三维荧光技术结合化学计量学检测青贮微生物生长量
张微微,张 静,孟 德,吕日琴,顾海洋,孙艳辉
三维荧光技术结合化学计量学检测青贮微生物生长量
张微微,张 静,孟 德,吕日琴,顾海洋,孙艳辉※
(滁州学院生物与食品工程学院,滁州 239000)
青贮中微生物的数量是影响青贮料质量的关键因素。为了高效监控青贮微生物的生长情况,该研究以青贮乳酸菌、乙酸菌和丁梭菌等作为指示菌株,考察菌株生长过程0、2、4、8、12、24和48 h共7个不同时间点共105个样本的三维荧光光谱、微生物菌落数和吸光值,通过平行因子法和BP神经网络等化学计量学建立微生物生长量预测模型。三维荧光光谱图显示指示菌株有2个荧光峰,波峰分别在225和275 nm附近,主要是微生物内源荧光酪氨酸和色氨酸类物质。随着微生物培养时间的增加,荧光强度逐渐增强,荧光波峰位置红移,峰宽增加。利用平行因子法对三维荧光光谱进行降维,获取组分数为6,特征波长差Δ为50 nm时,微生物生长荧光信息差异显著。以该二维光谱数据作为BP神经网络模型输入值,分别以微生物菌落数和吸光值作为模型输出值,对不同检测方法的微生物生长量进行建模训练。试验结果表明两种不同方法对应的训练集、验证集、测试集模型决定系数2均接近1.0,均方误差均很小,说明该模型能较好预测微生物生长量。研究结果显示三维荧光光谱技术结合化学计量学对青贮中微生物生长量监测是可行的,项目为快速判定青贮发酵阶段提供了一种新的技术途径。
三维荧光;青贮;微生物生长量;化学计量学;平行因子分析
0 引 言
青贮是一种能降低饲料成本、提高适口性同时还可以减少环境污染的储藏技术[1]。青贮技术主要利用乳酸菌发酵产酸使得有害微生物处于稳定的被抑制的状态,从而达到青绿饲料进行长期保存的目的[2-3],发酵过程中伴随着一系列微生物的繁殖代谢[4-5]。青贮过程中微生物生长对青贮品质起着决定性的作用,尤其有害微生物如梭菌、乙酸菌、酵母菌等的增殖,不仅直接影响青贮品质,浪费作物资源,还会对反刍动物生产造成威胁[6-8]。因此实时监控青贮微生物的生长至关重要。青贮微生物的测定主要是检测发酵过程乳酸菌、梭菌、酵母菌等菌落数量。目前,实验室中生长检测方法以平板计数法和比浊法使用最为广泛,此类方法具有步骤繁琐、耗时长和响应速率慢等缺点,不能及时准确表征青贮微生物生长状态[9-10],导致不良青贮发酵。因此,探究一种高效、便捷、实时监测微生物生长量的方法成为检测新需求。
荧光光谱技术作为一种新兴的无损检测技术,具有低能、高效、快捷等优点,在食品成分检测、掺假[11-12]与土壤、水环境有机质研究[13-15]等方面应用前景广阔。微生物体内固有的氨基酸,如色氨酸、酪氨酸等物质在紫外或可见光的激发下,会产生出特征的荧光反射[16-17],使得荧光光谱法检测微生物成为可能,目前已有相关研究。但这些内源氨基酸物质部分存在荧光峰重叠现象[18],区别与常规荧光光谱技术,三维荧光光谱技术具有较高的选择性。Dartnell等[19]基于对生物体内色氨酸荧光光谱的检测,开发了一个手持式微生物快速检测装备,可实现临床医疗保健环境或设备是否受细菌污染的快速甄别。许瑞等[20]指出利用三维荧光光谱技术结合平行因子法可以实时在线监测微生物净化黑臭水的治理情况。宋晓康等[21]研究指出三维荧光光谱结合平行因子分析法能够快速测定细胞培养基中多种代谢类荧光组分的含量,在细胞能量和物质代谢检测中具有良好的应用前景。为了进一步增强荧光光谱技术对目标物质预测精确度,充分发挥机器学习技术对利用荧光技术的定量分析起到了较好的支撑作用。BP神经网络属于机器学习领域中的一种技术,因其强大的非线性分析能力被广泛应用于物质定量分析[22-24]。
综上所述,三维荧光结合化学计量学方法是一种强有力的分析策略。本研究利用平行因子法对不同生长时间点的微生物三维荧光光谱图进行解析,获取特征光谱,联合BP神经网络建立微生物生长量预测模型,并使用模型进行样本预测,验证方法准确性。该项目的开展为快速判别青贮发酵阶段提供参考。
1 材料与方法
1.1 材料和仪器
青贮乳酸菌、乙酸菌、丁梭菌,本学院食品学院微生物实验室青贮料筛选备用;MRS培养基、MRS肉汤培养基、丁梭菌增殖培养基、醋酸菌基础培养基,青岛海博生物技术有限公司;无水乙醇、生理盐水,国药集团化学试剂(上海)有限公司。
UV-5500PC紫外可见分光光度计,上海元析仪器有限公司;Cary Eclipse荧光分光光度计,美国瓦里安有限公司;H1850台式高速离心机,湖南湘仪离心机仪器有限公司;DHP-9272B电热恒温培养箱,上海一恒科学仪器有限公司。
1.2 试验方法
1.2.1 样品制备
乳酸菌、丁梭菌和乙酸菌在对应液体培养中置于37 ℃,180 r/min恒温摇床培养至生长后期阶段。在生长过程中(0、2、4、8、12、24、48 h)定点无菌取样,每个时间点设置5个平行,用于光谱数据采集、吸光值测定(OD600)和微生物平板培养计数。
1.2.2 微生物光谱信息采集
参考Dartnell等[19]研究对本研究中菌悬液制备及光谱方法稍做改变,具体方法如下:
菌悬液制备:在0、2、4、8、12、24、48 h定点采集的样品,取5 mL各时间点菌液放入到10 mL离心管中,用离心机3 000 r/min离心10 min,无菌吸管吸除上层液体,加入5 mL生理盐水,获得菌悬液。菌悬液用于荧光光谱检测。
三维荧光光谱扫描条件:激发波长(Ex)为200~600 nm,增量为1 nm,通过同时扫描激发单色仪和发射单色仪,在10~180 nm范围内以10 nm恒定的波长间隔(Δ),扫描速度为1 200 nm/min,采集每个样品的同步荧光光谱。所有样品光谱采集记录3次并保存光谱数据,绘制荧光强度、Δ、激发波长三维图谱。
1.2.3 样品吸光值与菌落数测定
对采集荧光光谱数据的样品同时进行微生物吸光值和菌落总数测定。以空白样为对照,利用紫外分光光度计测定同一培养时间点的每一菌悬液的OD600值。将每一菌悬液稀释到适宜水平采用倾注法平行制作3个平板,倒置于37 ℃电热恒温培养箱,培养24 h后选取可计数范围稀释度进行平板菌落计数,并依据稀释倍数换算出菌液浓度,参照GB 4789.2—2016《食品安全国家标准食品微生物学检测菌落总数测定》[25]。
1.2.4 光谱数据处理方法
1)平行因子法(Parallel Factor analysis,PARAFAC)
使用PARAFAC分析时,必需预先创建样品数据集。设定Ex数为,Δ数为,分别采集个多组分样本的荧光光谱图,获得三维荧光光谱数据,多个样本数据次序叠加,获得××的三维响应矩阵,该法将分解为3个载荷矩阵、、,数学表达式如下
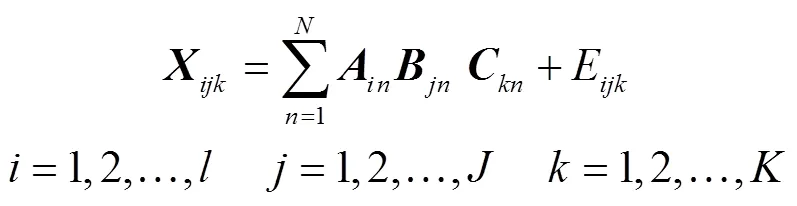
式(1)中X为三维数据矩阵的一个元素;A、B、C分别为中的元素;E为误差矩阵;代表模型因子数,也为对应模型的最佳组分数。
平行因子分析法求解过程是确定建模的组分数,对矩阵、和,采用交替最小二乘方法[26],且要残差平方和最小,逐次迭代重复直至收敛。该法在MATLAB 2014a中的DOMFluor工具箱环境下运行。
2)BP神经网络分析法
BP神经网络属于机器学习技术中的人工神经网络技术,在数据分析和处理中被广泛应用[27-28]。BP神经网络层主要包括输入层、隐藏层与输出层,使用BP神经网络建立拟合关系中,神经拟合应用程序将帮助选择数据,随机获取试验数据和目标数据,即输入数据和输出数据。按照比例划分训练集、校正集、验证集,通过创建和训练一个网格,使用Levenberg-Marquardt反向传播算法(trainlm)进行训练,并评估其性能使用均方误差和回归分析,直至选定高拟合能力模型,再进行数据仿真操作[29-30]。该方法利用神经网络拟合Neural Net Fitting工具箱在MATLAB 2014a环境下运行。
2 结果与讨论
2.1 微生物三维荧光光谱
青贮丁梭菌、乳酸菌和乙酸菌在0和24 h的菌悬液的原始三维荧光图谱如图1显示。细菌在不同时间点组分变化存在显著差异。微生物0和24 h在200~300 nm间有2个特征荧光峰,与Dartnell等[19]结果相同。第一个在200~250 nm(峰值在225 nm附近);第二个峰在250~300 nm(峰值在275 nm附近),此二峰的产生主要与微生物体内固有的类蛋白质有关,分别对应酪氨酸和色氨酸类物质[31-33]。荧光光谱颜色的鲜艳程度与荧光强度成正向相关。微生物培养24 h后,2个特征荧光峰的荧光强度显著增强,最强荧光峰位置稍向长波方向移动,峰宽变大,此现象主要是微生物生长过程菌体大量繁殖,体内固有物质增多[17]。结果表明三维荧光光谱技术可以定性反馈不同时期微生物生长量,与平板计数法和比浊法测定结果呈现一致。
2.2 微生物光谱信息降维分析
PARAFAC法是处理多维多向数据集的有力工具,主要通过交替最小二乘法确定模型因子数实现三维荧光光谱矩阵的有效分解,提取微生物特征荧光光谱信息,解析样本显著信息[34]。图2中误差平方和大小明显显示组分6和组分7为PARAFAC中较适合的成分数,基于模型计算过拟合现象问题考虑,选定组分6。当组分数为6时,样本不同Δ的载荷值见图3。Δ的载荷值越高,说明该波长下对应样本间的差异越显著,区分效果越好[26]。由图可知,产生样本间差异显著的最高载荷值的Δ为50 nm,该波长为微生物特征荧光光谱。

图1 乙酸菌、丁梭菌和乳酸菌0和24 h三维荧光光谱图
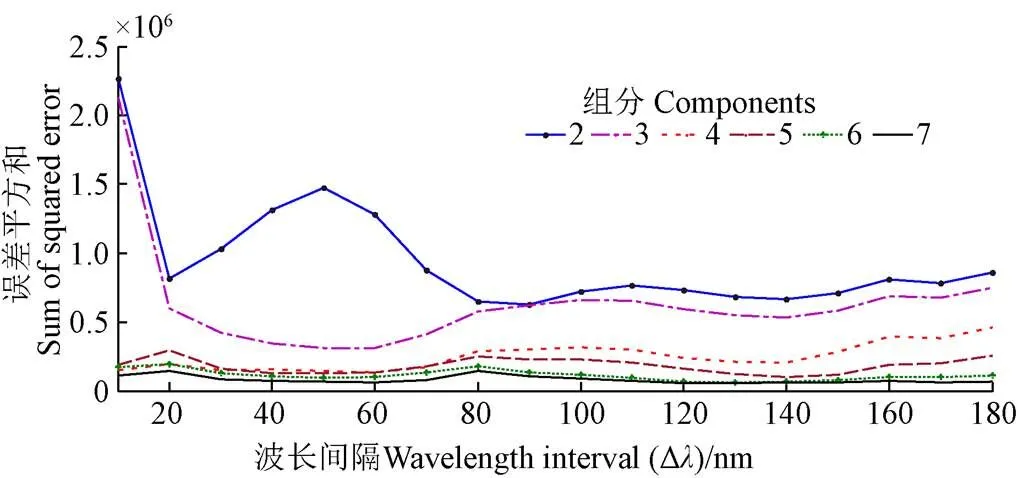
图2 不同组分数误差平方和
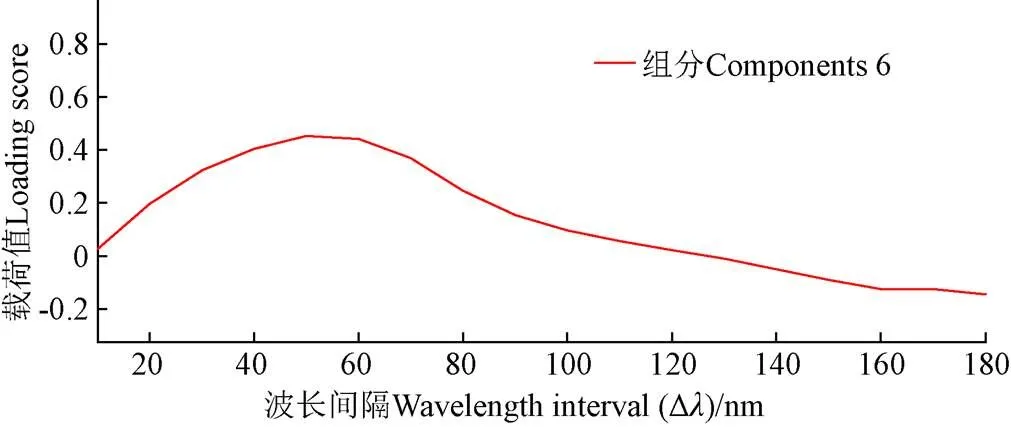
图3 不同波长间隔(Δλ)载荷值
Δ为50 nm对应特征波长下的微生物生长二维荧光光谱如图4所示。图4显示微生物在生长过程中在250~300 nm呈现特征荧光峰(峰值275 nm附近),且随着培养时间增长,总体荧光强度呈现显著增强,峰宽变大。该现象与三维荧光光谱现象一致,说明平行因子分析法能较好解析三维荧光光谱,且方法是适当的。在310~360、370~390 nm处出现2个微弱的荧光峰(峰值340和380 nm附近),研究发现是微生物代谢产物或者某种带有荧光基团的酸类物质[35-36],且380 nm附近的自然荧光峰的强度与培养时间呈现正相关。由此可知,平行因子分析获取的二维荧光光谱能更多的获取荧光组分信息[16],更准确的说明微生物生长过程物质的变化。
2.3 微生物生长量判定模型建立
基于判定利用三维荧光光谱预测微生物生长量的合理性,项目利用PARAFAC法选取菌株对应Δ为50 nm波长光谱数据作为BP神经网络模型输入层神经元,比浊法和平板计数法的结果分别作为模型输出层神经元,对青贮微生物生长量进行数据建模训练[37]。以随机抽取的方式,所有的数据按照60∶20∶20分别作为训练集、验证集与测试集,隐含层为1,隐含层神经元数量为10,得到BP神经网络预测模型,模型参数见表1。表1显示,这两种方法与特征波长荧光强度通过BP神经网络拟合,获得决定系数(2)值> 0.99(接近1),均方误差(Mean Square Error,MSE)值均很小,表明该方法建立模型相关性较好。

图4 不同微生物生长时间的二维荧光光谱
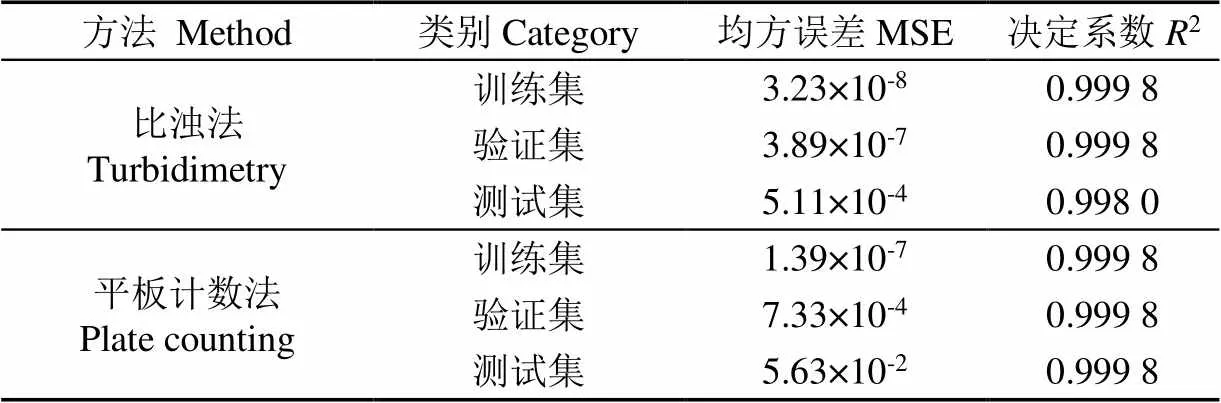
表1 微生物生长量智能预测模型相关性分析
为了更好检验建立模型对样本的预测能力,重新采集青贮乳酸菌、丁酸菌、乙酸菌、酵母菌等共91个样本,并使用建立模型进行预测,不同方法BP神经网络预测结果如图5所示。由图5可清晰看到BP神经网络具有较高的拟合能力。综上可知,三维荧光光谱法结合平行因子及BP神经网络法监测青贮过程中微生物的生长情况是可行的,且操作便捷,数据可靠。
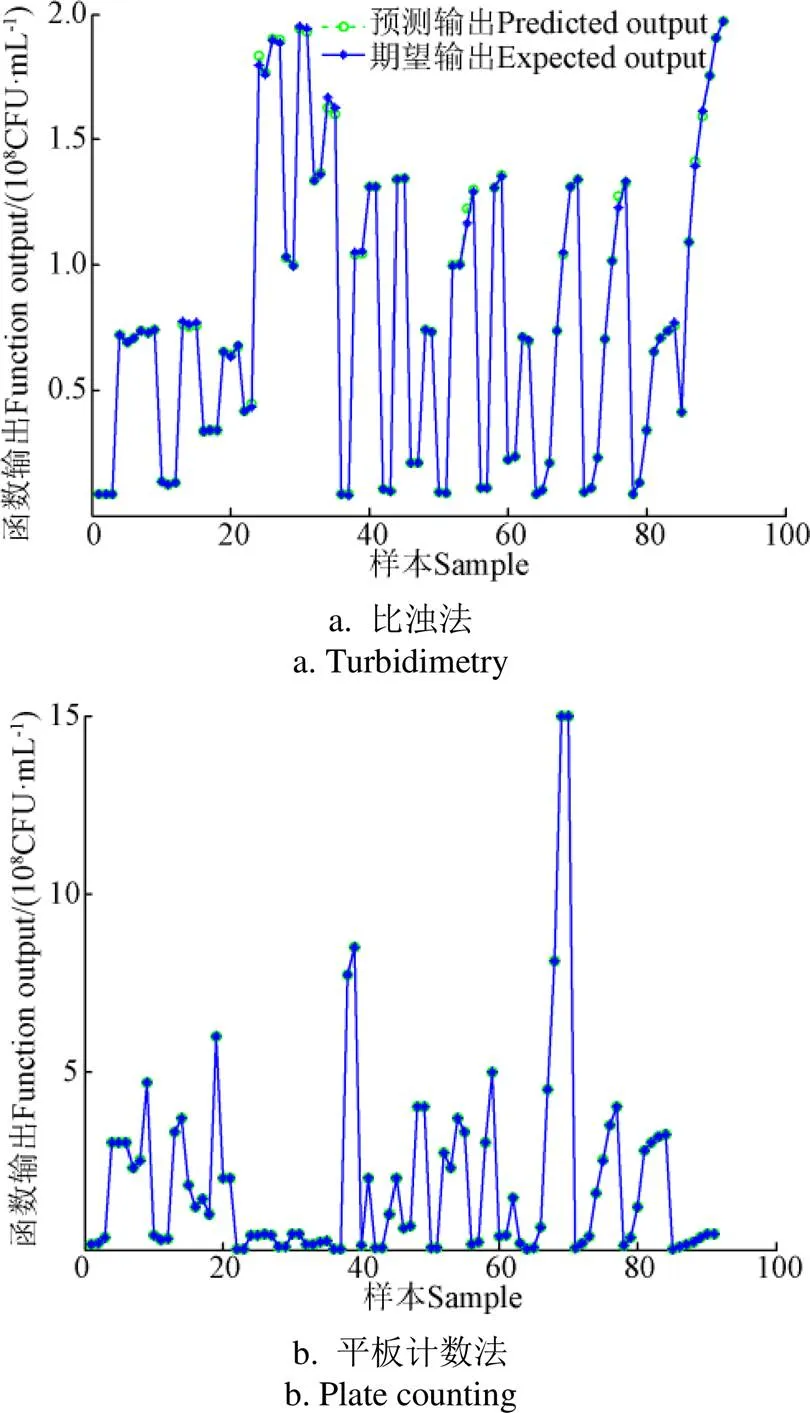
图5 不同方法BP神经网络预测结果
3 结 论
本文以青贮细菌乳酸菌、乙酸菌、丁梭菌为研究对象,采集不同生长时间点的微生物三维荧光光谱数据,利用比浊法和平板计数法测定微生物生长量,基于平行因子法和BP神经网络方法构建预测模型。结果表明:
1)利用三维同步荧光光谱测定3种指示菌株在225和275 nm左右呈现特征高强度波峰,主要是类蛋白物质相关,分别为酪氨酸和色氨酸。
2)利用平行因子法解析三维同步荧光光谱数据,得到菌株的特征波长差Δ值(50 nm),以此对应光谱数据通过BP神经网络建立预测模型,通过相关系数和均方误差都说明BP神经网络具有较强的拟合能力,可快速预测微生物生长量,判别微生物生长状态。
本研究利用三维荧光光谱结合化学计量学建立微生物生长量预测模型,为青贮发酵微生物生长量检测提供了新思路与方法。该模型与传统方法相比较大幅度降低劳动时间,提高了操作效率,但针对于青贮质量评定,需要更为特异性组分指标进行相关解析。下一步工作计划可以扩充青贮品质指标,在数据分析和模型构建部分引入机器学习模型进行进一步的信息挖掘和提取,从而增强模型的预测能力,为提高青贮质量提供更为直观的针对性策略。
[1] 苏嘉琪,辛杭书,张广宁,等. 国内外青贮饲料原料来源、品质评价及影响因素的研究进展[J/OL]. 动物营养学报:1-10[2022-10-08]. http: //kns. cnki. net/kcms/detail/11. 5461. S. 20220926. 1830. 012. html
Su Jiaqi, Xin Hangshu, Zhang Guangning, et al. Research progress on sources, quality evaluation and influencing factors of silages feed at home and abroad[J]. Chinese Journal of Animal Nutrition: 1-10[2022-10-08]. http: //kns. cnki. net/kcms/detail/11. 5461. S. 20220926. 1830. 012. html (in Chinese with English abstract)
[2] Gollop N, Zakin V, Weinberg Z G. Antibacterial activity of lactic acid bacteria included in inoculants for silage and in silages treated with these inoculants[J]. Journal of Applied Microbiology, 2005, 98(3): 662-666.
[3] Park R S, Stronge M D. Silage production and utilisation[C]. Belfast, Northern,Ireland: Wageningen academic, 2005.
[4] 黄峰,张露,周波,等. 青贮微生物及其对青贮饲料有氧稳定性影响的研究进展[J]. 动物营养学报,2019,31(1):82-89.
Huang Feng, Zhang Lu, Zhou Bo, et al. Research process in silage microorganism and its effect on silage aerobic stability[J]. Chinese Journal of Animal nutrition, 2019, 31(1): 82-89. (in Chinese with English abstract)
[5] Li M H, Shan G L, Zhou H Y, et al. CO2production, dissolution and pressure dynamics during silage production: Multi-sensor-based insight into parameter interactions[J]. Scientific Reports, 2017, 7(1): 181-188.
[6] Ogunade I M, Jiang Y, Kim D H, et al. Fate ofO157:H7and bacterial diversity in corn silage contaminated with the pathogen and treated with chemical or microbial additives[J]. Journal of Dairy Science, 2017, 100(3): 1780-1794.
[7] 张适,常杰,胡宗福,等. 青贮饲料有害微生物及其抑制措施[J]. 动物营养学报,2017,29(12):4308-4314.
Zhang Shi, Chang Jie, Hu Zongfu, et al. Harmful microorganism in silage and their suppression measures[J]. Chinese Journal of Animal nutrition, 2017, 29(12): 4308-4314. (in Chinese with English abstract)
[8] Wambacq E, Vanhoutie I, Audenaert K, et al. Occurrence, prevention and remediation of toxigenic fungi and mycotoxins in silage: A review[J]. Journal of the Science of Food and Agriculture, 2016, 96(7): 2284-2302.
[9] Shao J, Xiang J D, Axner O, et al. Wavelength-modulated tunable diode-laser absorption spectrometry for real-time monitoring of microbial growth[J]. Applied Optics, 2016, 55(9): 2339-2345.
[10] 项金冬. 基于光谱技术的微生物生长检测研究[D]. 金华:浙江师范大学,2016.
Xiang Jindong. Research on Microbial Growth Detection Based on Spectral Technique[D]. Jinhua: Zhejiang Normal University, 2016. (in Chinese with English abstract)
[11] 李杨,孙禹凡,赵城彬,等. 体外模拟消化过程中大豆分离蛋白拉曼光谱和荧光光谱分析[J]. 中国食品学报,2019,19(2):266-272.
Li Yang, Sun Yufan, Zhao Chengbin, et al. Analysis of raman spectroscopy and fluorescence spectroscopy for soy protein isolate during vitro simulated digestion process[J]. Journal of Chinese Institute of Food Science and Technology, 2019, 19(2): 266-272. (in Chinese with English abstract)
[12] Olgun C, Cihat I N, Zeki D M. Rapid detection of adulteration of milks from different species using Fourier Transform Infrared Spectroscopy (FTIR)[J]. The Journal of Dairy Research, 2018, 85(2): 222-225.
[13] Chai L W, Huang M, Fan H, et al. Urbanization altered regional soil organic matter quantity and quality: Insight from excitation emission matrix (EEM) and parallel factor analysis (PARAFAC)[J]. Chemosphere, 2019(220): 249-258.
[14] Liu D P, Yu H B, Gao H J, et al. Applying synchronous fluorescence and UV-vis spectra combined with two-dimensional correlation to characterize structural composition of DOM from urban black and stinky rivers[J]. Environmental Science and Pollution Research, 2021, 28(15): 19400-19411.
[15] 陈营营,郑昭佩,杨芳,等. 同步荧光结合主成分与二维相关研究盐碱性土溶解性有机质组成与结构特征[J]. 光谱学与光谱分析,2020,40(2):489-493.
Chen Yingying, Zheng Zhaopei, Yang Fang, et al. The composition and structure of dissolved organic matter in saline soil were studied by synchronous fluorescence spectroscopy combined with principal components and two-dimensional correlation[J]. Spectroscopy and Spectral Analysis, 2020, 40(2): 489-493. (in Chinese with English abstract)
[16] 苏良湖,陈梅,孙旭,等. 谷类秸秆接种瘤胃液的厌氧消化性能和三维荧光光谱特征[J]. 生态与农村环境学报,2018,34(11):1034-1041.
Su Lianghu, Chen Mei, Sun Xu, et al. The anaerobic digestion performance of cereal straw inoculated with rumen fluid and its three-dimensional excitation emission matrix fluorescence spectroscopic characteristics[J]. Journal of Ecology and Rural Environment, 2018, 34(11): 1034-1041. (in Chinese with English abstract)
[17] 刘璐. 微生物代谢产物的三维荧光光谱分析[J]. 化学工程与装备,2010(2):144-146,137.
Liu Lu. Three-dimensional fluorescence spectrometric analysis of microbial metabolism[J]. Chemical engineering and equipment, 2010(2): 144-146, 137 (in Chinese with English abstract)
[18] 张为,曹玉珍,刘振宇,等. 平行因子算法用于酪氨酸、色氨酸和苯丙氨酸的同时定性与定量测定[J]. 化学通报,2002(6):418-421.
Zhang Wei, Cao Yuzhen, Liu Zhenyu, et al. Parallel factor algorithm for simultaneous qualitative and quantitative determination of tyrosine, Tryptophan and L-Phenylalanine[J]. Chemistry, 2002(6): 418-421. (in Chinese with English abstract)
[19] Dartnell L R, Roberts T A, Moore G, et al. Fluorescence characterization of clinically-important bacteria[J]. Plos One, 2013, 8(9): 1-13.
[20] 许瑞,王胜楠,陈乐,等. 基于三维荧光光谱技术解析不同微生物法净化黑臭水体的效果[J]. 环境工程学报,2020,14(1):123-132.
Xu Rui, Wang Shengnan, Chen Le, et al. Effect of different microbial methods on purifying black-odor water based on three-dimensional fluorescence spectroscopy[J]. Chinese Journal of Environmental Engineering, 2020, 14(1): 123-132. (in Chinese with English abstract)
[21] 宋晓康,赵强,张元志,等. 利用三维荧光光谱与平行因子分析法测定细胞培养基中多类代谢成分的含量[J]. 中国激光,2022,49(9):28-39.
Song Xiaokang, Zhao Qiang, Zhang Yuanzhi, et al. Utilizing three dimensional fluorescence spectra and parallel factor analysis algorithm to quantify the concentration of multiple metabolic fluorophores in the cell culture medium[J]. Chinese Journal of Lasers, 2022, 49(9): 28-39. (in Chinese with English abstract)
[22] 王书涛,陈东营,侯培国,等. 基于荧光光谱技术和GA-BP神经网络的对羟基苯甲酸甲酯钠含量的测定[J]. 光谱学与光谱分析,2015,35(6):1606-1610.
Wang Shutao, Chen Dongying, Hou Peiguo, et al. Determination of the sodium methylparaben content based on spectrum fluorescence spectral technology and GA-BP neural network[J]. Spectroscopy and Spectral Analysis, 2015, 35(6): 1606-1610. (in Chinese with English abstract)
[23] 陈东营. 基于优化BP神经网络的防腐剂荧光检测技术研究[D]. 秦皇岛:燕山大学,2016.
Chen Dongying. Theoretical Study on Fluorescence Detection Technology of Preservative Based on Optimized BP Neural Network[D]. Qinhuangdao: Yanshan University, 2016. (in Chinese with English abstract)
[24] 张艳. BP神经网络结合FANWE与三维荧光光谱法测量美白面膜中的荧光增白剂[D]. 秦皇岛:燕山大学,2020.
Zhang Yan. BP Neural Network Combined with FANWE and Three-dimensional Fluorescence Spectroscopy to Measure the Fluorescent Whitening Agent in Whitening Mask[D]. Qinhuangdao: Yanshan University, 2020. (in Chinese with English abstract)
[25] 国家食品药品监督管理总局,国家安全和计划生育委员会. 食品安全国家标准食品微生物学检测菌落总数测定:GB 4789. 2-2016[S]. 北京:中国标准出版社,2016:1-7.
[26] Murphy K R, Stedmon C A, Graeber D, et al. Fluorescence spectroscopy and multi-way techniques PARAFAC[J]. Anal Methods, 2013, 5: 6557-6566.
[27] 张驰,郭媛,黎明. 人工神经网络模型发展及应用综述[J]. 计算机工程与应用,2021,57(11):57-69.
Zhang Chi, Guo Yuan, Li Ming. Review of development and application of artificial neural network models[J]. Computer Engineering and Application, 2021, 57(11): 57-69. (in Chinese with English abstract)
[28] 孙少杰,吴门新,庄立伟,等. 基于CNN卷积神经网络和BP神经网络的冬小麦县级产量预测[J]. 农业工程学报,2022,38(11):151-160.
Sun Shaojie, Wu Menxin, Zhuang Liwei, et al. Forecasting winter wheat yield at county level using CNN and BP neural networks[J]. Transactions of the Chinese Society of Agricultural Engineering (Transactions of the CSAE), 2022, 38(11): 151-160. (in Chinese with English abstract)
[29] Sen J, Das A. K. Artificial neural network model for forecasting the stock price of indian IT company[J]. Advances in Intelligent Systems and Computing, 2014, 236: 1153-1159.
[30] 邢旋旋. 基于BP神经网络及其改进算法的基坑变形预测研究[D]. 开封:河南大学,2022.
Xing Xuanxuan. Research on Deformation Predicition of Foundation Pit Based on BP Neural Network and Its Improved Algorithm[D]. Kaifeng: Henan University, 2022. (in Chinese with English abstract)
[31] Leblanc L, Dufour E. Monitoring the identity of bacteria using their intrinsic fluorescence[J]. FEMS Microbiology Letters, 2002, 211(2): 147-153.
[32] 刘雪茹,李欣,殷勇,等. 黄瓜贮藏中微生物信息三维荧光判别及其数量监控模型构建[J]. 食品科学,2021,42(5):32-38.
Liu Xueru, Li Xin, Yin Yong, et al. 3D fluorescence discrimination of microbial information and monitoring model establishment of the microbial quantity in cucumber storage[J]. Food Science, 2021, 42(5): 32-38. (in Chinese with English abstract)
[33] Pons M N, Swbatien L B, Potier O. Spectral analysis and fingerprinting for biomedia characterisation[J]. Journal of Biotechnology, 2004, 113(1): 211-230.
[34] Airado-Rodríguez D, Dura Nmera S I, Diaz Galeano T, et al. Front-face fluorescence spectroscopy: A new tool for control in the wine industry[J]. Journal of Food Composition and Analysis, 2010, 24(2): 257-264.
[35] Estes C, Duncan A, Wade B, et al. Reagentless detection of microorganisms by intrinsic fluorescence[J]. Biosensors and Bioelectronics, 2003, 18(5): 511-519.
[36] Dartnell L R, Storrie-Lombardi M C, Ward J M. Complete fluorescent fingerprints of extremophilic and photosynthetic microbes[J]. International Journal of Astrobiology, 2010, 9(4): 245-257.
[37] Gu H Y, Sun Y H, Liu S L, et al. A Feasibility study of the rapid evaluation of oil oxidation using synchronous fluorescence spectroscopy[J]. Food Analytical Methods, 2018, 11(12): 3464-3470.
Detection of silage microbial growth by using three-dimensional fluorescence coupled with chemometrics
Zhang Weiwei, Zhang Jing, Meng De, Lyu Riqin, Gu Haiyang, Sun Yanhui※
(239000,)
Silage is a type of storage fodder from green foliage crops to reduce the cost of feed and environmental pollution. The silage can be preserved by fermentation to the point of acidification. Among them, microbial growth can dominate in the silage quality. Especially, the proliferation of harmful microorganisms has also posed a great threat to crop resources, and ruminantia production, such as clostridium, acetic acid bacteria, and yeast. However, the commonly-used plate counting and turbidimetry for microbial growth in the laboratory cannot accurately characterize the growth state of silage microorganisms in time, due to tedious steps, time-consuming, and slow response rate. This study aims to effectively monitor the growth of silage microorganisms (lactic acid bacteria, acetic acid bacteria, and clostridium butyricum) separating from the silage as the indicator strains. A systematic investigation was made for the three-dimensional fluorescence spectra, the number of microbial colonies, and the absorption of 105 samples at the seven growth time points (0, 2, 4, 8, 12, 24 and 48 h). The chemometrics analysis and spectroscopic techniques were combined for the rapid screening of microbial growth. Parallel factor analysis was applied to resolve the three-dimensional fluorescence data. Back Propagation (BP) neural network was also used in the material quantitative analysis in the field of machine learning, due to its powerful nonlinear ability. The three-dimensional Synchronous Fluorescence Spectra (SFS) showed that there were two strong fluorescence peaks at about 225 and 275 nm, respectively. The main fluorescence peaks were the microbial endogenous tyrosine and tryptophan. The fluorescence intensity increased gradually with the increasing culture time, where the position of the fluorescence peak shifted the red. Meanwhile, the width of the fluorescence peak increased significantly. The parallel factor analysis showed that there was a significant difference in fluorescence information, where the characteristic wavelength Δwas 50 nm with six components. In addition to the two characteristic peaks, there were two weak fluorescence peaks at 310-360 and 370-390 nm. The two wave peaks at 340 and 380 nm were the microbial metabolism products or acids. There was a positive correlation between the intensity of natural fluorescence peak at 380 nm during culture time. Outstandingly, there was more information on fluorescence components in the two-dimensional fluorescence spectra from the parallel factor analysis. In terms of two-dimensional spectral data, the number of microbial colonies, and the absorbance were taken as the input or the output values of the BP neural network model, respectively. The modeling was constructed for the microbial growth of different detection. The experimental results showed that the correlation coefficients of the two models were close to 1.0, and the Mean Square Error (MSE) was all very small. A very reliable model was achieved in the neural network with a high fitting ability. Therefore, the three-dimensional fluorescence spectroscopy combined with the chemometrics was feasible to monitor the microbial growth in the silage. The finding can also provide a new technical approach for the rapid determination of the fermentation silage stage.
three-dimensional fluorescence; silage; microbial growth; chemometrics; parallel factor analysis
10.11975/j.issn.1002-6819.2022.18.033
O433.4;S816.11
A
1002-6819(2022)-18-0302-06
张微微,张静,孟德,等. 三维荧光技术结合化学计量学检测青贮微生物生长量[J]. 农业工程学报,2022,38(18):302-307.doi:10.11975/j.issn.1002-6819.2022.18.033 http://www.tcsae.org
Zhang Weiwei, Zhang Jing, Meng De, et al. Detection of silage microbial growth by using three-dimensional fluorescence coupled with chemometrics[J]. Transactions of the Chinese Society of Agricultural Engineering (Transactions of the CSAE), 2022, 38(18): 302-307. (in Chinese with English abstract) doi:10.11975/j.issn.1002-6819.2022.18.033 http://www.tcsae.org
2021-07-01
2022-09-01
国家自然科学基金项目(31701685);安徽省重点研究与开发计划项目(202004a06020039);滁州学院博士后基金项目(2020BSH002);滁州市科技局指导性计划(2021ZD025)
张微微,博士,副教授,研究方向为微生物,快速检测。Email:249541998@qq.com
孙艳辉,博士,教授,研究方向为快速检测。Email:1647608982@qq.com

