MutL homolog 1 methylation and microsatellite instability in sporadic colorectal tumors among Filipinos
Loraine Kay D Cabral,Cynthia A Mapua,Filipinas F Natividad,Caecilia H C Sukowati,Edgardo R Cortez,Ma Luisa D Enriquez
Loraine Kay D Cabral,Cynthia A Mapua,Filipinas F Natividad,Ma Luisa D Enriquez,Research and Biotechnology Group,St.Luke's Medical Center,Quezon City 1112,Philippines
Loraine Kay D Cabral,Caecilia H C Sukowati,Centro Studi Fegato,Fondazione Italiana Fegato ONLUS,Trieste 34149,Italy
Edgardo R Cortez,Department of Surgery,St.Luke's Medical Center,Quezon City 1112,Philippines
Ma Luisa D Enriquez,Center for Natural Science and Environmental Research,De La Salle University,Manila 1004,Philippines
Abstract BACKGROUND Colorectal cancer (CRC) ranks third in terms of incidence and second in mortality worldwide.In CRC,the silencing of mismatch repair genes,including the mutL homolog 1 (hMLH1) has been linked to microsatellite instability (MSI),the lengthening or shortening of microsatellite repeats.Very limited data have been presented so far on the link of hMLH1 methylation and MSI in Southeast Asia populations with sporadic CRC,and on its clinical significance.AIM To investigate the significance of the MSI status and hMLH1 methylation in CRC Filipino patients.METHODS Fifty-four sporadic CRC patients with complete clinical data were included in this study.Genomic DNA from CRC tumor biopsies and their normal tissue counterparts were profiled for MSI by high resolution melting (HRM) analysis using the Bethesda Panel of Markers (BAT25,BAT26,D2S123,D5S346,and D17S250).hMLH1 methylation screening was performed using bisulfite conversion and methylation specific polymerase chain reaction.Statistical analysis was conducted to calculate their associations to clinicopathological characteristics and survival relevance (Kaplan-Meier curves and the log-rank test).RESULTS hMLH1 methylation was observed in 9% and 35% of CRC and normal samples,respectively.Higher incidence of consistently methylated hMLH1 found in both normal and CRC was noticed for relation to location of tumor (P<0.05).As for MSI status,D2S123 the most common unstable microsatellite and MSI-high (MSIH) was the most common MSI profile,counted for 46% and 50% of normal and CRC tissues,respectively.The presence of MSI-low (MSI-L) and microsatellite stable (MSS) was 43% and 11% for normal,and 31% and 19% for CRC samples.The mean month of patients’ survival was shorter in patients whose normal and tumor tissues had methylated compared to those with unmethylated hMLH1 and with MSI-H compared to those with MSI-L/MSS (P<0.05).This was supported by significant difference in Kaplan-Meier with log-rank analysis.This data indicated that hMLH1 methylation and high MSI status have prognostic value.CONCLUSION This study showed the clinical significance of hMLH1 methylation and MSI status in sporadic CRC Filipino patients,especially in the normal part of the tumor.
Key Words:Sporadic colorectal cancer;DNA methylation;Microsatellite instability;Population genetic;Colorectal cancer
INTRODUCTION
Colorectal cancer (CRC) occurs when malignant tumors form in the lining of the large intestine,which includes the ascending,transverse,and descending colon,and the rectum.CRC ranks third in terms of incidence and second in mortality worldwide,for both sexes.Over 1.9 million new CRC cases and 935000 deaths were estimated to occur in 2020,accounting for about 1/10 cancer cases and deaths[1].In the Philippines,CRC is currently the third leading site of malignancy.The incidence of CRC had almost doubled from 2010 to 2015 with survival rates under 50%[2,3].In terms of mortality rate,Filipinos (and Chinese) ethnicity had significantly decreased risk of death compared with Caucasians[4].
CRC,like many other cancers,is a malignancy caused by DNA changes causing abnormal behavior of cells.DNA deletions,duplications,substitutions,mutations,and rearrangements can either activate or inactivate a gene or several genes and consequently affect cellular function[5].Depending on the origin of the mutation,CRC can be classified as sporadic (70%),inherited (5%),and familial (25%)[6].Sporadic CRC is characterized by the carcinogenesis pathway,derived from point mutations which can occur in different genes[7].
In an inherited CRC,the hereditary non-polyposis CRC (HNPCC),germline mutation is involved in one or two of the mutator or mismatch repair (MMR) genes,including the mutL homolog 1 (hMLH1).Mutations inhMLH1have been linked to microsatellite instability (MSI),the lengthening or shortening of microsatellite repeats(sequences with 1-6 repeating nucleotides).When instability is left unrepaired,it may accumulate as mutations and can affect other genes that have microsatellite repeats in their coding regions[8,9].MSI is found in 85%-90% of HNPCC patients[10].
Recent studies demonstrated that in a subset of sporadic CRC,DNA methylation,the transfer of a covalent methyl group to the C5 position of the cytosine to form 5-methylcytosine by DNA methyltransferases,may cause the loss of function of a MMR gene.Likewise,it leads to the accumulation of MSI[11,12].Tumors with methylatedhMLH1and high levels of MSI present with a distinct characteristic from other CRC tumors.CRC with methylatedhMLH1had a delayed onset and was associated with female gender[13].This linkedhMLH1and MSI in the development of sporadic CRC,suggesting that the two molecular profiles are closely related[14].
So far,very limited data have been presented on the link ofhMLH1methylation and MSI in Southeast Asia populations with sporadic CRC.This study presents the detection and characterization of MSI status andhMLH1methylation in CRC and its paired non-tumoral adjacent tissues in Filipino patients.
MATERIALS AND METHODS
Patients and samples
Fifty-four sporadic CRC patients with complete clinical data were included in this study.Diagnosis of CRC was based on the presence of malignancy in the initial biopsy.Patients should be Filipino by descent with no family history of cancer.From each patient,paired tumor and its corresponding normal tissue were obtained from surgical resection at the St.Luke’s Medical Center,Quezon City,Philippines.Normal tissues were collected approximately 6 inches away from the margin of the tumor.Upon pathological confirmation,fresh frozen sections were stored in -80°C.The project of the Colorectal Cancer Study Group was approved by the Institutional Ethics Review Board of St.Luke’s Medical Center (Project Code No.06-015).All patients enrolled in the study signed an Informed Consent Form allowing the use of their tissues and clinical data in the Colorectal Cancer (CRC) Databank of St.Luke’s Medical Center.
CRC cell lines SW480 (ATCC®CCL-228) and SW48 (ATCC®CCL-23) were purchased from American Type Culture Collection (ATCC) as controls for MSI andhMLH1methylation.SW480 is a CRC cell line that has stable microsatellite and unmethylatedhMLH1,while SW48 is high MSI and methylatedhMLH1.Lymphocytes from patients who underwent colonoscopy and who were found to be free of cancer and polyp were used as additional controls for stable microsatellite.
Isolation of genomic DNA
Genomic DNA (gDNA) extraction was performed using the QIAamp®DNA Mini Kit(Qiagen),according to the manufacturer's instructions.Briefly,tissues were finely minced and put in lysis solution and proteinase K until completely lysed.After DNA precipitation with ethanol,DNA extract was washed twice and eluted with appropriate buffer.DNA quality and quantity were assessed using Nanodrop®v1000 spectrophotometer (Thermo Fisher Scientific).Final working concentration of 50 ng of gDNA was used for each sample in the succeeding analysis.
Bisulfite conversion and methylation specific polymerase chain reaction for hMLH1
gDNA from normal and tumor specimens was subjected to bisulfite treatment to differentiate methylated cytosines from unmethylated ones.Bisulfite chemically modifies non-methylated cytosines into uracil,which is then converted to thymidine in polymerase chain reaction (PCR) cycles.
Bisulfite treatment of the DNA sample was performed using EZ DNA Methylation Lightning Kit (Zymo Research) according to the manufacturer’s suggestion.Briefly,200-500 ng gDNA was incubated in the conversion reagent and then treated with binding buffer in a spin column.Converted DNA was then subjected to desulphonation and clean-up using washing buffer.DNA (approximately 10 μL) was eluted and collected for methylation specific PCR (MS-PCR)[15].Reaction was carried out using Qiagen Taq Core Kit in a reaction volume of 25 μL with 10x PCR Buffer,2.5 mmol/L of MgCl2,50 pmol of primer,1.25 mmol/L of dNTPs and 1.25 units of Taq DNA polymerase added to 1.5-2 μL of converted DNA.Primer sets used forhMLH1MS-PCR are taken from published work of Foxet al[15].
MSI test by high resolution melting analysis
MSI test was carried out using PCR for five markers from the Bethesda panel that included BAT25,BAT26,D2S123,D5S346,and D17S250,as in a previous study[16].PCR conditions of each marker were optimized and validated in our earlier study on HNPCC (Evangelista &Enriquez,unpublished work).
In brief,PCR was performed using Qiagen Taq PCR core kit with EvaGreen dye(Biotium) in a 25 μL reaction volume containing 50 ng of gDNA.All reactions were done in triplicates.PCR and high resolution melting (HRM) analysis were carried out using Rotor-Gene™ 6000 (Qiagen) system with data collected over the range from 55°C to 95 °C with ramp rising at 0.1 °C/s[17].Melting curve was analyzed using Rotor-Gene Q (RGQ) Scanning Software version 2.0.2 (Qiagen).Raw melting-curve data were normalized by manual adjustment of linear regions before (pre-;100%fluorescence) and after (post-;0% fluorescence) the melting transition.The melting curve of DNA from SW480 cells which exhibits microsatellite stable (MSS) phenotype[18] was used as the normal/stable control.The RGQ software assigned the profile of each sample in reference to the stable control.The confidence percentage (cut-off),was optimized by analyzing non-cancer patients’ DNA,with values not lower than 60%.Therefore,the confidence value of ≥ 60% was regarded as MSS,while any confidence value of<60% was regarded as unstable.
MSI score was defined as MSI-high (MSI-H) where HRM instability was observed in≥ 2 markers;MSI-low (MSI-L) where instability was only in one marker;and MSS if no instability was observed in any of the markers.For validation,amplified PCR products of the stable and unstable control were subjected to Sanger sequencing to verify their microsatellite repeats.
Statistical analysis
Statistical analysis was constructed using software GraphPrism version 5.01(GraphPad Software,Inc.,La Jolla,CA,United States).Associations between clinicopathological characteristics (age,sex,tumor location,grade and stage),MSI status and methylation status were determined using Pearson’s chi-square test or Fisher’s exact test.Survival relevance was analyzed using Kaplan-Meier curves and the logrank test based on thehMLH1methylation and MSI status grouping.All tests were two-tailed and values were considered significant atPvalue of less than 0.05.
RESULTS
Patients’ demographic and tumor features
This study analyzed 54 Filipino CRC patients with comparable male to female ratio(30M:24F).The age mean was 56.9 ± 11.8 years old,with 5 patients under 40 years old and 49 patients above 40 years old.Regarding tumor parameters,CRC was mostly found in the distal part of the colon (38;70%),followed by the rectum (10;19%) and proximal part of the colon (6;11%).By histology,moderately-differentiated grade CRC was noticed in 44 (81%),poorly-differentiated in 8 (15%),and well-differentiated in 2(4%) patients.By disease stage,stage 1 CRC was observed in 6 (11%),stage 2 in 14(26%),stage 3 in 30 (56%),and stage in 4 (7%) of patients.None of the patients had family history of CRC.
hMLH1 methylation status in Filipino CRC patients
We analyzed thehMLH1DNA methylation by MS-PCR.From 54 paired CRC and its normal adjacent tissue,most of the samples were unmethylated (91% and 65%,respectively).Representative MS-PCR gel electrophoresis is shown in Figure 1.hMLH1methylation was noticed only in 5 (9%) and 19 (35%) of CRC and normal samples,respectively.It is interesting to note that the methylatedhMLH1in tumor tissues was accompanied by methylation in its paired normal tissues (4/5,80%).Only 1 sample showed tumoralhMLH1methylation without methylation in its normal tissue(Figure 2).

Figure 1 Determination of hMLH1 methylation by methylation-specific polymerase chain reaction and microsatellite instability status by high resolution melting analysis.
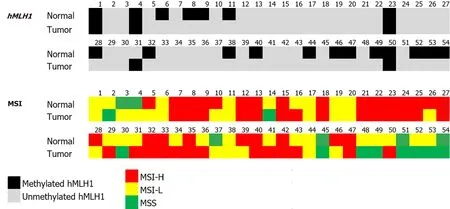
Figure 2 hMLH1 methylation and microsatellite instability analysis results of paired normal and tumor tissues.
The distribution ofhMLH1methylation status in patients’ demography and tumor parameters is shown in Table 1 (left panel).In normal tissues,the incidence ofhMLH1methylation was slightly higher in female compared to male (42%vs30%),and in tumors located in the proximal colon compared to distal/rectum location (67%vs31%).The same pattern was found forhMLH1methylation in tumor tissues.Methylation was higher in females compared to males (13%vs7%),and in tumors located in the proximal colon compared to those in distal colon or rectum (33%vs6%,P=0.031).
The signature ofhMLH1status in paired normal and tumor tissue is presented in Table 1 (right panel).Although the incidence was not statistically significant,a higher incidence of methylatedhMLH1in both normal and tumor tissues (M/M) was noticed in females,in > 40 years old patients,and in poorly differentiated cancer.A significant difference was found for the tumor location (P=0.029),where M/M signature was noticed in 33% of proximal tumors,while U/U was in 67% of distally located or rectal tumors.

Table 1 hMLH1 methylation status in the Filipinos colorectal cancer patients
MSI status in Filipino sporadic CRC patients
MSI status was assessed using HRM of the Bethesda panel that included BAT25,BAT26,D2S123,D5S346,and D17S250.As controls,cell lines SW480 and SW48 were used to represent a MSS and MSI-H,respectively.MSS feature was also checked in lymphocytes DNA from a normal individual.A representative HRM analysis of BAT26 gene and its direct Sanger sequencing are shown in Figure 1.
From the Bethesda panel markers,the microsatellite with the highest instability was D2S123 and this was found in 32 (59%) and 33 (61%) samples of normal and tumor,respectively.On the other hand,BAT26 instability was observed only in 1 (2%) sample of each normal and tumor tissues.In normal tissues,the instability of BAT25,D17S250,and D5S346,were 2 (4%),14 (26%),and 29 (54%),respectively.Among the CRC tissues,instability was found in BAT25,BAT26,D17S250,and D5S346 accounting for 5(20%),12 (22%),and 25 (46%),respectively.There was no statistical difference in the instability of these markers between normal and tumor tissues.
The distribution of MSI status in patients’ demography and tumor parameter is shown in Table 2 (left panel).In normal tissue samples,25 (46%),23 (43%),and 6 (11%)samples were MSI-H,MSI-L,and MSS,respectively.In female,the highest percentage was for MSI-H (54%),followed by MSI-L (33%),and MSS (13%).Similar pattern was observed in tumoral tissues with the following rates:MSI-H with 27 (50%),MSI-L with 17 (31%),and MSS with 10 (19%).Instability rates of microsatellites in tumors tissues in the female group were the exactly the same as in the normal tissues.
The signature of MSI status in paired normal and tumor tissue is presented in Table 2 (right panel).Analysis of data using showed that the MSI status between normal and tumor tissues was not statistically significant.Nonetheless,a slightly higher incidence of MSI-H inhMLH1was observed in both normal and tumor tissues(H/H) among females,similar to methylatedhMLH1.
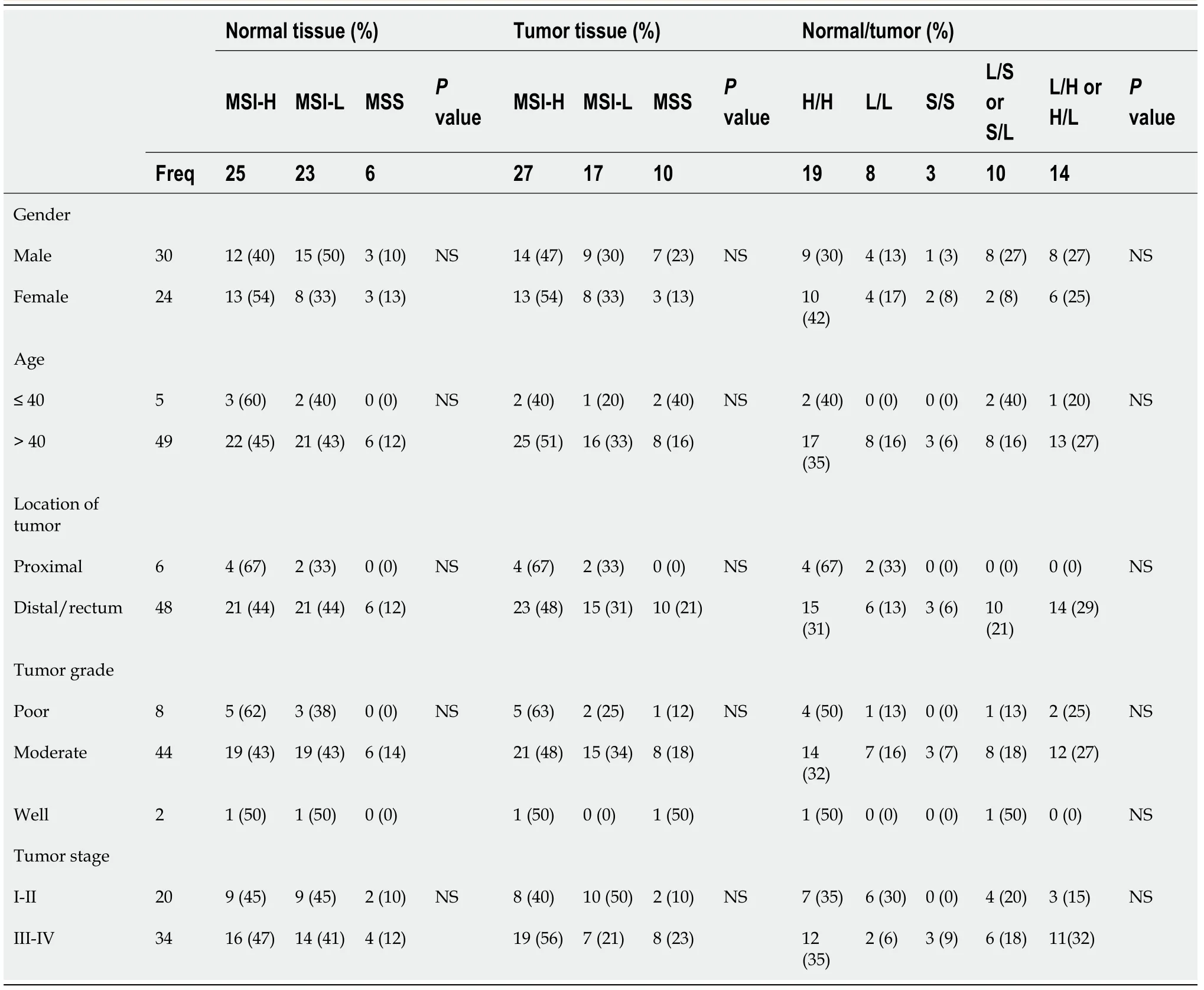
Table 2 Microsatellite instability status in the Filipinos colorectal cancer patients
Correlation of MSI and hMLH1 methylation status
Table 3 shows the MSI and methylation status of each sample according to the paired groupings described previously.There was no statistically significant correlation between MSI andhMLH1methylation status of all the sample pairs.Thirteen paired samples (24%) were unmethylated with MSI-H (H/H and U/U).The clinicopathological profile of this cancer group showed that 11 samples came from the distal colon/rectum (85%),12 (92%) were > 40 years old,10 (77%) were moderately differentiated,and 8 (62%) were stage III/IV cancers.
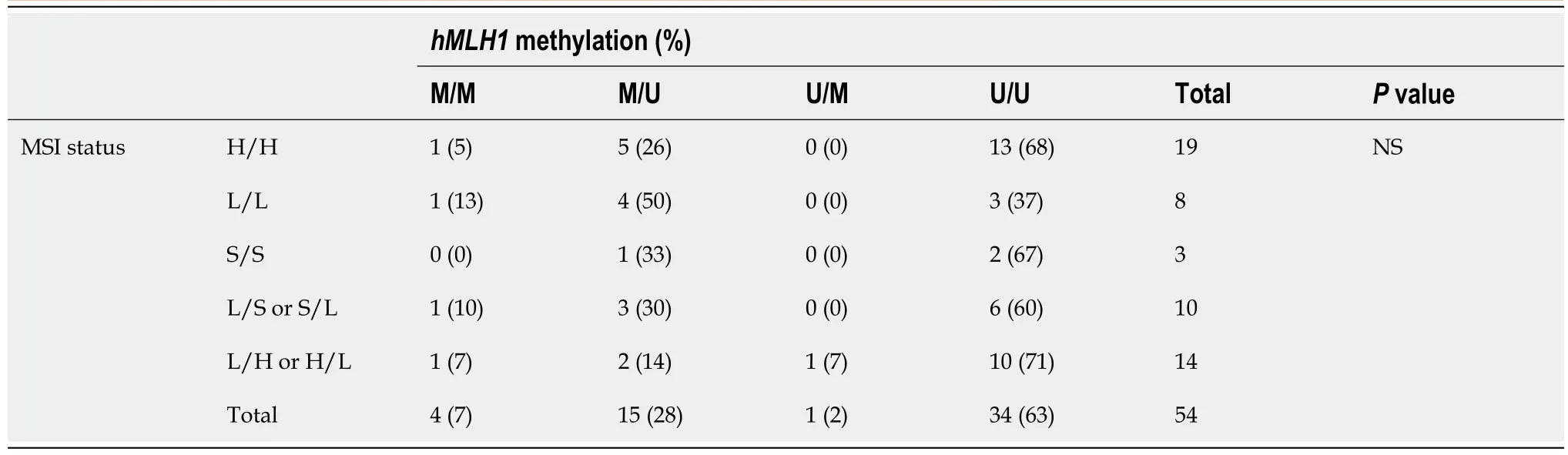
Table 3 Correlation of hMLH1 methylation and microsatellite instability status of paired samples
Correlation of MSI and hMLH1 methylation status with prognosis
To determine the prognostic values of bothhMLH1methylation and MSI status,we performed Kaplan-Meier analysis on patient survival for independent groups.The survival was defined as death event (in months) post-surgery.Patients were followed up until 60 mo after surgery.
Our results show that the mean survival was relatively shorter in patients with methylated compared to those with unmethylatedhMLH1,both in tumor [18 (6-29) movs44 (34-54) mo] and in normal tissues [34 (21-46) movs44 (33-54) mo],expressed as mean (95% confidence interval).A significant difference was also observed in tumor tissues with MSI-H compared to those with MSI-L/MSS [27 (19-36) movs56 (44-67)mo].For the MSI status in normal tissues,again a shorter survival was found for MSIH compared to MSI-L/MSS [27 (18-36) movs54 (39-68) mo].
Kaplan-Meier analysis was performed to associate thehMLH1methylation and MSI status with patients’ survival in both cohorts.In concordance with the mean survival above,we found the association of MSI-H status in both normal and tumor tissues with shorter probability of survival (P<0.05),compared to that of MSI-L/MSS(Figure 3).Our data indicated thathMLH1methylation and high MSI status might have a prognostic value.
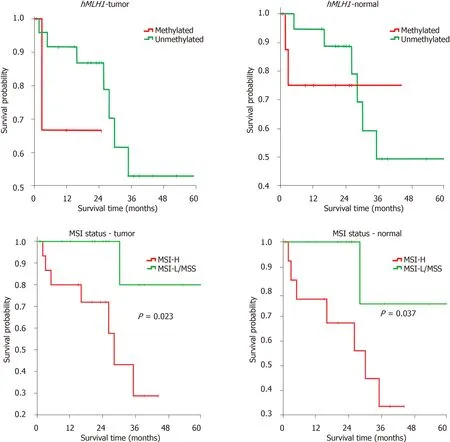
Figure 3 hMLH1 methylation and microsatellite instability analysis for patients’ prognosis.
DISCUSSION
CRC is a classic example of disease progression accompanied by molecular changes.Its carcinogenesis is composed of stage-specific molecular changes in gene expression and the accumulation of mutations.A mutation in the MMR genes may lead to accumulation of DNA damage.Consequently,the tumor suppressor genes or oncogenes are affected,leading to the development of the cancer[8].
DNA methylation has been linked to tumor suppressor gene silencing[19],including the silencing ofhMLH1MMR gene[20].In a subset of sporadic CRC,the loss of function ofhMLH1gene led to the accumulation of instability in microsatellite regions of the DNA[11,12].
In this study,there was a notable higher incidence of methylation ofhMLH1in normal tissue as compared to its paired tumor tissue (35%vs9%).This is in agreement with other studies showing elevatedhMLH1methylation in the normal gastric epithelia in patients with stomach cancer as compared to those of non-cancer patients[21].Methylation in the normal mucosa was proposed as a marker of risk for the development of CRC[22].As demonstrated recently,the acceleration of ageing-related DNA methylation drift in normal mucosa was correlated with an earlier age of diagnosis[23].
hMLH1methylation in non-neoplastic epithelia was shown also to exhibit MSI[24].It is interesting to note that the methylatedhMLH1in tumor was accompanied by methylation in its paired normal tissue.This finding warrants further investigation on the clinical utility ofhMLH1methylation in normal tissue as a useful marker not only in CRC but in also in other gastrointestinal cancers.
However,in this study,a high incidence of non-methylatedhMLH1in MSI-H samples (both normal and tumor) suggests thathMLH1methylation did not bring about MSI.We assumed,at least in our cohort,that unstable microsatellites can be attributed to methylation in other MMR genes,such as hSMH2[25,26] and mutation in hSMH6[27].It was also noted that MSI may also result from other defects in the component of base excision repair of the proteins (DNA glycosylase,AP endonuclease,DNA polymerase,etc.)[28].MSI tumors were also grouped to Type A (≤ 6 bp change)and Type B (≥ 8 bp change).In a report on MMR gene knock-out animals,no Type B MSI was observed,suggesting that a kind of MSI group may involve malignancies apart from MMR deficiency[29].
Significant differences forhMLH1methylation were found for the location of the tumor.Colon is divided into two anatomical regions:the proximal/right colon and the distal/left colon and each with very unique features[30-32].This may be partially attributed to their difference in embryological development and physiological circumstances.The proximal colon develops from the hindgut while the distal colon originates from the midgut[33].Proximal and distal colon are also different in the genetic patterns[34].
Since methylation is affected by environmental exposure,the difference might also be related to function.The proximal colon is the site where breakdown of complex carbohydrates occurs while the undigested dietary proteins are broken down in the distal colon.This process of digestion in the distal colon generates by-products such as ammonia,phenols,indoles and sulfurs,that may inhibit DNA methyltransferase activity[35].Among African-American race,higher vegetable intake diet was shown being associated with greater odds for highhMLH1methylation[36].Because methylation is less inhibited in proximal region,this may explain the higher incidence of methylated normal and tumor tissue (M/M),in contrast to high percentage of unmethylation in distal region.
In this study,we observed that from both the average of survival data and log-rank test of Kaplan-Meier analysis,hMLH1methylation in normal tissues and MSI status in both normal and CRC tissues had prognostic value.UnmethylatedhMLH1and MSIL/MSS status were positively associated with better patients’ survival.Our data may support earlier findings where expression ofhMLH1is considered an independent prognostic and predictive factor stage II-III CRC in Chinese population[37].Data from this study,however,in contrast with previous studies showing the association between MSI-H with improved overall and disease-free survival[38,39],showed that population genetics and the disease stages might influence the prognosis of CRC patients.
Although there was no statistical difference,the incidence of MSI andhMLH1methylation showed a similar trend in association with sex and grade.MSI andhMLH1methylation were more common in females and in poorly differentiated tumors.This non-significant correlation may be attributed to the small sample size,thus studies using larger number of samples that are well distributed in terms of tumor clinicopathological parameters will be needed.Moreover,significance of both markers in relation to patients' prognosis,especially for Southeast Asian population,needs to be validated.Investigation of the methylation status of two or more MMR genes and their correlation with MSI status is also recommended,since this study only looked into one MMR gene out of the six.A recent data from metastatic CRC showed that a novel epigenetic signature of eight hypermethylated genes was able to identify CRC with poor prognosis.These genes were characterized with CpG-island high methylator and MSI-like phenotype[40].
The establishment of molecular biomarkers in diseases is essential for an effective management and treatment protocol for cancer patients.For the precision treatment in the future,especially for immunotherapy,it was shown that patients with MSI tumors exhibited significant response to anti-PD-1 inhibitors after failure of conventional therapy[41,42].The role of MSI andhMLH1methylation is shedding light in the management of sporadic CRC in clinical practice in other countries[43,44] but not yet in the Philippines.
CONCLUSION
To conclude,we showed the clinical significance ofhMLH1methylation and MSI status in sporadic CRC Filipino patients,especially in the normal part of the organ.This study is one of the few attempts to establish the molecular profile of Filipino CRC patients in the local setting,to highlight the importance of epigenetic modification.Understanding the diverse molecular and genetic key players involved in cancer development will ultimately translate to improvement in patient care.
ARTICLE HIGHLIGHTS
Research background
A distinct molecular signature marks a particular subset of sporadic colorectal cancer(CRC).It involves the mismatch repair (MMR) genes silencing due to DNA methylation,leading to microsatellite instability (MSI).
Research motivation
To improve the management of the CRC patients based on their distinct molecular subtypes.
Research objectives
To examine the association of mutL homolog 1 (hMLH1) methylation (MMR gene) and the MSI phenotype in relation to cancer characteristics and patient survival among Filipino sporadic CRC patients.
Research methods
Paired tissues (normal and tumor) from sporadic CRC patients was screened for hMLH1 methylation using methylation specific polymerase chain reaction.Subsequent MSI typing was done by high resolution melting analysis.
Research results
The results of this study showed that hMLH1 methylation was mostly noticed in proximal tumors.Low overall survival was observed in methylated hMLH1 and MSI tumors.
Research conclusions
The epigenetic silencing of hMLH1 as well as MSI may present a distinct pattern of CRC in Filipino patients.
Research perspectives
This is an initial attempt to characterize sporadic CRC in Filipino population.It may shed light in understanding molecular epigenetic modification in CRC as well as its role in tumor development and management.
ACKNOWLEDGEMENTS
The members of the Colorectal Cancer Study Group of the St.Luke's Medical Center actively and consistently participated in the study.
 World Journal of Gastrointestinal Oncology2021年12期
World Journal of Gastrointestinal Oncology2021年12期
- World Journal of Gastrointestinal Oncology的其它文章
- Management of obstructive colon cancer:Current status,obstacles,and future directions
- Role of endoscopic ultrasound in anticancer therapy:Current evidence and future perspectives
- Mesenchymal stem cell-derived exosomes for gastrointestinal cancer
- Gender differences in the relationship between alcohol consumption and gastric cancer risk are uncertain and not well-delineated
- Pancreatic intraductal papillary mucinous neoplasms:Current diagnosis and management
- Combined treatments in hepatocellular carcinoma:Time to put them in the guidelines?
