujita M, Satoh R,.AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in[J]., 2005, 17(12): 3470-3488.
[33] Choi H I, Hong J H, Ha J O,.ABFs, a Family of ABA-responsive element binding factors [J]., 2000, 275(3): 1723-1730.
Bioinformatics analysis ofgene family of
WANG Si-jia1, SUN Jia-ying1, LIU Mei-qi1, REN Wei-chao1, YU Xin-xin1, LIU Xiu-bo2, MA Wei1, 3
1.College of Pharmaceutical Sciences, Heilongjiang University of Chinese Medicine, Harbin 150040, China 2.College of Jiamusi, Heilongjiang University of Chinese Medicine, Jiamusi 154007, China 3.Key Laboratory of Basic and Application Research of Beiyao (Heilongjiang University of Chinese Medicine), Ministry of Education, Harbin 150040, China
To analyzegene family bioinformatics of Renshen (), so as to provide theoretical basis for the development and utilization of ginseng functional genes.Using PlantTFDB database to predict ginsenggene; The genetic family information and characteristics of ginsengwere obtained through ExPASy website; The conserved gene sequences ofgene family were analyzed by MEME website; The phylogenetic relationship ofgene family were established by using MEGA software;expression analysis were conducted by TBtools software.The ginseng genome contained 157 bZIP gene family members, of which 152 were located in the nucleus, and the remaining five were located in the chloroplast and endoplasmic reticulum, respectively.The relative molecular mass was between 14 009.93 and 83 440.38, and the isoelectric point was in the range of 4.53—10.05, the number of amino acids was between 120 aa—760 aa, except for, all were hydrophilic proteins.The expression level ofunder drought stress was 83.47; the expression level ofwas the highest under root (119.82) and salt stress (117.86) conditions, respectively; the expression level ofin leaves was 48.54; the highest expression level ofin flowers was 79.55; The highest expression level ofin mature fruits was 119.32; the highest expression level ofin immature fruits was 65.32.It is speculated thathas similar functions to the homologous gene, and can regulate root meristem activity;has the highest expression under drought stress conditions, and it is speculated thathas an important regulatory effect on drought stress; A subfamily members,,/is involved in the regulation of drought stress and salt stress.It is speculated that,,and, which have a homologous relationship with members of subfamily A, have similar regulatory effects under drought stress and salt stress.
C.A.Meyer;; bioinformatics; gene family; salt stress; drought stress
R282.12
A
0253 - 2670(2022)09 - 2786 - 09
10.7501/j.issn.0253-2670.2022.09.022
2021-11-09
黑龙江省“头雁”团队项目(黑龙江省头雁行动领导小组文件 [2019] 5号);黑龙江中医药大学科研基金项目(中药健康相关产品研发及产业化专项)(2019BJP06)
王思嘉,女,硕士研究生,研究方向为药用植物生物工程研究。Tel: (0451)87266988 E-mail: 2077737519@qq.com
通信作者:马 伟,研究员,博士生导师,主要从事药用植物生物工程研究。Tel: (0451)87266988 E-mail: 88788891@qq.com
刘秀波,教授,硕士生导师,主要从事中药资源与中药化学。Tel: 13796353268 E-mail: 358270831@qq.com
[责任编辑 时圣明]

