大脑中动脉栓塞模型大鼠海马组织circRNA-miRNA-mRNA三元转录网络分析*
陈博威, 唐荣梅, 徐雅倩, 易健, 刘柏炎△
大脑中动脉栓塞模型大鼠海马组织circRNA-miRNA-mRNA三元转录网络分析*
陈博威1, 唐荣梅1, 徐雅倩2, 易健2, 刘柏炎1△
(1湖南中医药大学,湖南 长沙 410208;2湖南中医药大学第一附属医院,湖南 长沙 410007)
探讨脑缺血模型大鼠海马组织中差异表达的环状RNAs(circRNAs)及信使RNAs(mRNAs),并进行circRNA-微小RNA(miRNA)-mRNA三元转录网络的构建。将雄性SD大鼠随机分为对照组和模型组,每组8只,其中每组用于神经行为学评分8只,尼氏染色4只,对照组基因芯片检测4只,模型组基因芯片检测3只。模型组大鼠使用大脑中动脉栓塞法(MCAO)复制脑缺血模型。7 d后采用神经行为学评分和尼氏染色验证造模成功,Agilent竞争性内源RNA(ceRNA)芯片筛选差异表达的circRNAs及mRNAs,通过GO功能和KEGG通路富集分析差异基因参与的主要生物学过程,RT-qPCR验证基因芯片结果,最后构建circRNA-miRNA-mRNA转录网络。与对照组相比,模型组大鼠神经行为学评分显著升高(<0.01),并出现海马神经元损伤。ceRNA芯片筛选出脑缺血大鼠海马组织中有18个差异表达的circRNAs和836个差异表达的mRNAs(FC≥1.5,<0.05)。RT-qPCR验证结果显示,随机挑选的RNO_CIRCpedia_136、RNO_CIRCpedia_5686、RNO_CIRCpedia_9127及α-血红蛋白稳定蛋白(Ahsp)在模型组中表达下调(<0.05或<0.01),分泌型磷蛋白1(Spp1)及清道夫受体1(Msr1)在模型组中表达上调(<0.01),与基因芯片结果的变化趋势一致。构建了由3个circRNAs、4个miRNAs及9个mRNAs组成的三元转录网络。生物信息学分析显示这些差异基因可能通过血管内皮生长因子信号通路、Toll样受体信号通路、Janus激酶-信号转导及转录激活因子信号通路、磷脂酰肌醇3-激酶-蛋白激酶B信号通路及Hedgehog信号通路等调控脑缺血损伤。脑缺血大鼠海马组织存在差异表达的circRNAs和mRNAs,这些差异基因可能通过circRNA-miRNA-mRNA三元转录网络系统调控脑缺血损伤。
大脑中动脉栓塞;基因微阵列;环状RNA;竞争性内源RNA
缺血性脑血管病(ischemic cerebrovascular disease, ICD)具有高发病率和高致残率的特点,造成了极大的医疗负担[1]。对于缺血性脑卒中,静脉溶栓是目前主要的治疗方法。然而,因溶栓严格的时间窗限制,故在临床运用中仍存在诸多限制,脑梗死仍是目前尚未攻克的疑难疾病之一。
与常规的线性RNA不同,环状RNA(circular RNA, circRNA)特有的共价闭合环状结构使其具有良好的稳定性,更难以被降解,这一特性也显示了circRNA作为生物标志物具有良好的潜力[2]。研究显示,circRNA虽不能直接编码翻译蛋白质,但是其能充当“微小RNA(microRNA, miRNA)海绵”,通过海绵吸附miRNA的方式,间接降低miRNA与下游靶向信使RNA(messenger RNA, mRNA)的结合,从而影响靶基因的表达,即竞争性内源RNA(competing endogenous RNA, ceRNA)调控机制[3]。有证据表明,circRNA参与的ceRNA调控轴在脑梗死的早期诊断和有效治疗中发挥着重要作用,如circ_HECTD1 (HECT domain E3 ubiquitin protein ligase 1)可通过miR-27a-3p/卵泡抑素样蛋白1(follistatin-like protein 1,FSTL1)轴调控ICD后神经损伤[4];circRNA TTC3 (tetratricopeptide repeat domain 3)能够竞争性结合miRNA-372-3p,从而影响Toll样受体4(Toll-like receptor 4, TLR4)表达,参与脑缺血后的神经炎症反应[5]。目前,由多个ceRNA轴组成的circRNA-miRNA-mRNA转录网络已成为研究脑缺血病理生理机制的热点[6]。
研究表明,哺乳动物大脑海马区域存在静默的神经干细胞。当脑缺血时,由于微环境变化激活了神经干细胞,促进其增殖并移行至损伤部位,分化成熟为各类神经细胞,重建大脑的组织结构[7]。因此,进一步明确脑缺血后海马区的分子机制变化,有利于寻找到治疗脑缺血的新靶点。故本实验拟采用脑缺血模型大鼠,运用ceRNA芯片分析脑缺血大鼠海马组织差异表达的circRNAs及mRNAs,并进行circRNA-miRNA-mRNA三元转录网络的构建,为进一步寻找到治疗脑缺血的靶点提供参考资料。
材料和方法
1 实验动物
SPF级雄性SD大鼠,周龄6~8周,体重(230±10) g,共20只,购自湖南斯莱克景达实验动物有限公司,实验动物生产许可证号为SCXK(湘)2019-0004。喂养于湖南中医药大学第一附属医院SPF级动物房,实验单位使用许可证号为SYXK(湘)2020-0010。室温22~26 ℃,湿度45%~55%,通风,昼夜规律照明,适应性饲养1周,大鼠自由饮水进食。本实验已通过湖南中医药大学第一附属医院实验动物伦理委员会批准(伦理号:ZYFY20201215-1)。
2 主要试剂
动脉栓线(货号:2432A2)购自北京西浓科技有限公司;RNAwait非冻型组织保存液(货号:MA0208)购自大连美仑生物技术有限公司;Trizol试剂(货号: 10296010)购自Life Technologies;ceRNA基因芯片由Agilent定制;PCR引物由北京擎科新业生物合成。
3 主要仪器
2100型生物分析仪和G2505C型芯片扫描仪(Agilent);9700型PCR仪(ABI);5418型高速离心机(Eppendorf);Vectra3智能组织切片成像系统(PerkinElmer)。
4 方法
4.1模型的制备与评价采用Longa等[8]报道的线栓法复制大鼠大脑中动脉栓塞(middle cerebral artery occlusion, MCAO)脑缺血模型:术前禁食禁水12 h,0.3%戊巴比妥钠麻醉,取颈正中切口,钝性分离左侧颈总、颈内、颈外动脉,将线栓经颈总动脉送入颈内动脉,当线栓上黑色标记点恰好位于颈总分叉时固定线栓,消毒并缝合皮肤。术后2 h进行神经行为学评分,参照Longa等[8]报道的方法,1~3分表明造模成功,入选模型(model)组。共造模12只大鼠,实验期间死亡4只,剩余8只,对照(control)组8只大鼠无死亡。
4.2神经行为学评估造模后7 d,应用Ayelet Levy 14分评分法对每组所有大鼠神经行为学进行评估[9]。
4.3尼氏染色神经行为学评分完成后,每组选取4只大鼠。麻醉后首先用预冷的生理盐水及4%多聚甲醛进行灌注处理,后断头取脑。4%多聚甲醛固定脑组织后,常规脱水、透明、包埋及切片,以1%甲苯胺蓝染色。使用Vectra3智能组织成像系统扫描整个切片,并从海马齿状回(dentate gyrus, DG)和CA3区提取100倍视野。
4.4ceRNA芯片检测神经行为学评分完成后,每组选择4只大鼠。麻醉后直接断头取脑,冰上操作迅速分离缺血侧海马组织,放入预先盛满RNAwait液的冻存管中,严格按照操作流程保存送检。采用Trizol法提取总RNA,检测RNA浓度及质量后,共有7个样本进行后续基因芯片分析(其中正常组4个,模型组3个)。扩增RNA并转录成荧光cRNA。接着纯化标记的cRNA,将50 μL杂交溶液分布在ceRNA微阵列载玻片上。将载玻片放在安捷伦杂交炉中于65 ℃温育17 h。洗涤、固定和扫描混合阵列,扫描完成后通过Feature Extraction软件进行数据的抽提,生成的原始数据文件经GeneSpring软件标准化后进行后续的数据分析。
4.5基因本体(gene ontology, GO)功能及京都基因与基因组百科全书(Kyoto Encyclopedia of Genes and Genomes, KEGG)通路富集分析将差异表达的circRNAs母源基因和mRNAs进行GO功能及KEGG通路富集分析。
4.6RT-qPCR验证将芯片检测后的剩余样本进行RT-qPCR验证。随机选取了3个差异表达的circRNAs(RNO_CIRCpedia_136、RNO_CIRCpedia_5686及RNO_CIRCpedia_9127)和3个差异表达的mRNAs[α-血红蛋白稳定蛋白(alpha-hemoglobin stabilizing protein, Ahsp)、分泌型磷蛋白1(secreted phosphoprotein 1, Spp1)及清道夫受体1(macrophage scavenger receptor 1, Msr1)]。实验过程严格参照试剂盒步骤,内参照选用β-actin,使用2-ΔΔCt法计算基因的相对表达量,各引物序列详见表1。
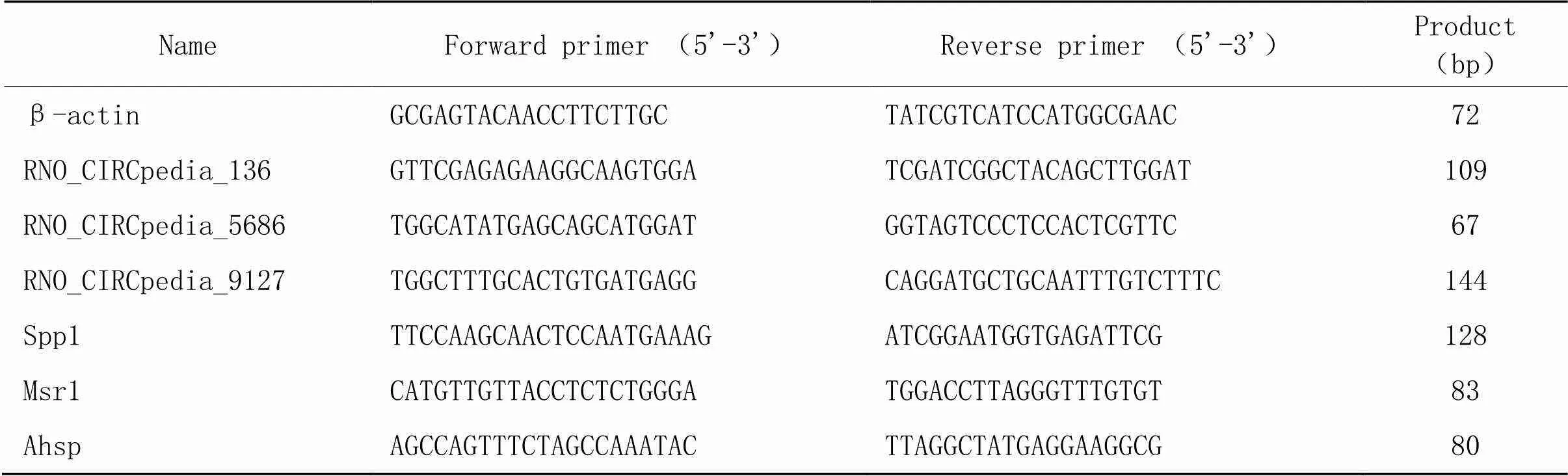
表1 RT- qPCR引物序列
4.7circRNA-miRNA-mRNA转录网络的构建根据先前报道的方法及筛选条件[10],首先借助欧易生物云平台共表达相关性分析模块(https://cloud.oebiotech.cn/task/),输入参数(相关系数阈值<0.8,相关系数值<0.05,分析算法为Pearson),筛选与差异mRNAs显著正相关的差异circRNAs,随后应用miRBase 22.0数据库预测circRNA-miRNA和miRNA-mRNA关系对,最后以miRNA为桥梁构建circRNA-miRNA-mRNA三元转录网络。
5 统计学处理
采用差异倍数(fold change, FC)≥1.5及<0.05的标准筛选差异基因[10]。计量资料以均数±标准差(man±SD)进行表示。采用单因素方差分析比较多组间均数。以<0.05为差异有统计学意义。应用GraphPad Prism 8作图软件分析并处理。
结果
1 脑缺血大鼠模型的建立
与对照组相比,模型组大鼠神经行为学评分显著升高(<0.01),见图1A。另外尼氏染色显示,与对照组相比,模型组大鼠海马齿状回区和CA3区神经元排列不规则,细胞间隙增宽,细胞核固缩,见图1B。这提示脑缺血大鼠模型建立成功,并且脑缺血大鼠缺血侧海马组织有病理损伤。
2 差异circRNAs与富集分析
基于ceRNA芯片,识别出模型组大鼠缺血侧海马组织相比对照组有18个差异表达的circRNAs,其中3个上调,15个下调,详见图2A、B。对这些差异circRNAs的母源基因进行GO功能及KEGG通路富集分析,显示GO功能的生物过程主要为兴奋性突触后电位、脑形态发生及对组织重塑的调节等,细胞组成主要为突触及细胞连接等,分子功能主要为PDZ结构域绑定、蛋白激酶C结合及蛋白质结合等,详见图2C。KEGG通路富集分析结果主要为血管内皮生长因子(vascular endothelial growth factor, VEGF)信号通路、核因子κB(nuclear factor-κB, NF-κB)及Toll样受体信号通路等,详见图2D。

Figure 2.Differentially expressed (DE) circRNAs and functional enrichment analysis. A: the scatter plot of the expression profiles of DE circRNAs between model and control groups (red dots: up-regulated genes; blue dots: down-regulated genes); B: heat map about DE circRNAs; C: GO enrichment analysis of maternal genes of DE circRNAs; D: KEGG enrichment analysis of maternal genes of DE circRNAs.
3 差异mRNAs与富集分析
同样,本研究识别出模型组相比对照组有836个差异表达的mRNAs,其中537个上调,299个下调,详见图3A、B。对这些差异mRNAs进行GO功能及KEGG通路富集分析,显示生物过程主要为对病毒的防御反应、吞噬作用及炎症反应等,细胞组成主要为细胞外空间及血红蛋白复合物,分子功能主要为趋化因子活性、IgG结合及氧载体活性等,详见图3C。KEGG通路富集分析结果主要为Toll样受体信号通路、肿瘤坏死因子(tumor necrosis factor, TNF)信号通路及酪氨酸蛋白激酶/信号转导及转录激活因子(Janus kinase/signal transducer and activator of transcription, JAK/STAT)信号通路等,详见图3D。
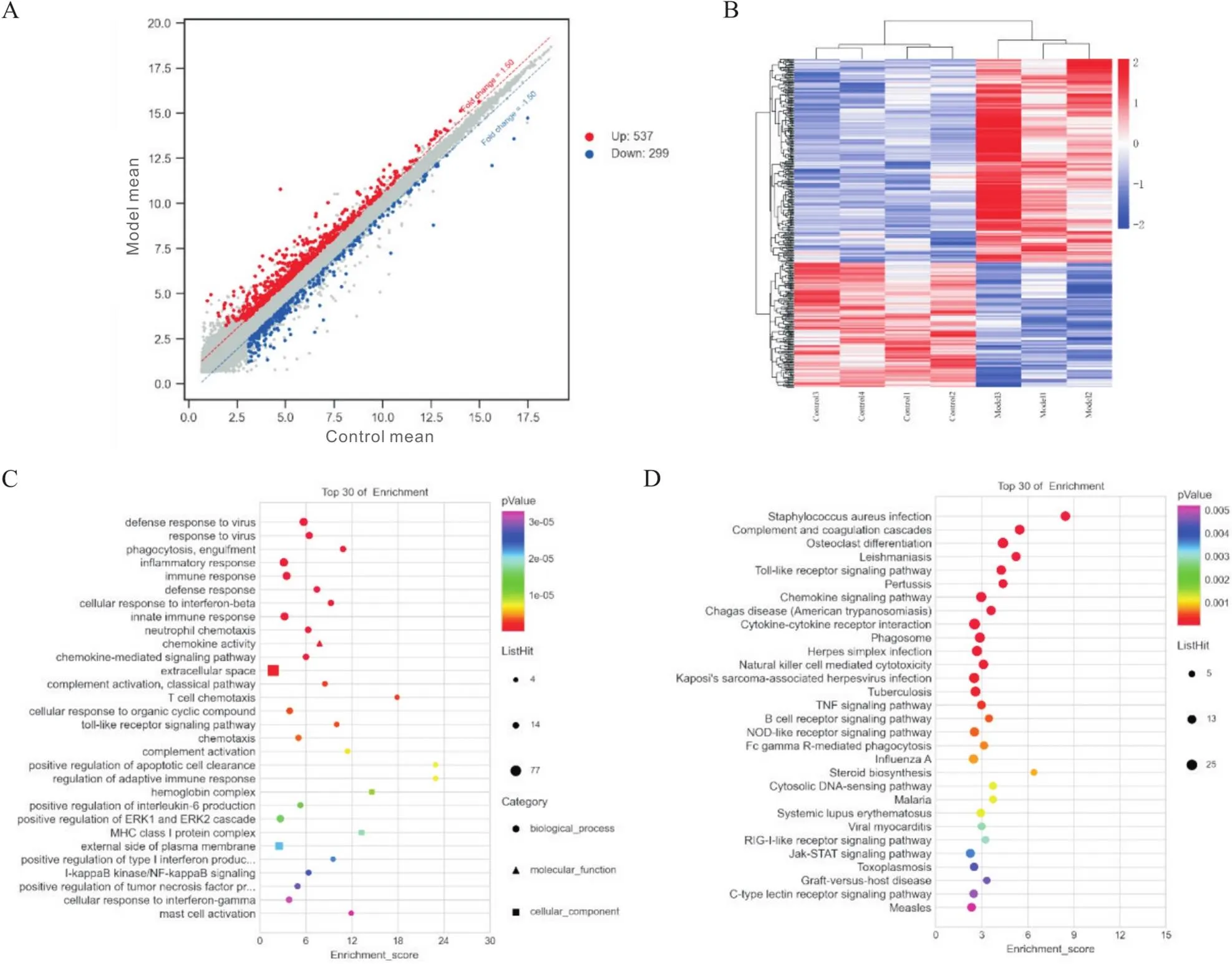
Figure 3.Differentially expressed (DE) mRNAs and functional enrichment analysis. A: the scatter plot of the expression profiles of DE mRNAs between model and control groups (red dots: up-regulated genes; blue dots: down-regulated genes); B: heat map about DE mRNAs; C: GO enrichment analysis of DE mRNAs; D: KEGG enrichment analysis of DE mRNAs.
4 差异表达基因的验证
随机挑选3个差异表达的cricRNAs和3个mRNAs,通过RT-qPCR验证基因芯片结果,其中RNO_CIRCpedia_136、RNO_CIRCpedia_5686、RNO_ CIRCpedia_9127及Ahsp在模型组中表达下调,Spp1及Msr1在模型组中表达上调(<0.05或<0.01),与基因芯片结果的变化趋势一致,见图4,证实了基因芯片结果的准确性。因此将所有差异表达的基因均列入后续分析。

Figure 4.Validation of differentially expressed genes. Mean±SD. n=3. *P<0.05, **P<0.01 vs control group.
5 circRNA-miRNA-mRNA三元转录网络的构建
为了进一步阐明差异circRNAs的生物学功能,课题组首先筛选出与差异mRNAs显著正相关的circRNAs,接着以miRNA为核心,预测具有结合位点的circRNA-miRNA和miRNA-mRNA关系对,最终构建出由3个circRNAs、4个miRNAs及9个mRNAs组成的circRNA-miRNA-mRNA三元转录网络,见图5。对该网络中的效应分子mRNAs进行富集分析,显示生物过程主要为调节血管内皮细胞迁移及小胶质细胞激活的正调控等,细胞组成主要为Wnt信号体及自噬泡,分子功能主要为组蛋白脱乙酰基酶结合及激酶活性等,见图6A;KEGG通路富集分析结果主要为磷脂酰肌醇3-激酶/蛋白激酶B(phosphatidylinositol 3-kinase-protein kinase B, PI3K-Akt)信号通路及Hedgehog信号通路等,见图6B。
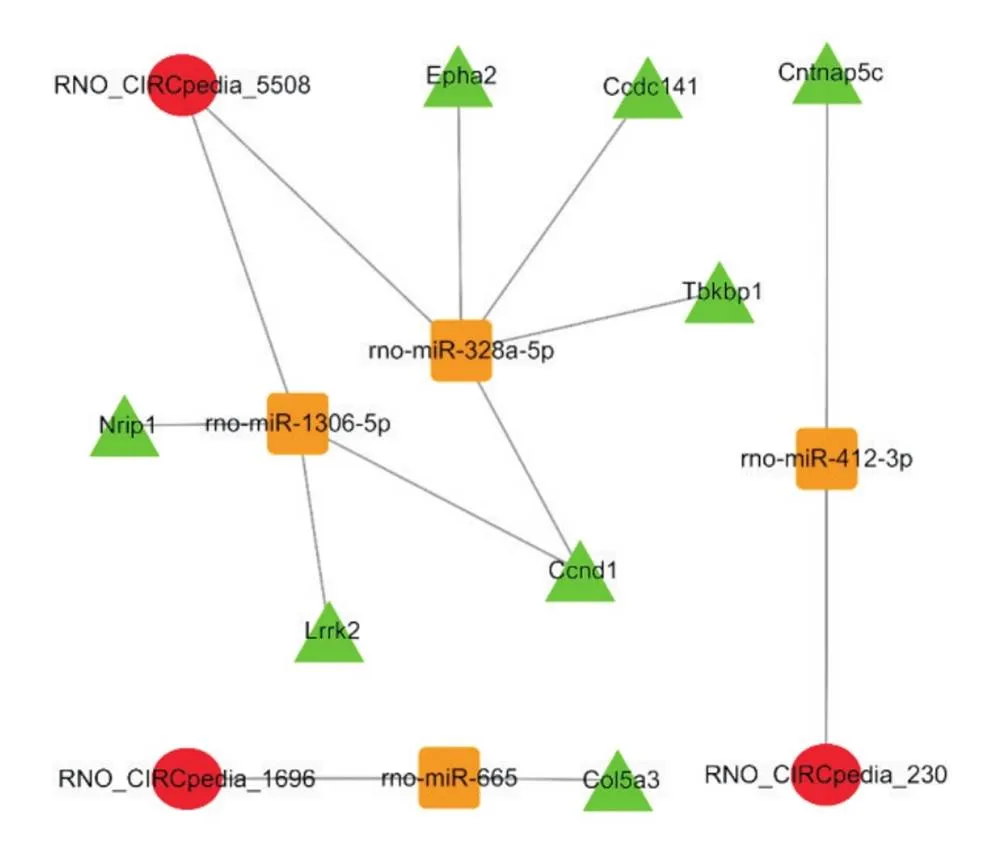
Figure 5.circRNA-miRNA-mRNA ternary transcriptional network. Circle: circRNA; square: miRNA; triangle: mRNA. Lines indicate a targeted regulatory relationship between the two genes.
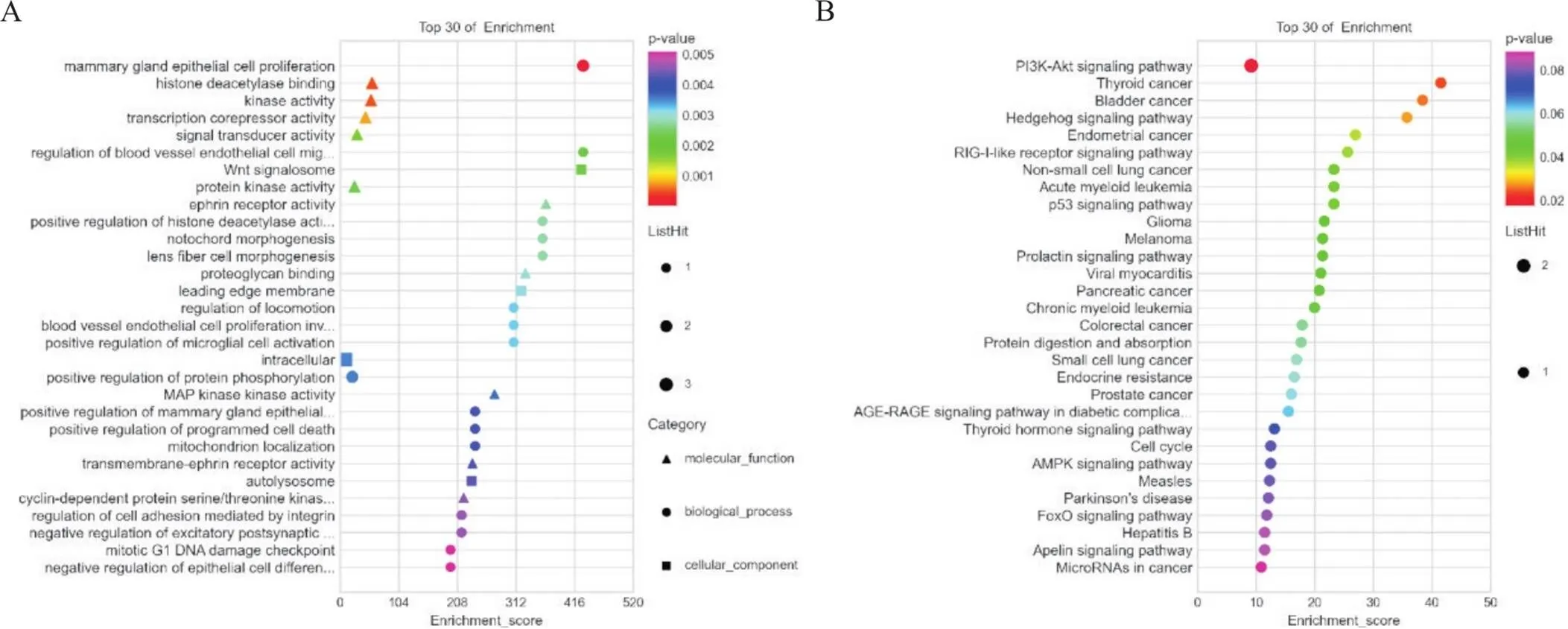
Figure 6.Enrichment analysis of mRNAs in the ternary transcriptional network. A: GO enrichment analysis of mRNAs; B: KEGG enrichment analysis of mRNAs.
讨论
circRNA最初被认为是异常RNA剪接或剪接错误的产物,生物学功能的潜力很小[11]。然而,随着高通量测序、基因芯片技术和生物信息学的迅速发展,越来越多的证据表明,circRNA在大脑内有着丰富且保守的表达[12],且参与了脑梗死、多系统萎缩和阿尔茨海默病等中枢神经系统疾病的病理过程[13-14]。此外,目前已有脑缺血后circRNA表达谱的研究[6],但circRNA在海马组织中的表达谱及其ceRNA机制研究尚未见报道。
首先,本研究识别出脑缺血大鼠海马组织中有21个差异表达的circRNAs,其中4个上调,17个下调。其中经过验证的RNO_CIRCpedia_9127亦被称为circ-ATXN1 (ataxin 1),研究表明circ-ATXN1可作为ceRNA海绵,形成circ-ATXN1/miR-526b-3p/基质金属蛋白酶2(matrix metalloproteinase 2, MMP2)轴,影响血管内皮生长因子A(vascular endothelial growth factor A, VEGFA)的表达,敲除-后MMP2与VEGFA表达下降,而circ-ATXN1的过表达逆转了MMP2和VEGFA的表达降低,并可促进血管新生[15]。此外,我们通过GO功能和KEGG通路富集分析研究了差异circRNA的潜在功能,GO功能富集分析主要为兴奋性突触后电位、脑形态发生、对组织重塑的调节、细胞突触、细胞连接、蛋白激酶C结合及蛋白质结合等。而KEGG通路富集主要为VEGF信号通路、NF-κB信号通路及Toll样受体信号通路等。VEGF信号通路是调控脑缺血后血管生成的经典通路,VEGF是胚胎发生和血管生成过程中血管形成的关键调节因子,研究表明VEGF可在细胞膜上与其受体血管内皮细胞生长因子受体(vascular endothelial cell growth factor receptor, VEGFR)结合,触发磷酸化细胞外信号调节激酶(phosphorylated extracellular signal-regulated kinases, p-ERK)和内皮型一氧化氮合酶(endothelial nitric oxide synthase, eNOS)等多个下游信号,从而促进血管生成[16]。NF-κB信号通路及Toll样受体信号通路亦被证实与脑缺血后的炎症级联反应关系密切[17]。另外,作为circRNA分子功能的效应器,本研究识别出脑缺血后共有836个差异表达的mRNAs,其中537个上调,299个下调。进一步的GO功能及KEGG通路富集分析发现,这些差异表达的mRNAs主要参与了对病毒的防御反应、吞噬作用、炎症反应、趋化因子活性、IgG结合及氧载体活性等生物功能,参与的信号通路为Toll样受体信号通路、TNF信号通路及JAK-STAT信号通路等。TNF信号通路及JAK-STAT信号通路均已被证明与脑缺血后的细胞存活、细胞增殖和细胞周期等密切相关[18-19]。大脑功能的正常运行与充足的糖氧供应密切相关,脑缺血后导致的过氧化应激、炎症反应及代谢失调等均会导致大脑神经元功能异常[20]。这些差异基因的富集结果与先前的报道一致[21],表明脑缺血的病理机制可能涉及氧化应激,微血管损伤及炎症反应等。
基于共表达关系及生物信息学预测,本研究进一步构建了由3个circRNAs、4个miRNAs及9个mRNAs组成的三元转录网络,以求进一步从整体的角度揭示脑缺血后海马组织的分子变化。相关研究亦证实了上述部分靶点的作用,如有研究观察到在脑梗死患者外周血中,miR-665表达显著升高[22],miR-665的过表达可减轻缺氧/复氧诱导的小胶质细胞凋亡和炎症反应[23];miR-1306可作为ceRNA轴的成员,抑制下游细胞凋亡相关分子的表达,发挥神经细胞保护作用[24-25];miR-328是评估脑卒中预后的重要生物标志物,其高表达会显著加重脑缺血再灌注损伤[26];细胞周期蛋白D1(cyclin D1, CCND1)属于细胞周期家族,能够促进细胞周期正向进行,与脑缺血后的神经再生密切相关[27]。进一步的富集分析显示该网络主要与PI3K-Akt信号通路及Hedgehog信号通路密切相关。PI3K-AKt信号通路是参与细胞增殖、分化及存活的重要通路,目前已有研究观察到脑缺血后PI3K-AKt信号通路被激活,能促进多种生长刺激因子的释放,诱导神经再生[28]。Hedgehog信号通路亦被证实与中枢神经系统的神经发生密切相关[29],有研究表明Hedgehog通路的激活能够保护脑缺血后的神经突触及神经干细胞,诱导神经再生并促进神经功能恢复[30-31]。此外,本研究显示该网络中RNO_CIRCpedia_5508和miR-328a-5p分别在circRNA类别及miRNA类别中节点度值最高,结合上述报道,推测RNO_CIRCpedia_5508可能作为ceRNA海绵靶向调控miR-328a-5p,影响CCND1的表达,进而调控PI3K-Akt信号通路及Hedgehog信号通路,影响脑缺血后海马组织的病理过程。但值得注意的是,针对该ceRNA轴,还需要进一步的验证。
综上所述,本研究探讨了脑缺血后海马组织circRNA及mRNA的表达谱,并初步构建了circRNA-miRNA-mRNA三元转录网络,这些差异基因与大脑缺血缺氧后的炎症反应、血管新生及神经发生等病理生理过程密切相关,还预测RNO_CIRCpedia_5508/miR-328a-5/CCND1可能是该网络中的重要ceRNA调控轴,可为后续研究提供了基础。
[1] Mendelson SJ, Prabhakaran S. Diagnosis and management of transient ischemic attack and acute ischemic stroke: a review[J]. JAMA, 2021, 325(11):1088-1098.
[2] Hanan M, Soreq H, Kadener S. CircRNAs in the brain[J]. RNA Biol, 2017, 14(8):1028-1034.
[3]刘燕芳, 逯丹, 徐安定. 脑血管病环状RNA的转录后调控及展望[J]. 中国病理生理杂志, 2018, 34(4):760-763.
Liu YF, Lu D, Xu AD. circRNAs: post-transcription regulation in cerebrovascular diseases[J]. Chin J Pathophysiol, 2018, 34(4):760-763.
[4] Zhang Z, He J, Wang B. Circular RNA circ_HECTD1 regulates cell injury after cerebral infarction by miR-27a-3p/FSTL1 axis[J]. Cell Cycle, 2021, 20(9):914-926.
[5] Yang B, Zang L, Cui J, et al. Circular RNA TTC3 regulates cerebral ischemia-reperfusion injury and neural stem cells by miR-372-3p/TLR4 axis in cerebral infarction[J]. Stem Cell Re Ther, 2021, 12(1):125.
[6] Li S, Chen L, Xu C, et al. Expression profile and bioinformatics analysis of circular RNAs in acute ischemic stroke in a South Chinese Han population[J]. Sci Rep, 2020, 10(1):10138.
[7]马晓娇, 承欧梅, 校欢, 等. 急性脑缺血通过激活EphB2/ephrin-B1/NMDA受体信号通路促进小鼠海马神经发生[J]. 中国病理生理杂志, 2020, 36(8):1389-1395.
Ma XJ, Cheng OM, Xiao H, et al. Acute cerebral ischemia activates EphB2/ephrin-B1/NMDA receptor signaling pathway to promote hippocampal neurogenesis in mice[J]. Chin J Pathophysiol, 2020, 36(8):1389-1395.
[8] Longa EZ, Weinstein PR, Carlson S, et al. Reversible middle cerebral artery occlusion without craniectomy in rats[J]. Stroke, 1989, 20(1):84-91.
[9] Levy A, Bercovich-Kinori A, Alexandrovich AG, et al. CD38 facilitates recovery from traumatic brain injury[J]. J Neurotrauma, 2009, 26(9):1521-1533.
[10] Zhang Z, Yue L, Wang Y, et al. A circRNA-miRNA-mRNA network plays a role in the protective effect of diosgenin on alveolar bone loss in ovariectomized rats[J]. BMC Complement Med Ther, 2020, 20(1):220.
[11] Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: from the bench to the clinic[J]. Pharmacol Ther, 2018, 187:31-44.
[12] Piwecka M, Glažar P, Hernandez-Miranda LR, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function[J]. Science, 2017, 357(6357):eaam8526.
[13] Chen BJ, Mills JD, Takenaka K, et al. Characterization of circular RNAs landscape in multiple system atrophy brain[J]. J Neurochem, 2016,139(3):485-496.
[14] Li Y, Lv Z, Zhang J, et al. Profiling of differentially expressed circular RNAs in peripheral blood mononuclear cells from Alzheimer's disease patients[J]. Metab Brain Dis, 2020, 35(1):201-213.
[15] Liu X, Shen S, Zhu L, et al. SRSF10 inhibits biogenesis of circ-ATXN1 to regulate glioma angiogenesis via miR-526b-3p/MMP2 pathway[J]. J Exp Clin Canc Res, 2020, 39(1):121.
[16] Hatakeyama M, Ninomiya I, Kanazawa M. Angiogenesis and neuronal remodeling after ischemic stroke[J]. Neural Regen Res, 2020, 15(1):16-19.
[17] Eltzschig HK, Eckle T. Ischemia and reperfusion: from mechanism to translation[J]. Nat Med, 2011, 17(11):1391-1401.
[18] Wu Y, Xu J, Xu J, et al. Study on the mechanism of JAK2/STAT3 signaling pathway-mediated inflammatory reaction after cerebral ischemia[J]. Mol Med Rep, 2018, 17(4):5007-5012.
[19] Chen AQ, Fang Z, Chen XL, et al. Microglia-derived TNF-α mediates endothelial necroptosis aggravating blood brain-barrier disruption after ischemic stroke[J]. Cell Death Dis, 2019, 10(7):487.
[20] Gong C, Zhou X, Lai S, et al. Long noncoding RNA/Circular RNA-miRNA-mRNA axes in ischemia-reperfusion injury[J]. Biomed Res Int, 2020, 2020:8838524.
[21] Huang Q, Li C, Xia N, et al. Neurochemical changes in unilateral cerebral hemisphere during the subacute stage of focal cerebral ischemia-reperfusion in rats: anH-1 magnetic resonance spectroscopy study[J]. Brain Res, 2018, 1684:67-74.
[22] Greco R, Demartini C, Zanaboni A, et al. Characterization of CB2 receptor expression in peripheral blood monocytes of acute ischemic stroke patients[J]. Transl Stroke Res, 2021, 12(4):550-558.
[23] Zhang X, Feng Y, Li J, et al. MicroRNA-665-3p attenuates oxygen-glucose deprivation-evoked microglial cell apoptosis and inflammatory response by inhibiting NF-κB signaling via targeting TRIM8[J]. Int Immunopharmacol, 2020, 85:106650.
[24] Chen X, Li C, Li J, et al. Upregulation of miR-1306-5p decreases cerebral ischemia/reperfusion injury by targeting BIK[J]. Biosci Biotech Bioch, 2019, 83(12):2230-2237.
[25] Huang Y, Deng L, Zeng L, et al. Silencing of H19 alleviates oxygen-glucose deprivation/reoxygenation-triggered injury through the regulation of the miR-1306-5p/BCL2L13 axis[J]. Metab Brain Dis, 2021, 36(8):2461-2472.
[26] Wang S, Jun J, Cong L, et al. miR-328-3p, a predictor of stroke, aggravates the cerebral ischemia-reperfusion injury[J]. Int J Gen Med, 2021, 14:2367-2376.
[27] Lee Hc, Ahn SM, Pak ME, et al. Positive effects of α-asarone on transplanted neural progenitor cells in a murine model of ischemic stroke[J]. Phytomedicine, 2018, 51:151-161.
[28] Beker MC, Caglayan B, Caglayan AB, et al. Interaction of melatonin and Bmal1 in the regulation of PI3K/AKT pathway components and cellular survival[J]. Sci Rep, 2019, 9(1):19082.
[29] Wilson NH, Stoeckli ET. Sonic Hedgehog regulates Wnt activity during neural circuit formation[J]. Vitam Horm, 2012, 88:173-209.
[30] Yu P, Wang L, Tang F, et al. Resveratrol-mediated neurorestoration after cerebral ischemic injury - Sonic Hedgehog signaling pathway[J]. Life Sci, 2021, 280:119715.
[31] Yin S, Bai X, Xin D, et al. Neuroprotective effects of the sonic hedgehog signaling pathway in ischemic injury through promotion of synaptic and neuronal health[J]. Neural Plast, 2020, 2021:9762592.
Analysis of circRNA-miRNA-mRNA ternary transcriptional network in hippocampus of a rat model of middle cerebral artery occlusion
CHEN Bo-wei1, TANG Rong-mei1, XU Ya-qian2, YI Jian2, LIU Bai-yan1△
(1,410208,;2,410007,)
To investigate the differentially expressed (DE) circular RNAs (circRNAs) and messenger RNAs (mRNAs) in hippocampus of cerebral ischemia model rats, and to construct circRNA-microRNA (miRNA)-mRNA ternary transcription network.Male SD rats were randomly divided into control group and model group with 8 rats each, of which 8 rats in each group were used for neurobehavioral scores, 4 rats for Nissl staining, 4 rats for microarray in control group, and 3 rats for microarray in model group. Middle cerebral artery occlusion (MCAO) was used to replicate cerebral ischemia model in model group. Seven days later, neurobehavioral scoring and Nissl staining were performed to verify the success of modeling. Agilent competing endogenous RNA (ceRNA) microarray was used to screen DE circRNAs and mRNAs. The main biological processes involved in DE genes were analyzed by GO function and KEGG pathways, and the results of gene microarray were validated by RT-qPCR. Finally, the circRNA-miRNA-mRNA transcription network was constructed.Compared with control group, the neurobehavioral scores of MCAO rats increased significantly (<0.01), and hippocampal neuronal cell damage appeared. Furthermore, 18 DE circRNAs and 836 DE mRNAs were detected in hippocampal tissues of rats with MCAO by ceRNA microarray (FC≥1.5,<0.05). RT-qPCR showed that RNO_CIRCpedia_136, RNO_CIRCpedia_5686, RNO_CIRCpedia_9127 and alpha hemoglobin stabilizing protein (Ahsp), selected randomly for validation, were down-regulated in model group (<0.05 or<0.01). The expression of secreted phosphoprotein 1 (Spp1) and macrophage scavenger receptor 1 (Msr1) was up-regulated in model group (<0.01), which was consistent with the change trend of gene microarray results. A ternary transcription network consisting of 3 circRNAs, 4 miRNAs and 9 mRNAs was constructed. Bioinformatic analysis showed that these DE genes may regulate cerebral ischemia injury through vascular endothelial growth factor signaling pathway, Toll-like receptor signaling pathway, Janus kinase-signal transducer and activator of transcription signaling pathway, phosphatidylinositol 3-kinase-protein kinase B signaling pathway and Hedgehog signaling pathway.There are DE circRNAs and mRNAs in the hippocampus of cerebral ischemia rats, and these DE genes may regulate cerebral ischemia injury through circRNA-miRNA-mRNA ternary transcription network system.
Middle cerebral artery occlusion; Gene microarray; Circular RNA; Competing endogenous RNA
R743; R363.2
A
10.3969/j.issn.1000-4718.2022.03.012
1000-4718(2022)03-0479-08
2021-10-11
2021-12-10
[基金项目]国家自然科学基金资助项目(No. 82074251);湖南中医药大学中医学一流学科开放基金资助项目(No. 2021ZYX38); 湖南中医药大学研究生创新课题(No. 2021CX20)
Tel: 0731-88536925; E-mail: liubaiyan@126.com
(责任编辑:林白霜,宋延君)

