AAV8 transduction capacity is reduced by prior exposure to endosome-like pH conditions
Jeffrey A. Lowell, Kar Men Mah, John L. Bixby, , Vance P. Lemmon, *
Abstract Adeno-associated virus (AAV) is an essential instrument in the neuroscientist’s toolkit, which allows delivery of DNA to provide labeling with fluorescent proteins or genetic instructions to regulate gene expression. In the field of neural regeneration, the transduction of neurons enables the observation and regulation of axon growth and regeneration, and in the future will likely be a mechanism for delivering molecular therapies to promote sprouting and regeneration after central nervous system injury. Traditional formulations of AAV preparations permit efficient viral transduction under physiologic conditions, but an improved understanding of the mechanistic limitations of AAV transduction may facilitate production of more resilient AAV strains for investigative and therapeutic purposes. We studied AAV transduction in the context of prior exposure of AAV serotype 8 (AAV8) to environmental pH within the range encountered during endosomal endocytosis (pH 7.4 to pH 4.4), during which low pH-triggered structural and autoproteolytic changes to the viral capsid are believed to be necessary for endosome escape and virus uncoating. Due to the fundamental nature of these processes, we hypothesized that premature exposure of AAV8 particles to acidic pH would decrease viral transduction of HT1080 cells in vitro, as measured by fluorescent reporter gene expression using high-content imaging analysis. We found that increasingly acidic incubation conditions were associated with concomitant reductions in transduction efficiency, and that quantitative levels of reporter gene expression in transduced cells were similarly decreased. The biggest decrease in transduction occurred between pH 7.4 and pH 6.4, suggesting the possible co-occurrence of a pH-associated event and viral inactivation within that range. Taken together, these findings indicate that exposure of AAV8 to acidic pH for as little as 1 hour is deleterious to transduction ability. Future studies are necessary to understand the pH-associated causative mechanisms involved. This study was approved by the University of Miami Institutional Animal Care and Use Committee, USA (Protocol #18-108-LF) on July 12, 2018.
Key Words: adeno-associated virus; autolytic; capsid; low pH; protease; proteolytic; temperature; trafficking; transduction; viral inactivation
Introduction
Viral gene delivery systems have achieved widespread adoption as safe and effective tools for introduction of exogenous nucleic acids to express fluorescent markers or to regulate gene expression. Clinical trials using viral vectors also indicate their potential use in treatment of neurodegenerative diseases and central nervous system injuries that are intractable to current therapies, potentially delivered as combinatorial therapy with adjunct pharmaceuticals. Drug solubility is a common consideration in the development of new therapeutics and an estimated 40% of currently marketed drugs and 75% of new drugs in development have poor solubility under physiologic conditions (Williams et al., 2013), raising the question of how viral vectors perform in combinatorial therapy when diluted in non-physiologic solvents such as low pH that may be necessary to solubilize certain compounds. To optimize viral transduction efficiency,it is necessary to have a more complete understanding of the interaction of viral particles with potential drug formulations to ensure effective transduction strategies.
The most commonly used vectors of viral gene delivery include lentivirus and adeno-associated virus (AAV), the latter of which lacks a phospholipid envelope and exposes the viral capsid directly to environmental challenges (Naso et al.,2017). Without an envelope, AAV particles cannot enter cells via fusion with the cell membrane and must instead be initially internalized via endocytosis (Smith and Helenius, 2004;Agosto et al., 2006). During endocytosis, the cell’s membrane invaginates to create an internal partition of extracellular material containing membrane-bound AAV particles (Marsh and Helenius, 2006). Progressive maturation of the virusbound endosome to a lysosome is associated with a gradual acidification of the endosome contents (Huynh and Grinstein,2007), in addition to increasing protease activity and proximity to the nucleus (Authier et al., 1996; Rink et al., 2005). To avoid lysosomal degradation and promote release in the vicinity of the nucleus, timing of escape from the endosome is crucial to maximizing the chance of successful delivery of AAV cargo to the nucleus. Thus, AAV and other endosome-dependent viruses rely on the gradual pH decrease during endosome maturation to time their escape (Chandran and Nibert, 2003).
Escape from the endosome at an optimal time occurs through the conformational changes of pH-sensitive endosomolytic viral proteins. An acidic pH leads to formation of small pores that disrupt the local endosomal membrane in association with phospholipase A2 activity on the viral protein 1 unique region of the viral capsid (Helenius et al., 1980; Marsh and Helenius, 1989, 1991; Girod et al., 2002). It has been suggested that a low pH-activated autolytic protease activity of the AAV capsid may also aid in this escape process (Salganik et al., 2012). Despite knowledge that AAVs, other nonenveloped viruses, and many enveloped viruses are pH-sensitive, the effect of environmental pH on AAV transduction efficiency has received little scrutiny. We undertook this study to determine if AAV would remain functional in a situation in which it is co-injected with a compound requiring an acidic solvent necessary for solubilization, such as in a theoretical therapy of an AAV carrying instructions for an axon growthpromoting transcription factor (Blackmore et al., 2012)delivered alongside an axon growth-promoting small molecule(Al-Ali et al., 2015). We hypothesized that premature exposure of AAV to an acidic environment would reduce transduction capacity. Specifically, we examined the effect of lowering solvent vehicle pH on transduction by AAV serotype 8 (AAV8)and found that decreases in incubation pH were associated with decreases in AAV transduction ability in a temperatureindependent manner.
Materials and Methods
Viral vector construction
A recombinant replication-deficient AAV8 containing an expression cassette for the eGFP (AAV8-GFP) gene was produced in-house using previously developed methods(Zolotukhin et al., 2002). Briefly, we used the pAAV-UbC-eGFP which was a gift from Drs. Jae Lee & Pantelis Tsoulfas(Addgene plasmid #62518; http://n2t.net/addgene:62518;RRID: Addgene 62518) as the transfer plasmid for the viral production. The pAAV-UbC-eGFP plasmid was transfected together with AAV2/8 RepCap and pHelper into HEK293T cells grown to 70–80% confluence. Recombinant AAVs were isolated from media harvested on day 5 and day 8 and from the HEK293T cells harvested and lysed on day 8. Culture media were subjected to PEG precipitation (8% final PEG-8000, 500 mM NaCl) and the cells collected in lysis buffer(150 mM NaCl and 50 mM Tris Cl pH 8.0 in ddH2O). The harvested cells were combined with the PEG precipitate and this was lysed by three cycles of freeze-thaw between a 37°C water bath and a dry ice-ethanol bath. The lysate was then digested with 50 U/mL of Benzonase (Sigma, E1014) at 37°C for 30 minutes. Recombinant AAV (rAAV) was purified by ultracentrifugation using an iodixanol step gradient (12 mL lysate, 6 mL 15% iodixanol, 4 mL 25% iodixanol, 6 mL 40% iodixanol, 4 mL 60% iodixanol) on Beckman Coulter ultracentrifuge tubes (OptiSealTM, Polyallomer, 32.4 mL, 26 ×77 mm, Cat# 361625) at 489,000×gfor 1.15 hours. The rAAVs were collected by puncturing the OptiSealTMtubes with an 18 gauge needle attached to a 10 mL syringe. The needle was introduced into the tube and placed beveled side up to collect 4 mL of the 40% fraction and then turned beveled side down to collect 1 mL of the 40–60% interphase. After the iodixanol gradient ultracentrifugation, the collected iodixanol fraction was loaded onto an FPLC system (Akta Explorer) with a HiTrap Q HGPE HP 5 mL column (GE Cat# 17-1154-01). The column was equilibrated with 5 column volumes of 20 mM Tris Cl,15 mM NaCl pH 8.0 (Buffer A), 5 column volumes of 20 mM Tris Cl, 500 mM NaCl pH 8.0 (Buffer B) and then 5 column volumes of Buffer A. The unbound sample was washed with 10 column volumes of Buffer A and the AAVs were eluted with 25 mL of 40% buffer B. The eluted rAAVs were loaded onto Millipore Amicon Ultra-15 100 kDa (Cat# UFC910024),washed three times with Hanks’ Balanced Salt Solution and finally concentrated until a final volume of 200–250 μL and aliquots were stored at –80°C. Titers of virus stocks were measured by quantitative polymerase chain reaction (qPCR)using SYBR green technology. Briefly, primers binding to the hGHPolyA signal were used to measure the titer with qPCR.Before releasing the viral DNA from the particles, all extra-viral DNA was removed by digestion with DNase I. Then, the viral DNA was extracted using the QIAamp® MinElute®Virus spin kit(Qiagen, 57704). The qPCR was performed using the Qiagen RT2 SYBR® Green FAST Mastermix (cat No. 330600), and primers against the hGHPolyA sequence (hGH PolyA Forward:CTA TTG GGA ACC AAG CTG GA, hGH PolyA Reverse: GTG AAA CCC CGT CTC TAC CA). The extracted viral DNA and a serial dilution of the pAAV-UbC-eGFP plasmid as a standard were measured using the Applied Biosystems QuantStudio 3 Real-Time PCR machine.
Unilateral pyramidotomy
All animal-related work was performed in accordance with the University of Miami Institutional Animal Care and Use Committee (Protocol #18-108-LF, approved on July 12, 2018).The University of Miami follows the Guide for the Care and Use of Laboratory Animals, 8thEdition and is an Association for Assessment and Accreditation of Laboratory Animal Care International approved institution. Mice (5/cage) were grouphoused in vivariums with temperature and humidity control,kept on a 12:12 light cycle. Animal health and weight were monitored by both scientists and veterinary staff within the University of Miami, and there were no adverse reactions during the course of experiments. Adult C57BL/6J mice (8–10 weeks old) were anesthetized (100 mg/kg Ketamine, 10 mg/kg Xylazine, intraperitoneally). The head and ventral surface of the neck were shaved and wiped down with Nolvasan disinfectant solution. An incision was made 5 mm lateral to the midline and the underlying muscle was bluntly dissected to expose the surface of the ventrocaudal portion of the occipital bone. The caudal portion of the occipital bone was removed using laminectomy forceps to expose the right medullary pyramid. A feather microscalpel (15°; Electron Microscopy Sciences, 72045-15) was used to puncture the dura and lesion the right pyramidal tract. Muscle was repositioned following the lesion and skin was closed using 4-0 Vicryl Sutures (Patterson Veterinary, 07-891-0083).
AAV8-GFP in vivo injection
After the unilateral pyramidotomy, the head of the animal was placed on a stereotaxic frame. A midline incision was made to expose the skull and a craniotomy was performed over the left sensorimotor cortex. AAV8-GFP (2.5 × 1013GC/mL)previously incubated in 4°C PBS either pH 7.4 or pH 6.0 for 24 hours was injected (500 nL/injection) into five sites: –1.0 mm anteroposterior (AP) and +1.0 mm lateral, –0.25 mm AP and +1.0 mm lateral, +0.5 mm AP and +1.5 mm lateral, +1.25 mm AP and +1.5 mm lateral, +2.0 mm AP and +1.5 mm lateral(all coordinates in reference to bregma), at a depth of 0.5 mm below the surface of the brain, using a pulled glass pipet connected to a nanoliter injector (WPI, Nanoliter 2010 with Micro 4 controller). After injection, the pipet was left in place for 1 minute before being withdrawn.
Histology
Seven weeks after surgery and virus injection, animals were anesthetized with an overdose of Ketamine/Xylazine and transcardially perfused with 4% paraformaldehyde. Tissues were left to fix overnight at 4°C in 4% paraformaldehyde before transferring to PBS the next day. The brainstem and cervical spinal cord were isolated and embedded in 2%agarose dissolved in PBS. 100 μm sections were cut on a vibratome and mounted in Vectashield (with DAPI) (Vector Laboratories, H-1200-10). Spinal cord sections (levels C3–C5)were imaged on an Olympus FV1000 confocal microscope.
Viral dilution and transduction of HT1080 cells
To prepare the virus, a purified and concentrated AAV8-GFP viral stock (1.4 × 1014genome copies (GC)/mL) was diluted 1:100 for incubation in either 4°C or room temperature citrate-phosphate buffer at pH 7.4, pH 6.4, pH 5.4, or pH 4.4 supplemented with 150 mM NaCl. Virus diluted in 4°C buffer was incubated for 24 hours before addition to culture, and virus diluted in room temperature buffer was incubated for 1 hour before addition to culture. Freshly split HT1080 cells were plated at an initial density of 500 cells per well in 96-well plates (Falcon, 353075) in 100 μL DMEM (Gibco, 11996-040) supplemented with 10% (v/v) fetal bovine serum (Gibco,16000-044) and 1× antibiotic/antimycotic (Gibco, 15240-062). Plates were kept at 37°C in 5% CO2incubators for the duration of the experiment. Incubated virus was added to cells 24 hours after the HT1080 cells were plated. Each preparation of incubated AAV8-GFP was added to cells within each plate in quintuplicate, for a final total dilution of 1:2500(5.6 × 109GC/well). This titer dilution provided negligible levels of cell death at this cell concentration while providing efficient levels of transduction (30–40%) in basal conditions.Cells were grown for an additional 48 hours after addition of AAV8 to permit GFP expression.
Nuclear staining
After a total of 3 days in culture (two days after addition of AAV8), HT1080 cell plates were removed and immediately fixed in darkness at room temperature in 4%paraformaldehyde, 4% sucrose in PBS pH 7.4 for 20 minutes.Plates were washed three times with PBS and stained with Hoechst dye (Invitrogen, H3570) for nucleus visualization.
GFP quantification
Plates containing fixed AAV8-GFP-infected HT1080 cells were imaged using a Cellomics ArrayScan VTI (Thermo Scientific Cellomics, Pittsburgh, PA, USA) to automatically image nine adjacent fields in each well of the plates with a 5× objective lens for nuclear identification (Hoechst) and reporter gene(eGFP) expression. Nuclei and GFP expression were detected automatically using the Target Activation Bioapplication(version 3.5). Hoechst DNA staining was used to evaluate nuclear size and shape in order to trace a nucleus mask. A cell body mask was drawn by extending the nucleus mask 2 μm outward. The cell mask tracing algorithm was designed to prevent overlap of cell masks between neighboring cells. A cell mask was rejected if the cell had an irregular (stretched)shape that would have caused poor tracing, or if cell neighbor density caused significant overlap of cell mask traces. Cells were scored GFP+if their log-transformed intensity values were ≥ the mean plus 3 standard deviations of the logtransformed intensity values of cells in wells from the same plate with no virus added. Intensity measurements of GFP+cells were calculated as the average pixel value of pixel intensities within the cell mask trace.
Statistical analysis
One-way analysis of variance (ANOVA) was used to examine the effect of pH on the dependent variables %GFP+cells and GFP+intensities. F statistics andP-values are reported in the text, and findings of significance were followed by multiple comparison of means testing with Tukey’spost hoccorrection when relevant. The no virus control was not included during ANOVA calculations. Data are represented as individual biological replicate values with corresponding group means and 95% confidence intervals.
Results
AAV8 transduction capacity is decreased by prior exposure to low pH
Among the AAV serotypes, we chose to test AAV8 due to its widespread use in our lab and others to effectively transduce neuronsin vivoas well as in primary cell culture. In initial animal experiments designed to explore the use of a novel test compound, we observed that transduction by AAV8 injected into the sensorimotor cortex of mice was reduced when virus was prepared using acidic PBS as the diluent. In these pilot studies, mice received a unilateral pyramidotomy to transect the corticospinal tract on one side as part of a series of experiments examining axon sprouting in the spinal cord after a lesion. AAV8 packaged with an UbC promoter-driven eGFP recombinant plasmid (AAV8-GFP) was then injected into the contralateral cortex to visualize collateral axon sprouting in the spinal cord (Figure 1A). Mice that received AAV8-GFP prepared 24 hours previously in PBS at pH 7.4 showed excellent labeling of axons in spinal cord tissue sections (Figure 1B). However, mice that received AAV8-GFP prepared in PBS at pH 6.0 had dramatically reduced labeling of axons (Figure 1C), suggesting that a reduction in diluent pH was associated with an attenuation of viral transduction ability.
To investigate this hypothesis, we tested the ability of AAV8 to transduce HT1080 cells following incubation in acidic diluent, to simulate co-injection of AAV8 in a vehicle used to administer drugs that are soluble at low pH. AAV8 has high tropism for HT1080 cells, which are a common cell line for AAV particle stock titering and are frequently used to assess viral transduction before use in other cell types. In our experiments, freshly split HT1080 cells were plated in a 96-well plate in quintuplicate, and treated 24 hours later with prior-incubated AAV8-GFP at an MOI estimated at 5.6 × 106prior to pH treatment. Specifically, we incubated AAV8 in citrate-phosphate buffer diluent at pH 7.4, pH 6.4, pH 5.4, or pH 4.4 to assess a pH range relevant to endosomal trafficking and therefore inclusive of pH-activated endosomolytic mechanisms. Incubations at the various pHs were carried out either at room temperature for 1 hour or 4°C for 24 hours to evaluate effects of temperature and time on AAV8 transduction. The expression of exogenous GFP in each cell was used to assess transduction; some wells received pH 7.4 vehicle without virus as a negative control (see Methods). The cells were fixed 48 hours after addition of virus and imaged to quantify the expression of GFP using high-content analysis(Figure 2).
A one-way ANOVA with apost hocTukey’s correction revealed that HT1080 transduction by AAV8-GFP vectors was significantly affected by prior incubation at acidic pHs for 1 hour at room temperature (F(3,12)= 8.96,P= 0.00218) (Figure 3A). The greatest transduction efficiency occurred at pH 7.4,and increasingly acidic incubation pHs were associated with decreases in transduction efficiency (pH 7.4vs. pH 6.4,P=0.026; pH 7.4vs. pH 5.4,P= 0.008; pH 7.4vs. pH 4.4,P=0.002). The greatest decrease in transduction efficiency (35%)occurred between pH 7.4 and pH 6.4. Subsequent decreases in pH were associated with modest decreases in transduction efficiency, decreasing by an additional 11% and 14% from pH 6.4 to 5.4 and pH 5.4 to 4.4, respectively. Similar decreases in transduction efficiency were observed after prior incubation at 4°C for 24 hours, though a one-way ANOVA did not detect any significant differences (F(3,12)= 1.90,P= 0.184).
In addition to pH altering transduction efficiency, a previous report suggested that acidic pH can cause AAV particles to aggregate in high concentrations; this was hypothesized to cause an increase in the number of virus particles internalized per cell (Johnson and Bodily, 1975). To evaluate the possibility of AAV8 particles being internalized in aggregate during endocytosis, we assessed the intensity of GFP signal in GFP+cells as a proxy for the relative number of eGFP copies internalized per transduced cell. A one-way ANOVA revealed a significant effect of pH on soma intensity after room temperature incubation for 1 hour (F(3,12)= 3.89,P= 0.0375)(Figure 3B). However, instead of an increase in soma GFP fluorescence intensity, post-hoc testing revealed a significant decrease in intensity between pH 7.4 and pH 4.4 (P= 0.031).Similar intensity level changes were found after incubating AAV8-GFP at 4°C for 24 hours, though one-way ANOVA testing did not reveal differences between individual pHs (F(3,12)= 1.69,P= 0.223).
Taken together, these results suggest that the effective transduction by AAV8 particles is decreased by prior environmental exposure to acidic pH, with the sharpest drop in transduction efficiency occurring between pH 7.4 and pH 6.4.
Discussion
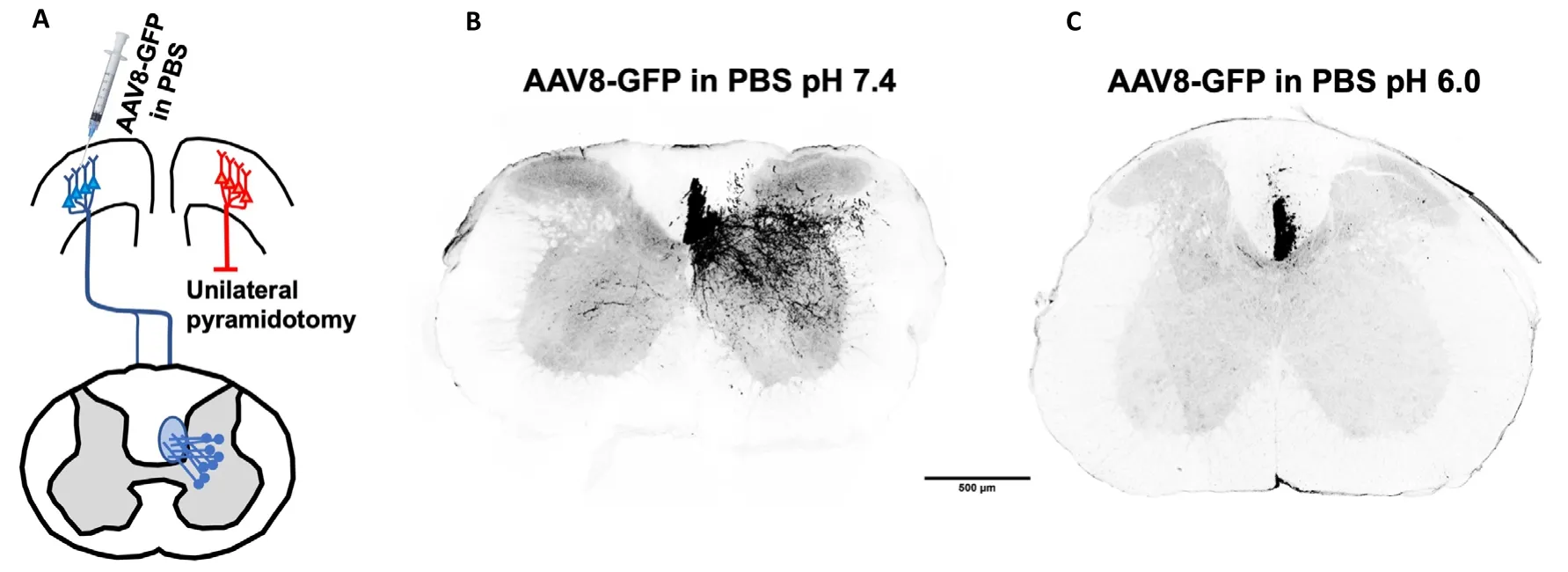
Figure 1 | Incubation of AAV8-GFP at pH 6.0 for 24 hours at 4°C attenuates fluorescent labeling of corticospinal tract sprouts in the spinal cord.
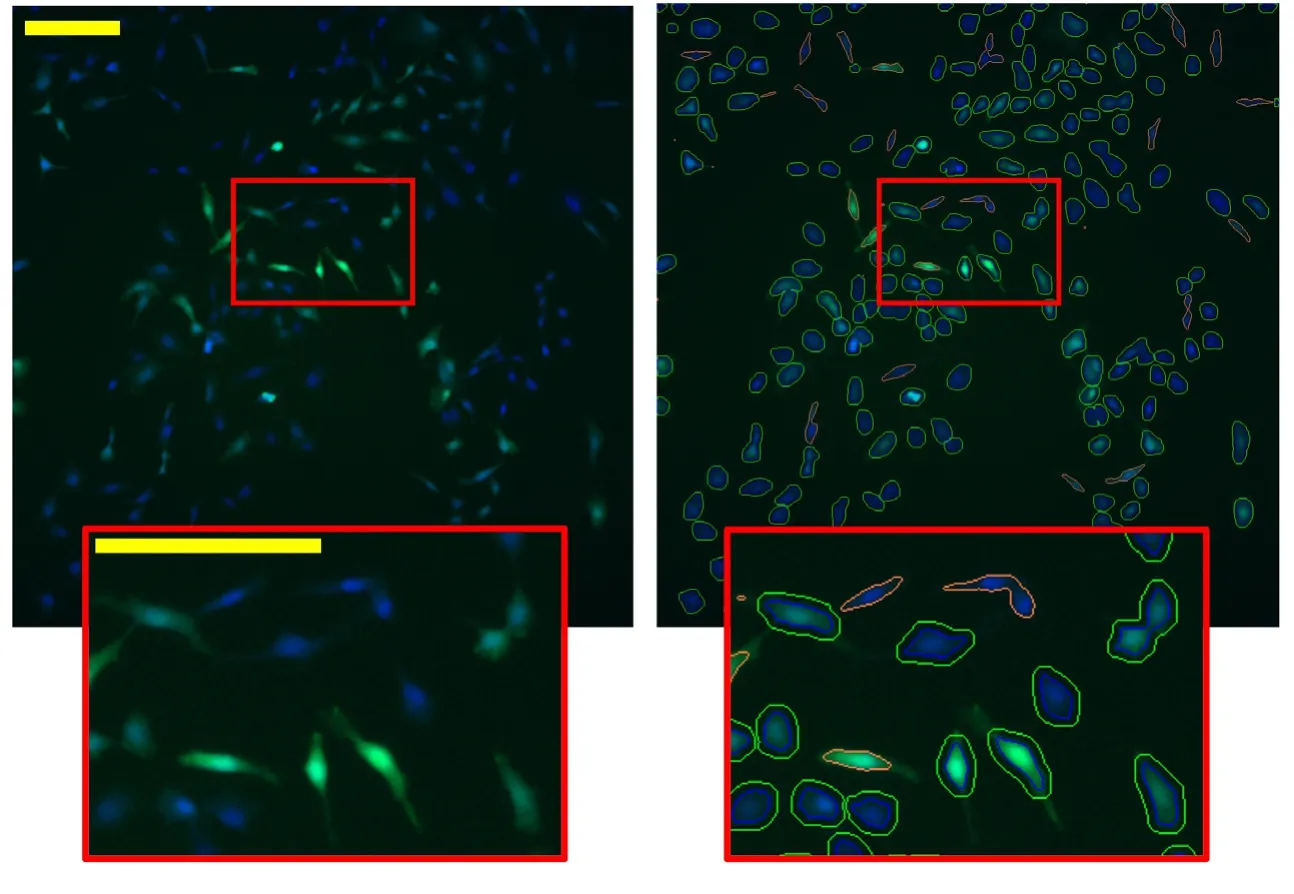
Figure 2|Demonstration of high-content analysis method of GFP expression in HT1080 cells transduced with AAV8-GFP.

Figure 3 | Incubation at lower pH reduces transduction efficiency of AAV8-GFP.
We initially observed that incubation of AAV8 in acidic PBS(pH 6.0) for 24 hours at 4°C rendered the virus particles less effective atin vivotransduction of mouse cortical neurons for axon tracing compared to incubation in neutral PBS (pH 7.4). This observation contrasted with a previous report that AAV1 transduction was unaffected when virus was admixed with a pH 5.5 solvent compared to PBS vehicle, and introduced via intramuscular administration in the mouse hindlimb (Gruntman et al., 2015). Since mixing of the AAV into the vehicle diluent in the previous study occurred just prior to injection, we hypothesized that AAV transduction is decreased by a period of incubation in an acidic diluent. In the current study with AAV8, we sought to investigate the effect of incubating AAV8 particles in endosomal-like acidic pHs at different temperatures for different durations.
Overall, we observed that decreasing the incubation pH for periods as short as 1 hour was deleterious to the transduction ability of AAV particles, as indicated by a significant reduction in transduction efficiency and fluorescence intensity of transduced cells. Additionally, reducing the incubation temperature to 4°C, but incubating for a longer period (24 hours), did not guard against reductions in transduction ability nor further diminish it. These results are in contrast to a previous report that decreases in pH affect AAV transduction beginning at pH 5.8, but required an incubation period of at least 24 hours at room temperature; no decline was observed in a period of 2 hours at any pH tested (Potter et al., 2014).During the preparation of the present manuscript, a new study was published, which showed that AAV8 transduction declines after prior incubation at room temperature and below pH 5.5,though no incubation period less than 24 hours was tested(Lins-Austin et al., 2020). In this recent study, transduction ability was temperature independent within the range of temperatures and pHs tested (Lins-Austin et al., 2020), which aligns with our observations that AAV8 transduction efficiency was similar with virus incubated for 24 hours at 4°C compared to 1 hour at room temperature.
Additionally, we observed a decrease in transduction efficiency of 35% that occurred with pre-incubation at pH 6.4, in contrast with previous reports (Bachmann et al., 1979; Hansen et al.,2001; Potter et al., 2014; Lins-Austin et al., 2020). It is possible that this pH triggers an event that facilitates reduction in transduction capacity of AAV8 particles when it occurs prematurely (i.e., outside an endosome), though it is unclear why a similar decrease in AAV transduction was not observed in previous studies. It is known from previous studies of AAV capsids that starting by pH 6.0 and continuing to pH 4.0,there is a pH-triggered change in AAV capsid conformation that is necessary, but not sufficient, to enable endosomal escape (Sonntag et al., 2006; Nam et al., 2011). Similarly,low pH-triggered structural changes in AAV capsid proteins are believed to activate an autolytic function that has been hypothesized to be associated with eventual virus uncoating and endosomal escape mechanisms (Salganik et al.,2012). pH-dependent conformational changes and proteolytic processing of capsid or capsid-related proteins are features common to the lifecycle of many types of viruses (Johnson and Vogt, 2010). It is plausible that these mechanisms are necessary aspects of the AAV virus lifecycle but partially disable viral transduction when prematurely activated outside the endosome, even if potentially reversible (Venkatakrishnan et al., 2013).
Further study of the nature of AAV transduction is necessary to determine causes of inactivation after extra-endosomal encounter of low-pH environments, and to identify potential mechanisms to limit AAV inactivation at acidic pHs. Until a strategy to prevent low pH inactivation of AAV particles is developed, we suggest that diluting AAV in acidic solvents should be minimized to the extent possible. In general,investigators should consider the possible ramifications of diluting AAV in non-physiologic solvents when preparing it for experimental or therapeutic administration.
Acknowledgments:We acknowledge and appreciate the excellent technical assistance provided by Yania Martinez (Miami Project Viral Vector Core), Yan Shi (Miami Project High Content Screening Core), and Melissa Muñoz.
Author contributions:Experiments were conceived by jAL, KMM, jLB, and VPL and performed by jAL and KMM. Data analysis was done by jAL and KMM.The manuscript was drafted by jAL, with editing and contributions from all authors. All authors have given approval to the final version of the manuscript.
Conflicts of interest:The authors have no conflicts of interest to declare.
Financial support:This work was supported by grants to jLB and VPL from the National Institutes of Health (NS100531), the Craig H. Neilsen Foundation(598684), and the Miami Project to Cure Paralysis. VPL holds the Walter G.Ross Distinguished Chair in Developmental Neuroscience. Funding sources had no role in the study design, conduct of research, or preparation and submission of this article.
Institutional review board statement:All animal-related work was performed in accordance with the University of Miami Institutional Animal Care and Use Committee, USA (Protocol #18-108-LF, approved on july 12, 2018).
Copyright license agreement:The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement:Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Plagiarism check:Checked twice by iThenticate.
Peer review:Externally peer reviewed.
Open access statement:This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak,and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
Open peer reviewer:Chengwen Li, University of North Carolina, USA.
Additional file:Open peer review report 1.
- 中国神经再生研究(英文版)的其它文章
- Therapeutic effectiveness of a single exercise session combined with WalkAide functional electrical stimulation in post-stroke patients: a crossover design study
- Enriched environment boosts the post-stroke recovery of neurological function by promoting autophagy
- Surgical intervention combined with weight-bearing walking training improves neurological recoveries in 320 patients with clinically complete spinal cord injury:a prospective self-controlled study
- Recognition of moyamoya disease and its hemorrhagic risk using deep learning algorithms: sourced from retrospective studies
- D-serine reduces memory impairment and neuronal damage induced by chronic lead exposure
- An integrative multivariate approach for predicting functional recovery using magnetic resonance imaging parameters in a translational pig ischemic stroke model