Complete Genome Sequencing and Analysis of Rehmannia Mosaic Virus Isolate from Shanxi Province
Wang De-fu, Zhang Xi-mei, Guo Shang, Shen Shao-fei, Long Dan-dan, Li Ling-yu, and Niu Yan-bing
1 College of Life Sciences, Shanxi Agricultural University, Taigu 030801, Shanxi, China
2 Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100193, China
3 Shanxi Institute for Functional Food, Shanxi Agricultural University, Taiyuan 030031, Shanxi, China
Abstract: Using double-stranded RNA (dsRNA) technology and sequence-independent amplification (SIA), the molecular identification on infected Rehmannia glutinosa in the field with mosaic symptoms was performed and the whole-genome of the Rehmannia mosaic virus (ReMV) Shanxi isolate (ReMV-SX) was sequenced.Sequencing analysis showed that the virus that infected Rehmannia glutinosa was Rehmannia mosaic virus (ReMV).The full-length of the obtained ReMV-SX sequence (GenBank accession no.JX575184) was 6 395 nt, containing four open reading frames (ORFs).The sequence homology analysis of the complete nucleotide sequence showed that ReMV-SX was 93.8%-97.0% homologous to ReMV in Tobamovirus subgroup I, while only 49.8%-58.9% homologous to the isolates in subgroups II and III of the same genus.Phylogenetic analysis showed that ReMV-SX and ReMV-Henan formed a separate branch and had the closest genetic relationship.The results laid the foundation for ongoing researches in the taxonomic status and evolution of ReMV and for further investigating the pathogenic mechanism of ReMV infecting Rehmannia glutinosa.
Key words: Rehmannia mosaic virus (ReMV), Rehmannia glutinosa, whole-genome amplification, sequence analysis
Introduction
Rehmannia glutinosais a perennial herbaceous plant of theScrophulariaceaefamily and is mainly produced in Henan, Shanxi, Shandong and Gansu provinces.It possesses a wide variety of roles, such as kidney tonifying, anti-ageing and anti-tumour effects (Wanget al., 2006; Zhanget al., 2010).In recent years, as the medicinal value and health benefits ofRehmannia glutinosaare demonstrated and utilised, the problems of viral infection and disease during its production have got growing attention.Rehmannia glutinosais mostly propagated vegetatively using tuberous roots.During long-term asexual propagation, viruses continue to accumulate and cause more damage toRehmannia glutinosa(Chenet al., 2007; Wenet al., 2000).Tian (1962) first isolate a dihuang degeneration virus (DDV) fromRehmanniaglutinosaand identify it as a member ofTobamovirus.Later, Puet al.(1983) obtain an isolate similar to tobacco mosaic virus (TMV) fromRehmannia glutinosa, and by comparing its biological characteristics with other TMV isolates, they find differences in the responses among the isolates in their hosts.Rehmannia glutinosaisolate causes no symptoms, but can infect the petunia and eggplant, while the tobacco isolate caused systemic mosaic symptoms on the leaf.Rehmannia glutinosaisolate causes systemic mosaic symptoms in napa cabbage, while the tobacco isolate causes the infection but not symptoms.Rehmannia glutinosaisolate leads to attenuated protection in the tomato.Currently, there are no studies that have comprehensively analyzed such isolates at the molecular level.Yu and Yang (1994) isolate four viruses from infectedRehmannia glutinosain Henan Province, among which a baculovirus resembled TMV, but no clear identification is performed.Houet al.(2007) obtain a TMV isolate fromRehmannia glutinosain Henan Province and find that this isolate causes necrosis spot symptoms in hosts includingNicotiana glutinosa,Nicotiana rusticaandDatura stramonium.It also exhibits serological reactions with antibodies of common strains of TMV.Zhanget al.(2004) later use RT-PCR to amplify the coat protein (CP) and movement protein (MP) from this isolate.
Sequence analysis shows that CP and MP of this isolate have low homology with published TMV strains and have major differences from other strains.Through the construction of a phylogenetic tree using amino acid sequences of CP and MP in different strains, this isolate is predicted to be a new TMV strain and it is named Rehmannia mosaic virus (ReMV).The complete genome sequence of ReMV is also obtained.Later, Kubotaet al.(2006) obtain a Japanese ReMV isolate, which is the second report of ReMV sequence.In October 2011, during a disease investigation inRehmannia glutinosaproduction region in Xinjiang County of Shanxi Province, it was found that manyRehmannia glutinosaplants exhibited mosaic symptoms.Molecular identification using dsRNA and sequenceindependent amplification (SIA) methods showed that the pathogen that infectedRehmannia glutinosawas ReMV.The complete genome sequence of ReMVSX was obtained and analyzed, which provided a foundation for studying the pathogenesis of ReMV inRehmannia glutinosaand for effectively controlling viral diseases inRehmannia glutinosa.
Materials and Methods
Plant materials
TwentyRehmannia glutinosasamples with suspected viral infections were collected atRehmannia glutinosacultivation base in Xinjiang County, Shanxi Province.Leaves of most infected plants exhibited typical mosaic symptoms (Fig.1).

Fig.1 Symptoms of infected Rehmannia glutinosa in field
Plant viral dsRNA extraction
InfectedRehmannia glutinosaleaves with typical mosaic symptoms were selected for the extraction of plant pathogen dsRNA as described by Niuet al.(2011) and Tzanetakis and Martin (2008).
SIA and RT-PCR detection
With extracted dsRNA as the template, polyT primer (M4-T) was used to amplify viral cDNA.SIA was performed using M4 primer and the detailed reaction system and procedures were followed by Niuet al(2015).Upon identification as ReMV, specific primers were designed based on the conserved region in ReMV isolates published in NCBI, and RT-PCR amplification of the viral complete sequence was conducted.Healthy plants were used as controls.The primer sequence used in this study are listed in Table 1.

Table 1 Primer pairs used in complete sequence amplification of ReMV-SX
Cloning of PCR product and sequence analysis
Amplified PCR product was recovered and inserted into pMD19-T vector, which was then transformed intoE.coliDH5α.After colony PCR identification, three positive clones were selected for sequencing at Sangon Biotech (Shanghai) Co., Ltd.The sequencing results were subjected to a BLAST search in NCBI to look for homologous sequences.DNAStar software was used for sequence assembly.DNAMAN 6.0 was used for comparing the nucleotide and amino acid sequence similarities between the obtained sequence and the representative strains from the three subgroups ofTobamovirusgenus.MEGA 7.1 was used to construct the phylogenetic tree with 1 000 replicates bootstrap analysis by the neighbour-joining method.
Results
SIA
Ten infectedRehmannia glutinosasamples were randomly selected for SIA with M4 as a random primer.cDNA synthesized from reverse transcription with extracted dsRNA was used as the template.A target band of 820 bp was obtained in eight samples (Fig.2), and 80% of samples tested positive.A band of the same size was not amplified from healthy samples.Sequence alignment using GenBank demonstrated that the sequencing result of the target fragment exhibited the highest identity, with 95.3%-98.5% identity with the nucleotide sequence of ReMV.This preliminarily confirmed thatRehmannia glutinosawas infected by ReMV, and it was named ReMV-SX isolate.
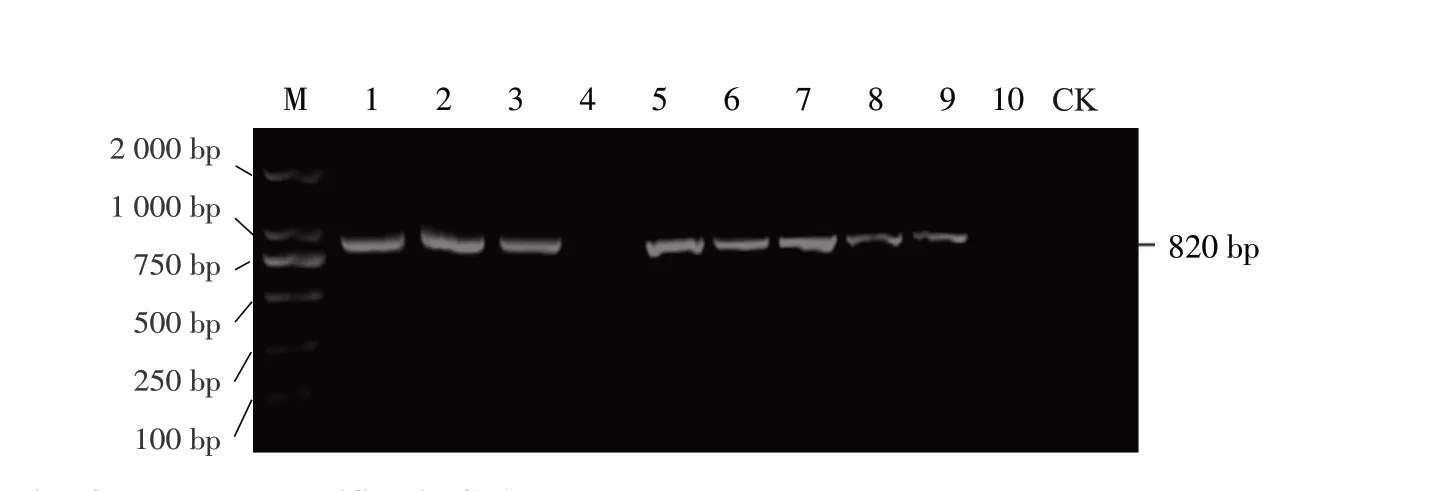
Fig.2 Electrophoresis of products amplified in SIA M, DL 2000TM DNA Marker; Lane 1-10, Diseased plants; Lane CK, Healthy plant.
ReMV-SX whole-genome sequence amplification and structural analysis
Took the fragment cloning approach, used the extracted viral dsRNA as a template, designed four pairs of specific primers using ReMV conserved sequence from GenBank (Table 1), and performed amplification.Among the primers used, ReMV-F1 and ReMV-R1 resulted in amplification of a 2 900 bp fragment (Fig.3A), ReMV-F2 and ReMV-R2 amplified a 2 600 bp fragment (Fig.3B), ReMV- F3 and ReMV-R3 amplified a 1 100 bp fragment (Fig.3C), and ReMV-F4 and ReMV-R4 amplified a 750 bp fragment (Fig.3D).5'- and 3'-RACE of ReMV-SX obtained 1 900 bp and 1 500 bp fragments at the 5' and 3' end, respectively (Fig.3E, F).DNAStar software was used for the assembly of the sequencing results of the six fragments obtained from amplification.A 6 395 nt complete genome sequence of ReMV-SX was obtained (GenBank accession No.JX575184).Sequence analysis showed that the genome sequence of ReMV-SX contained a 71 nt 5' UTR, 204 nt 3' UTR and four ORFs in the coding region.Among the ORFs, ORF1 encoded a 1 116 amino acid replicas (126 ku), ORF1a encoded a 1 615 amino acid replicas readthrough protein (180 ku), ORF3 encoded a 267 amino acid MP (30 ku), and ORF4 encoded a 159 amino acid CP (17.5 ku).ORF1a and ORF3 contained a 17 nt overlapping region (Fig.4).These results were consistent with the structure of the coding region in the genome of ReMV isolate fromTobamovirussubgroup I.
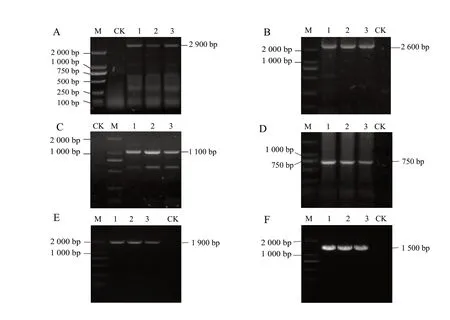
Fig.3 Electrophoresis of RT-PCR products for ReMV-SX A-D, Primer set ReMV-F* + ReMV-R*, where * represents primer sets 1 through 4 in panels A through D, respectively; E 5' RACE; F 3' RACE.M, DL 2 000TM DNA Marker; Lane 1-3, Diseased plants; Lane CK, Healthy plant.

Fig.4 Structural representation of ReMV-SX complete genome
Sequence homology between ReMV-SX and viral isolates in Tobamovirus genus
Complete nucleotide sequence homology analysis showed that ReMV-SX shared the highest homology with ReMV isolate in Henan (ReMV-Henan) inTobamovirussubgroup I (97%), followed by the Japanese ReMV isolate (ReMV-Japanese) (93.8%).The homology between ReMV-SX and TMV strains in subgroup I was above 80%.Among them, ReMV-SX exhibited the highest homology with Guizhou isolate (TMV-Zhenfeng) (83.8%).The homology between ReMV-SX and subgroup I tomato mosaic virus (ToMV) isolate in Australia (ToMV-Queensland) and in Xinjiang County (ToMV-XJT-1) was both 78.2%.The homology with ORSV, TMGMV and PMMV was all below 70%.The homology between ReMV-SX and subgroups II and III isolates was 49.8%-58.9%, among which the homology with subgroup II CGMMVKW was the lowest.Sequence homology analysis of the amino acid sequences of the 126 ku protein, 180 ku protein, MP and CP, encoded by the virus, was conducted.Results showed that these four proteins exhibited the highest homology with those in ReMVHenan: 97.6%, 97.7%, 98.9% and 99.4%, respectively.These four proteins exhibited the second highest homology with those in ReMV-Japanese.The 126 ku protein and 180 ku protein shared over 90% homology with TMV and ToMV strains, while CP shared over 90% homology with TMV, but only 81.1% homology with ToMV.MP had even lower homology with TMV and ToMV strains, indicating that MP had the highest variation.These four proteins were not closely related to the isolates inTobamovirussubgroups II and III, with subgroup II being the most distantly related (Table 2).
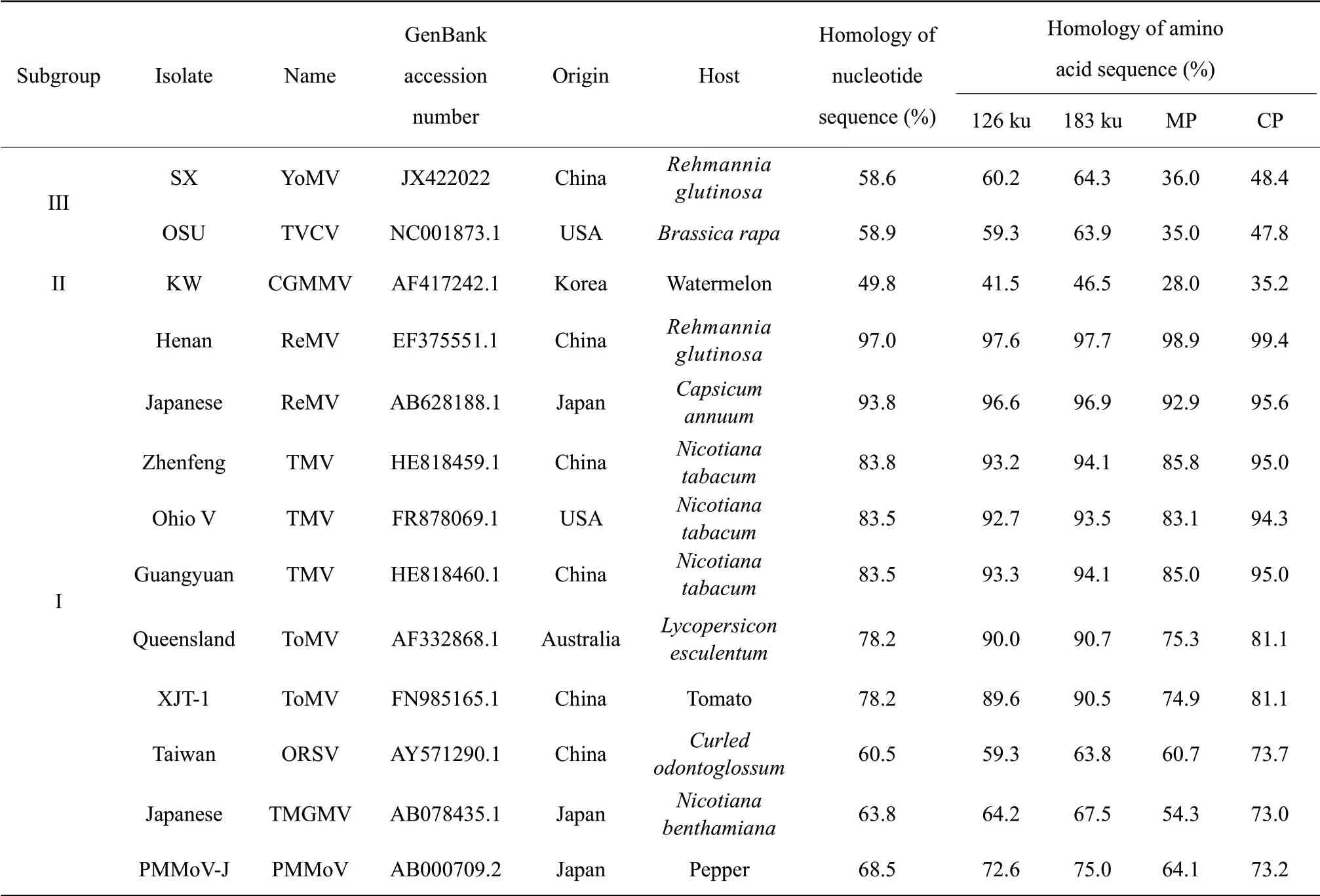
Table 2 Comparison of nucleotide and amino acid sequence homology between ReMV-SX and reported isolates in Tobamovirus
Phylogenetic analysis of ReMV-SX
MEGA 7.1 software was used to construct a phylogenetic tree with the amino acid sequence from the complete genome of ReMV-SX and reported isolates fromTobamovirusgenus.The phylogenetic tree (Fig.5) showed that ReMV-SX and ReMV-Henan were the most closely related, and they formed a separate small branch.ReMV-SX formed a big branch with TMV, ToMV, PMMoV and TMGMV strains in subgroup I, which were less closely related.They were distantly related to ORSV, CGMMV and YoMV isolates.These results further showed that ReMV-SX was a member of the subgroup I ofTobamovirus.
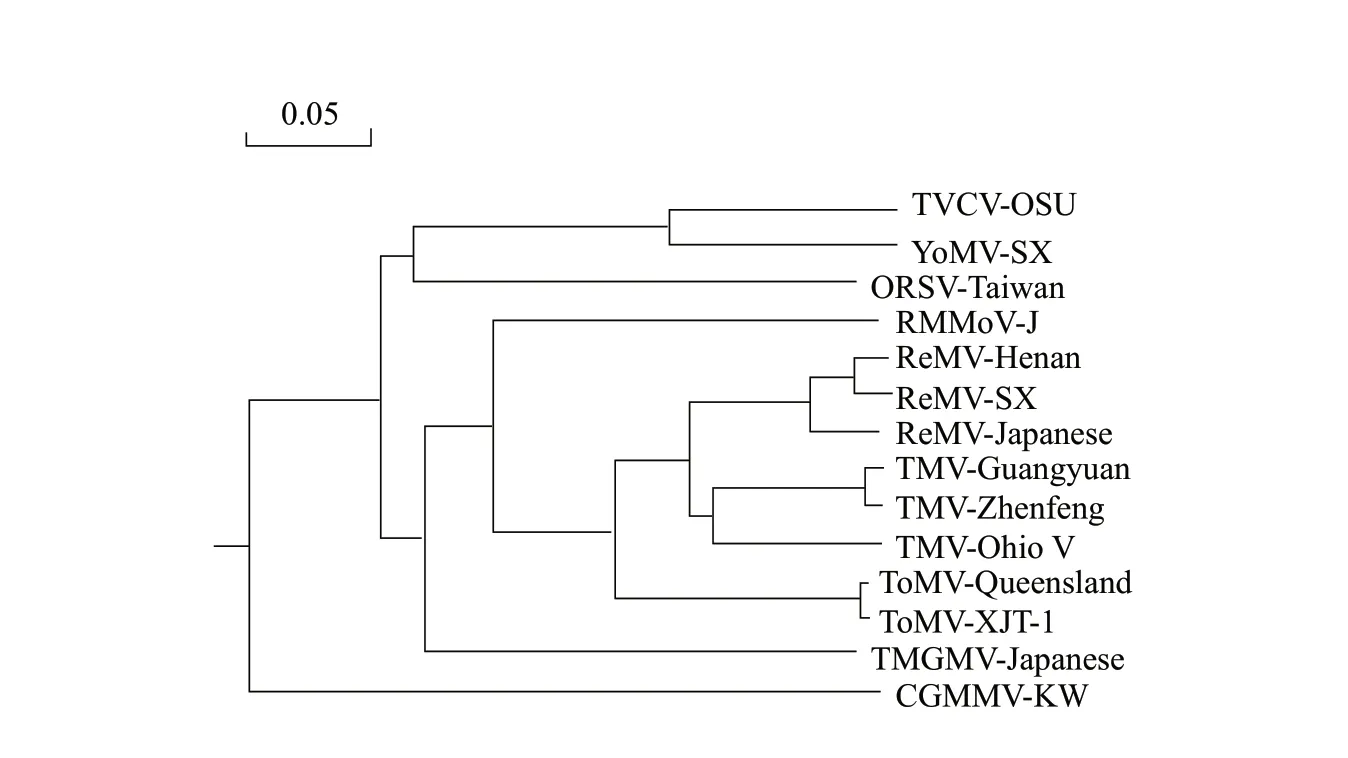
Fig.5 Phylogenetic tree constructed using ReMV-SX and reported isolates in Tobamovirus based on amino acid sequences
Discussion
Plant virus was known as the "plants cancer", which had caused enormous damage to crops (Tonget al., 2005).Almost every agricultural and economic crop had experienced damage from one or more viruses (Liu, 2005).Rehmannia glutinosawas an important raw material for traditional Chinese medicine preparation.The increasing demand forRehmannia glutinosawas forcing large-scale cultivation.However, during the process ofRehmannia glutinosacultivation, its limited propagation methods and improper agricultural practices resulted in several problems with diseases caused by viruses.So far, the major types of viruses that infectedRehmannia glutinosaincluded TMV (Duet al., 2013), ToMV (Duet al., 2013), ReMV (Houet al., 2007), broad bean wilt virus 2 (BBWV2) (Zhouet al., 2010), cucumber mosaic virus (CMV) (Zhouet al., 2010), carnation Italian ringspot virus (CIRV ) (Gaoet al., 2009), potato virus X (PVX) (Gaoet al., 2009) and Youcai mosaic virus (YoMV) (Wanget al., 2016).However, until now, the complete genome sequences of only two ReMV isolates had been reported, because the lack of ReMV sequence data had made it difficult to study the phylogenetics and evolution of ReMV.In this study, dsRNA technology and SIA were utilized to conduct molecular identification on infectedRehmannia glutinosa, and ReMV from infected leaves was detected.Comparison of amino acid sequence identity and phylogenetic analysis showed that ReMV isolate from Shanxi (ReMV-SX) belonged to the subgroup I ofTobamovirusand exhibited the highest homology with Henan ReMV isolate (ReMV-Henan) in the same subgroup.Meanwhile, the complete genome sequence of ReMV-SX was obtained.Structural analysis showed that ReMV-SX genome contained four ORFs that encoded four proteins.This result had established a foundation for future studies in the classification and evolution of ReMV in China.
This study took the approach of combining dsRNA and SIA to detect and identify plant viruses, which allowed researchers to avoid the disadvantages of using traditional RT-PCR, including the laborconsuming total RNA extraction and the interference caused by polyphenols and polysaccharides derived from the plant host.This method effectively solved the problems brought by low levels of viruses and unstable viral particles (Tzanetakisetal., 2005; Balijjaet al., 2008).SIA was fast, simple and highly sensitive; it produced reliable results and could be used in the detection of different virus species, as well as the identification of mixed infections by different strains of the same virus.
In this study, a 6 395 nt complete genome sequence of ReMV-SX was obtained.It contained four ORFs, with the second half of ORF3 (about 0.9-1.0 kb from the 3' end) containing GAA/GTT repeats.Combining this analysis with the description and reference for identifyingTobamovirusby Gibbs(1986), it was predicted that this could be the initiation site for viral particle assembly.A comparison between the 4 404-4 450 motif of ReMV-SX and other viruses inTobamovirusgenus showed that the specific conserved sites in the 4 404-4 450 motif of ReMV-SX was identical to the 4 404-4 450 motif of ReMV-Henan and ReMV-Japanese, indicating that ReMV-SX belonged toTobamovirus.Amino acid sequence homology analysis was conducted on the 126 ku protein, 180 ku protein, MP and CP of ReMV-SX, and the results showed that these four proteins exhibited over 97% homology with ReMV, while the 126 ku protein and 180 ku protein had over 90% homology with TMV and ToMV strains.CP was over 90% homologous with TMV, but only 81.1% with ToMV.MP had low homology with TMV and ToMV, indicating higher variations in MP.Therefore, MP might be the main factor that determined the differences in host range.The amino acid sequence of MP from ReMV-SX, which had the most variations, was aligned to those in the three closely related strains: ReMV-Henan, ReMV-Japanese and TMV-Zhenfeng.Results indicated that there was only three amino acids difference between ReMV-SX and ReMVHenan.There were more differences in amino acids from ReMV-Japanese and TMV-Zhenfeng, and they were mostly located at the 3' end of MP.Whether these different amino acids could lead to a wider range of infection in the host, or cause differences in its evolution, needed to be further investigated.
Conclusions
It was discovered that the diseasedRehmannia glutinosawas infected by aRehmanniamosaic virus, and the obtained viral genomic RNA was 6 395 nt in length.Sequence alignments and phylogenetic analysis showed that ReMV-SX shared the highest identity with ReMV-Henan isolate and had the closest evolutionary relationship, which might provide valuable information for ReMV genetic diversity analysis.
 Journal of Northeast Agricultural University(English Edition)2021年3期
Journal of Northeast Agricultural University(English Edition)2021年3期
- Journal of Northeast Agricultural University(English Edition)的其它文章
- Water Consumption Processes of Different Planting Models in Rice Production of Northeast China
- Optimum Combination Test of Erosion Gully in Black Soil Regions by Coal Gangue Reconstruction Soil
- Effect of Chilling Stress on Synthesis of Antioxidant Enzymes, Osmotic Adjustment Substances and Membrane Lipid Peroxide Levels in Two White Clover Cultivars
- Reversal Effects of Ivermectin and Moxidectin on Multidrug Resistance in C6/adr Cells in vitro
- Effects of DHRS3 in C2C12 Myoblast Differentiation and Mouse Skeletal Muscle Injury
- Effects of Lanthanum and Cerium on Seeds Fatty Acid Composition in Soybean
