Histone methylation in pancreatic cancer and its clinical implications
Xing-Yu Liu, Chuan-Hao Guo, Zhi-Yuan Xi, Xin-Qi Xu, Qing-Yang Zhao, Li-Sha Li, Ying Wang
Abstract Pancreatic cancer (PC) is an aggressive human cancer.Appropriate methods for the diagnosis and treatment of PC have not been found at the genetic level, thus making epigenetics a promising research path in studies of PC.Histone methylation is one of the most complicated types of epigenetic modifications and has proved crucial in the development of PC.Histone methylation is a reversible process regulated by readers, writers, and erasers.Some writers and erasers can be recognized as potential biomarkers and candidate therapeutic targets in PC because of their unusual expression in PC cells compared with normal pancreatic cells.Based on the impact that writers have on the development of PC, some inhibitors of writers have been developed.However, few inhibitors of erasers have been developed and put to clinical use.Meanwhile, there is not enough research on the reader domains.Therefore, the study of erasers and readers is still a promising area.This review focuses on the regulatory mechanism of histone methylation, and the diagnosis and chemotherapy of PC based on it.The future of epigenetic modification in PC research is also discussed.
Key Words: Pancreatic cancer; Epigenetics; Histone modification; Methylation; Demethylation; Clinical application
INTRODUCTION
Pancreatic cancer (PC) is a malignant tumor.The lack of adequate diagnostics for PC limits the efficacy of the few currently available treatment options.Current diagnostic methods include clinical biomarkers, imaging, biopsy,etc.To date, carcinoembryonic antigen 19 (CA-19) is the only PC clinical biomarker approved by the U.S.Food and Drug Administration[1], but the use of CA-19 is limited by its inadequate sensitivity and specificity[2,3].Percutaneous biopsy can result in micrometastases in younger patients who receive surgery, so it is only appropriate for inoperable patients[4].Current diagnostic methods are either inaccurate or limited.Conventional treatment methods for PC mainly include surgery, adjuvant chemotherapy, drug therapy, and radiation therapy[5].Surgery remains the most important treatment, followed by adjuvant chemotherapy[5].At present, only 15% to 20% patients can be surgically treated after diagnosis, and only 20% of the patients survive 5 years after receiving surgery[6,7].Regarding chemotherapy, gemcitabine and other drugs have proved effective for advanced and metastatic PC, but the development of drug resistance has limited the effectiveness[8].The survival rate of PC patients has not changed much in the past 40 years[8].The robust molecular biomarkers need to be developed for diagnosis and targeted therapies.
Cancer development is a complex process involving both genetic and epigenetic changes.Genome instability, regulated by both genetic mutations and epigenetic modifications, contributes to tumor progression[9].The concept of epigenetics itself is evolving with the increase of our knowledge of the molecular mechanism and regulation of gene expression.It is currently widely acknowledged that epigenetics is the study of alternations in gene expression patterns without changes in DNA sequences[10].Epigenetic modifications include DNA methylation, histone modification and non-coding RNAs.Epigenetic modifications present a new direction for cancer prevention, clinical diagnosis, and drug development.
Histone modification is one of the most important and complicated epigenetic regulatory mechanisms and is crucial in PC.Histone modification affects chromatin structure, transcription, and DNA repair process[11].Histone modification takes part in the regulation of chromatin architecture and specific loci regulation by recruiting cell-specific transcription factors and interacting with initiation and elongation factors[12].Histone modification also regulates the transcription process by influencing RNA processing[12].In terms of regulating chromatin structure, histone modification affects the higher-order chromatin structure by changing the interactions of histones with DNA, and/or by recruiting chromatin remodeling complexes indirectly[13-15].
Histone modifications include histone acetylation, methylation, phosphorylation,and ubiquitination.Histone methylation plays crucial roles in the development of PC.Therefore, this review focuses on histone methylation and its clinical applications.
HISTONE METHYLATION
Post-translational methylation in histone tails is a reversible dynamic chromatin modification.Methyl is dynamically added by methyltransferases-writers, removed by demethylase-erasers, and interpreted by effector proteins-readers[16].Readers recognize specific sites and promote the recruitment of transcription factors or chromatin-associated protein complexes and bind to histones to enable the localization of enzymes to specific targets[17].
Histone methylation takes place on the residues of arginine, lysine, and histidine.According to the amino acid residues modified, there are arginine residue methyltransferases and lysine residue methyltransferases[18].Histone arginine methylation is a universal post-translational modification, and aberrant histone arginine methylation is strongly associated with carcinogenesis and metastasis[19].Arginine residues may be differentially methylated by different types of protein arginine N-methyltransferases (PRMTs)[19].
The maintenance of the balance between histone methylation and demethylation is fundamental to normal cellular development and function[20,21].The break of the balance between histone methylation and demethylation results in oncogenesis and progression[21,22].Corresponding to writers, erasers can be divided into arginine residue demethylases and lysine residue demethylases.However, current research on histone arginine residue demethylases is limited, so we only discuss lysine residue demethylases.Based on their mechanism of action, lysine demethylases (KDMs) are classified into two families: Flavin adenine dinucleotide (FAD)-dependent and Fe(II)and 2-oxoglutarate (2OG)-dependent[23-25].
The appropriate localization of histone methyltransferase and histone demethylase is dependent on the readers that can recognize histone modifications[26].The reader can either be an independent polypeptide or a part of methyltransferase/demethylase[27-31].Some reader domains such as chromodomain[32,33], Tudor domain[34],tryptophan-aspartic acid 40 (WD40) domain[35,36] and plant homeodomain (PHD)finger[37,38] are well known.These reader domains all have their own specific structure[32-38].
HISTONE METHYLATION WRITERS IN PC
Among histone methylation, arginine and lysine methylation are the most widely studied in PC[39].Histone methylation is performed mainly by two types of writers:PRMTs and lysine methyltransferases (KMTs) (Table 1), with S-adenosyl-L-methionine(SAM) as the methyl donor[40].

Table 1 Histone methyltransferases play a major role in pancreatic cancer
PRMTs
PRMTs catalyze the transfer of a methyl group from SAM to a guanidino-nitrogen atom[41].Three types of methylated arginine residues are found in mammalian cells:Asymmetric dimethyl-arginine (ADMA), symmetric dimethyl-arginine (SDMA) and monomethyl-arginine (MMA)[41].Depending on their catalytic activity, PRMTs can be classified in three types[42].Type I PRMTs are responsible for producing ADMA,whose methyl groups are linked to the same guanidino nitrogen atom.Type II PRMTs add the methyl groups on each of the guanidino nitrogen atom of arginine symmetrically, producing SDMA[42].PRMT7 is the sole member of type III, exclusively catalyzing the formation of MMA[43].PRMT1 and PRMT5 function in PC[44,45].PRMT1 belongs to type I and PRMT5 belongs to type II[42].
PRMT1:PRMT1 is the founding member of the PRMT family, and PRMT1 can methylate histone H4 at arginine 3.This modification is associated with transcriptional activation[44].Upregulation of PRMT1 is found in various cancer types[46-49].PRMT1 is highly expressed in pancreatic ductal adenocarcinoma (PDAC) cells, and elevated PRMT1 levels predict a poor clinical outcome[44].PRMT1 promotes PC cell growthin vitroandin vivo[44].PRMT1 increases the β-catenin protein level in PC cells[44].Overactivation of β-catenin signaling promotes the growth, migration, and metastasis of PC cells[50-52].PRMT1 downregulation inhibits PC cell proliferation and invasion[53].GLI family zinc finger 1 (Gli1) is a substrate of PRMT1 in PDAC.Methylation of Gli1 at R597 by PRMT1 promotes its transcriptional activity by enhancing the binding of Gli1 to the promoters of its target gene[54].Interruption of Gli1 methylation attenuates oncogenic functions of Gli1 and sensitizes PDAC cells to gemcitabine treatment[54].
PRMT5:PRMT5 is a type II writer, responsible for symmetric demethylation[19,55].PRMT5 regulates the expression of a wide spectrum of target genes by modifying the chromatin structure or transcriptional machinery[56].Specifically, PRMT5 can catalyze the methylation of arginine 8 on histone H3 and arginine 3 on histone H4 (H4R3)[57].High expression of PRMT5 has been observed in various cancers.PRMT5 expression improves cancer cell survival, proliferation, migration and metabolism while inhibiting cancer cell apoptosis[55].PRMT5 expression is significantly upregulated in PC tissues[56].PRMT5 promotes tumorigenesis and PC cell proliferation[45].PRMT5promotes cell migration, invasion, and the epithelial-mesenchymal transition (EMT)viaactivating EGFR/AKT/β-catenin signaling in PC cells[45].PRMT5 knockdown reduces glucose intake and lactate levels in PC cells[56].PRMT5 can inhibit the expression of F-Box and WD repeat domain containing 7 (FBW7)[58,59].PRMT5 inhibits FBW7viasuppression ofFBW7gene promoter activity and elevation of cMyc stability, leading to tumorigenicity and aerobic glycolysis in PC cells[56].PRMT5 induces the phosphorylation of epidermal growth factor receptor (EGFR) at Y1068 and Y1172[45].Then PRMT5 activates phosphorylation of AKT and its downstream GSK3β[45].
KMTs
KMTs transfer one, two, or three methyl-groups to histone lysine residues[60].KMTs are categorized into two protein families based on catalytic domain sequence similarity and structural organization[61].Two major writers, SMYD3 (KMT3E) and EZH2 (KMT6), are related to PC[62,63].SMYD3 is a member of SET and MYNDdomain family[64].EZH2 belongs to the polycomb family[61].
SMYD3:SMYD3 belongs to the SET and MYND-domain family.SMYD3 can promote the proliferation, migration, and invasion of many types of cancer[64].SMYD3 is a protooncogene in liver, colon and breast tissue based on its high level of endogenous expression and cancer-related promoter polymorphism[65-70].SMYD3 is upregulated in PC.SMYD3 is positively associated with caspase-3 and MMP-2 expression in PC tissues[62].Active Src phosphorylates p300 in the nucleus, and then the complex binds to HMGA2 and SMYD3 genes.Therefore, HMGA2 and SMYD3 are regulated to promote PC cell migration and invasion[71].
EZH2:EZH2 is the enzymatic subunit of polycomb repressive complex 2 (PRC2), a complex that methylates lysine 27 of histone H3(H3K27) to promote transcriptional silencing[72].High expression of EZH2 protein has been associated with several cancers[73-75].EZH2 is overexpressed in PC[76].FBW7 interacts with EZH2 and downregulates EZH2viaubiquitination and degradation in PC cells[76].Downregulation of FBW7 induces high EZH2 protein expression and promotes tumor progression in PC[76].Long non-coding RNA (lncRNA)BLACAT1facilitates proliferation, migration, and aerobic glycolysis of PC cells by repressingCDKN1C viaEZH2-induced histone H3 lysine 27 trimethylation (H3K27me3)[77].EZH2 regulates the expression of miR-139-5pviaH3K27me3, and the EZH2/miR-139-5p axis participates in the progression of PC, whereby downregulation of EZH2 and upregulation of miR-139-5p repress the EMT and lymph node metastasis of PC[78].EZH2 can bind to the promoters of P15 and KLF2 to induce H3K27me3[79].LncRNASNHG15knockdown inhibits PC cell proliferation and tumorigenesis while inducing cell apoptosis, and theSNHG15-mediated oncogenic effect is partly by repressing P15 and KLF2 expressionviaEZH2-induced H3K27me3[79].
HISTONE METHYLATION ERASERS IN PC
The demethylation of arginine and lysine in histone tails is the two main forms of histone demethylation.Due to the large gaps in research on arginine demethylation,the main situation of KDMs in PC will be mainly described.KDMs can catalyze monomethyl, dimethyl or trimethyl labeling of histone lysine residues[12].There is some evidence that occurrence, development, and therapy of PC are all related to KDMs[80,81] (Table 2).

Table 2 Histone demethylase that plays a major role in pancreatic cancer
KDM1
Flavin-dependent KDMs are a subfamily of amine oxidases that catalyze the selective posttranslational oxidative demethylation of methyl lysine side chains within substrates[82].Two subtypes of KDMs, KDM1A and KDM1B, are related to PC[83,84].They are expressed at high levels in PC tissues.To date, the expression patterns and physiological functions of KDM1A/LSD1 in PC have not been fully elucidated.KDM1A and hypoxia inducible factor-1α (HIF1α) are the interaction partners of the homeobox protein PROX1[85,86].KDM1A acts synergistically with HIF1α in maintaining glycolysis[87].Compared with KDM1A, KDM1B/LSD2 lacks a "tower domain" and has a zinc finger domain in the N-terminal region, which makes KDM1B endowed with different biochemical properties[24,25,88].KDM1B is related to many important biological functions, including transcriptional regulation, genome imprinting, somatic cell reprogramming, DNA methylation, and signal transduction[89-92].The downregulation of KDM1B can inhibit PC cell proliferation and promote PC cell apoptosisin vitro[93,94].
JmjC domain-containing protein family
JmjC domain-containing (JMJD) protein family is a type of Fe (II) and α-ketoglutaratedependent dioxygenases.The JMJD protein family now consists of 33 members.There are 18 members with the ability to demethylate H3K4, H3K9, H3K27, H3K36, and H4K20[23,95-108].
KDM2B:KDM2B acts on H3K36 demethylation.KDM2B enhances the bypass of primary cell senescence by directly binding to tumor suppressor geneCDKN2Asites and demethylating histones, thereby guiding the recruitment of PRC2; thus, it plays an important role in cell cycle progression and senescence[109,110].KDM2B regulates cell proliferation, migration, and angiogenesis[111-113].KDM2B plays a crucial role in poorly differentiated PDAC, and there is an interaction between EZH2 and KDM2B[114].
KDM3A:KDM3A/JMJD1A, one member of the JMJD1 family, participates in transcriptional regulation by demethylating monomethyl or dimethyl H3K9[115,116].Since cells are heterogeneous in early PDAC tissues, new progress has been made in the study of PDAC morphology, which is specifically manifested by the upregulation ofDCLK1expression[117].KDM3A plays a key role in the upregulation ofDCLK1expression, and KDM3A expression inhibitors can inhibit the malignant properties of PDAC[118].
KDM4:The KDM4 subfamily consists of 12 demethylases including KDM4A, B, C,and D, which can catalyze the removal of inhibitory trimethyl marker of H3K9 and H3K36 related to transcription[98,119].KDM4A, B, and D play a role in PC mainly.The interaction between regulatory factor X-associated protein RFXAP and KDM4A can disrupt DNA damage repair[120].RFAXP is a key transcription factor for MHC II molecules[121,122].It can bind to the promoter ofKDM4Aand induce its expression[120].In PC, Fisetin can interact with RFXAP/KDM4A to inhibit PC tumor growthin vivoand cell proliferationin vitro[120].In PC, KDM4B shows the ability to downregulate E-cadherin[123].The high nuclear expression of KDM4D in the samples of pancreatic resection margins significantly and independently predicts an earlier recurrence in PC patients[124].
KDM5:KDM5 subfamily consists of four members, KDM5A, KDM5B, KDM5C, and KDM5D[125].The role of KDM5 family in PC is not completely clear.KDM5A is associated with the development of PC[126].KDM5A inhibits the expression of mitochondrial pyruvate carrier-1 (MPC-1) and controls the metabolites of pyruvate in mitochondria in PDAC[126].Upregulation of MPC-1 seems to inhibit the development of cancer.Therefore, it can be inferred that KDM5A promotes the development of PDAC.
KDM6:KDM6 subfamily is mainly composed of KDM6A/UTX, its paralogs UTY and KDM6B[127].They can demethylate the dimethyl and trimethyl groups of H3K27.They play important roles in the occurrence and development of many cancers.KDM6A/UTX has been the most frequently mutated epigenetic regulator in cancers including PC[128-133].In addition, KDM6A also antagonizes PRC2-mediated H3K27 trimethylation catalyzed by EZH2, thereby regulating development[99,104,134].KDM6A has not been found to function in PC tissues.Downregulation of KDM6B is widespread in many cancer cells[135,136].Almost all pancreatic epithelial tissues have been detectedKRASgene mutations before they become cancerous[137].KDM6B,which is located downstream of theKRASgene, is upregulated in the pre-tumor phase of pancreatic intraepithelial tumors[138].It is worth noting that the expression of KDM6B decreases with cancer development.
KDM7 (PHF and ZF protein subfamily):At present, the effect of KDM7 subfamily on PC has been seldom developed.According to relevant data, KDM7A may be related to the occurrence and development of PC[139].
READER DOMAIN IN WRITERS AND ERASERS
PHD fingers are central “readers” of histone post-translational modifications.They recognize specific histone modifications and bind to histone to ensure the different enzymes to locate in special targets[140,141].They are structurally conserved,represented by the canonical C4HC2C/H sequence coordinating two zinc ions.They present in many chromatin-modifying proteins, such as demethylases or methyltransferases, or act as scaffolding proteins that can connect multi-subunit enzymatic complexes with a particular genomic region[30,140,141].In this part, we will discuss how PHD fingers regulate histone methylation/demethylation and their binding substrates (Table 3).
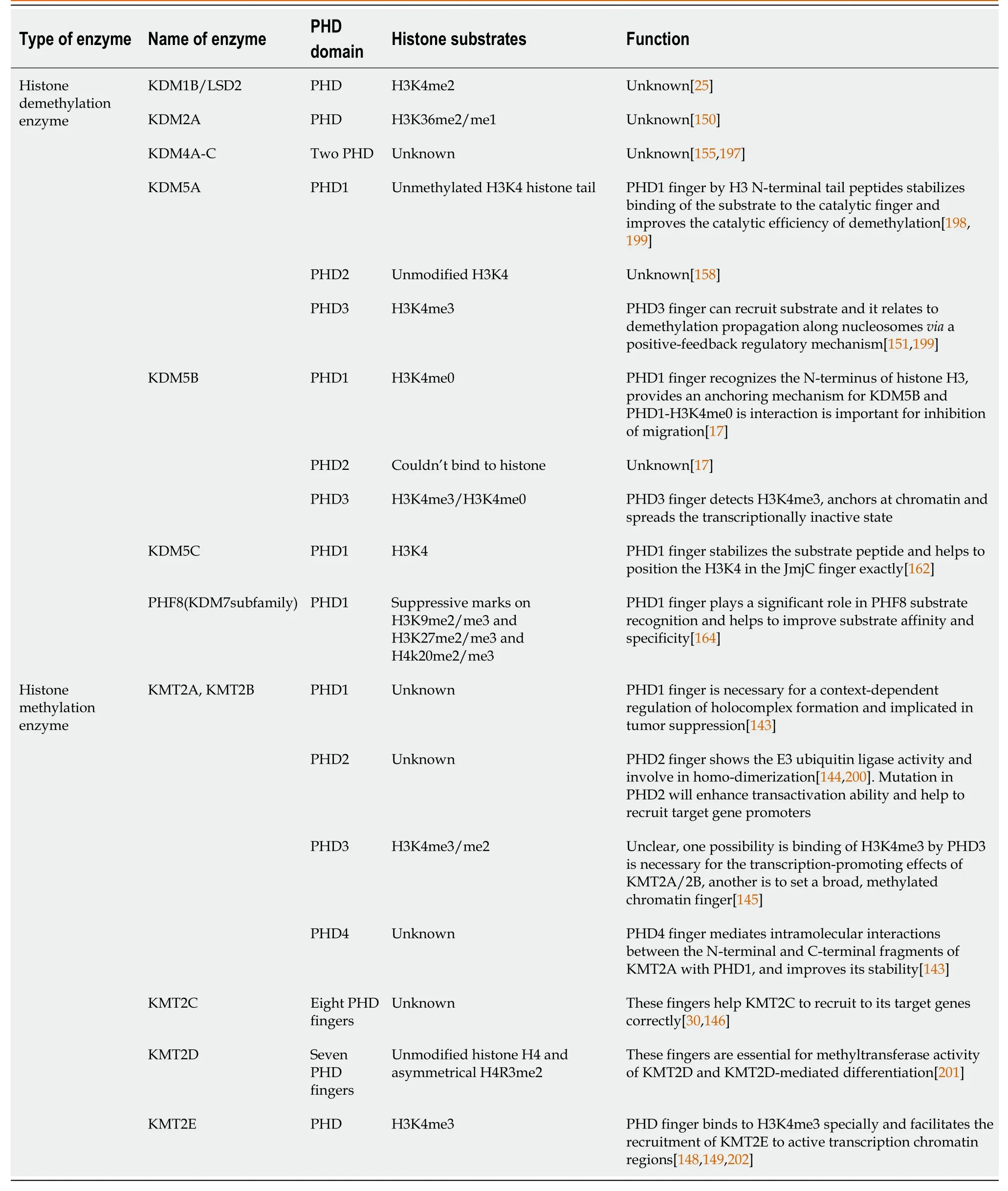
Table 3 Different enzymes and plant homeodomain finger domain
Regulation of writers by PHD finger
KMT2A-E all have PHD fingers, but the number of PHD fingers in these proteins is different.KMT2A and KMT2B have four PHD fingers, while KMT2C has eight PHD fingers and KMT2D has seven PHD fingers, but KMT2E only has one PHD finger.There are 24 PHD fingers in KMT2A-E[142].
Regulation of KMT2A and KMT2B by PHD finger:KMT2A and KMT2B have similar domain architecture and both contain three consecutive PHD fingers, PHD1-3.These consecutive PHD fingers are followed by a bromodomain and the fourth PHD4 finger[142].The precise function of PHD1 finger in KMT2A and KMT2B is unclear, but it can regulate the intramolecular interactions between N-terminal and C-terminal segments[143].PHD1 fingers are necessary for holocomplex formation and are implicated in tumor suppression[143].PHD2 finger has an E3 ubiquitin ligase in the presence of the E2-conjugating enzyme CDC34[144].Mutation of the PHD2 finger will cause increased transactivation ability of KMT2A and its recruitment to target genes[142], because of increased protein stability[144].PHD3 finger binds to H3K4me3/me2, but the affinity between PHD3 finger and H3K4me2 is eight times lower than the affinity between PHD3 finger and H3K4me3[145].Although PHD3 finger can recognize H3K4me3, the special function of KMT2A in transcriptional maintenance is unclear[145].One possibility is that binding of H3K4me3 by PHD3 finger is necessary for the transcription-promoting effects of KMT2A, and another possibility is that newly deposited H3K4me3 mark helps KMT2A slide along the gene to set a broad,methylated chromatin domain[145].The stability of KMT2A is dependent on its intramolecular interaction which is mediatedviaits PHD1 finger with PHD4 finger and the phenylalanine/tyrosine-rich domain of KMT2A[143].Therefore, PHD4 finger in KMT2A can improve the stability of KMT2A in case of hydrolysis.
Regulation of KMT2C by PHD finger:KMT2C contains eight PHD fingers while KMT2D contains seven PHD fingers[142].Although the function of PHD fingers in KMT2C is unclear, the functional extended PHD finger is important for KMT2C to be recruited to its target genes[146].PHD4, PHD5, and PHD6 in KMT2D are tandem and these tandem PHDs can bind to unmethylated or asymmetrically demethylated H4 arginine3[147].This connection is important for nucleosomal methylation activity and mediates stem cell differentiation by KMT2D[147].But this binding ability is repressed by symmetrical demethylation on arginine-3 of histone H4 (H4R3me2s), because H4R3me2s can hinder the histone binding ability and catalytic activity in PHD4-6[142,147].
Regulation of KMT2E by PHD finger:The binding of KMT2E and histone is based on its single PHD finger which can bind to H3K4me3, and this special spatial structure of KMT2E makes it possible to recognize H3K4me3[148].Although KMT2E can also bind to H3K4me2 and H3K4me1, the stability of binding of H3K4me2 and KMT2E is five times weaker than H3K4me3, while the stability of binding of H3K4me1 and KMT2E is sixteen times weaker than H3K4me3[148].This can facilitate the recruitment of KMT2E to active transcription chromatin regions[148,149].
Regulation of erasers by PHD finger
PHD fingers can be found in KDMs[150,151].These PHD fingers bind to the tail of H3 to enable the localization of enzymes to specific targets[152], and promote the recruitment of transcription factors or chromatin-associated protein complexes[17].
25. A little hut: Many fairy tales include huts or houses hidden in a forest for various reasons, such as in Hansel and Gretel, Snow White and the Seven Dwarfs, and Goldilocks and the Three Bears. The hut may be a place of danger or a safety zone for the heroine. Graham Andersen finds several strong relationships between Six Swans and Snow White and the Seven Dwarfs in his study of fairy tale origins in the ancient world (Anderson 2000).Return to place in story.
Regulation of KDM4 subfamily by PHD finger:PHD fingers can be found in KDM4 subfamily.KDM4A, KDM4B, and KDM4C have a catalytic histone demethylase domain, double PHD and Tudor domains, whereas KDM4D contains only a catalytic domain and lacks PHD and Tudor domains[153,154].Although KDM4A-C have PHD fingers, the function of PHD fingers is unclear[155].
Regulation of KDM5 subfamily by PHD finger:KDM5 subfamily, including KDM5A-D, catalyze demethylation of the transcriptionally activating trimethylated and demethylated lysine-4 mark on H3[100,103,156,157].KDM5A contains three PHD fingers (PHD1, PHD2, PHD3).Qualitative pull-down assays with isolated PHD1 domain of KDM5A show that it binds to unmodified H3K4 peptide[158].The PHD1 finger preferentially recognizes unmethylated H3K4 histone tail, which is a KDM5Amediated trimethylation products of H3K4 (H3K4me3) demethylation[151].The function of PHD2 finger is unknown.PHD3 finger has been studied in the context of its fusion with nucleoporin NUP98 and it specifically binds to the H3K4me3, with a decrease in affinity for lower methylation states[17,158].Since these preferred binding substrates are the products of KDM5A-mediated demethylation, a model in which demethylation can propagate along nucleosomesviaa positive-feedback regulatory mechanism, has been put forward[151].
The KDM5B PHD1 finger can recognize the N-terminus of H3, which is unmodified or methylated at Lys9[17].The KDM5B PHD2 finger cannot bind to histone.The KDM5B PHD3 finger prefers to bind to H3K4me3[17].The PHD1 finger specifically binds to H3K4me0, and the PHD3 finger is selective for H3K4me3.A combination of two ‘readers’ capable of recognizing distinctive epigenetic marks is likely to impact KDM5B activity.Binding of PHD1 to H3K4me0 may provide an anchoring mechanism for KDM5B to sense H3K4me3 through PHD3 and slide along the H3K4me3-enriched promoters, demethylating nearby methylated H3K4 and further spreading the transcriptionally inactive state of chromatin[17].In addition, abrogation of H3 tail recognition by point mutation in the PHD1 domain of KDM5B decreases H3K4 demethylation in cells, resulting in the repression of tumor suppressor genes[159].Therefore, the importance of interaction between PHD1 and H3 tail is proved.
Similarly, the PHD1 finger domain in KDM5C is close to the JmjC domain, and the linker of JmjC domain is 13 amino acids long and is expected to recognize and bind to H3K9me3[157,160].Although the PHD1 domain is not necessary for the demethylase activity, it helps to recognize the substrate peptide[157,161].The interaction between PHD1 domain and JmjC domain stabilizes the substrate peptide and the PHD1 domain can help precisely position H3K4 in the JmjC domain[162].
Regulation of KDM7 by PHD finger:PHF8 belongs to KDM7 subfamily and transcriptionally removes suppressive demethylation and monomethylation of lysine 9 and 27 on H3 and lysine 20 on H4[163].PHF8 has a PHD finger which is closed to the catalytic domain.PHD finger in PHF8 plays a significant role in PHF8 substrate recognition, because it helps to improve substrate affinity and specificity[164].PHF8 can be recruited to the promoters through the combination of its PHD finger and H3K4me2/3 during the cell cycle transition from G1 to S[107].Although the functions of PHD fingers can be found in gastric cancer[165], breast cancer[166], colorectal cancer[167], lung cancer[168],etc., the functions of PHD finger are still unclear in PC.
CLINICAL APPLICATION
Epigenetic genes play vital roles in maintaining structural stability and physiological functions of normal chromosomes and are deficient in some patients with PC, thereby serving as potential targets for correcting these deficiencies and precisely killing these aberrant PC cells[169].The discovery of histone methyltransferases, demethylases and their active sites has provided new insights in the diagnosis and treatment of PC.The active sites and mechanism of the inhibitors in PC treatment are shown in Table 4.

Table 4 Inhibitors for the treatment of pancreatic cancer
Histone modifications define the previously unrecognized subsets of PC patients with different epigenetic states and therefore represent the prognostic and predictive biomarkers that can be used to guide clinical decisions, such as the use of fluorouracil chemotherapy[170].H3K4me2, H3K9me2, or H3K18AC expressed at low levels are positively correlated with the poor prognosis of PC[170].EZH2 expression is higher in PC cells than in normal cells; thus, EZH2 can be used as a potential biomarker for early diagnosis of PDAC[171].High expression of KDM4D in benign cells near the edge of surgically resected PC tissues is predictive of early recurrence[124].The discovery of epigenetic biomarkers can provide a great reference for early diagnosis, drug selection and surgery prognosis of PC.
SMYD3 inhibitor piperidine-4-formamide-acetanilide compound, BCI-121, is a small molecule inhibitor that significantly inhibits proliferation in PC cell lines with high expression of SMYD3.BCI-121 and histone competitively bind to SMYD3; BCI-121 binds inside the lysine channel, which connects cofactor binding sites and histone peptide binding sites[173].
The PRMT5 inhibitor EZP015556 targetsMTAP(a gene commonly lost in PC)negative tumors, which indicates that it is an effective treatment for a subpopulation ofMTAPpositive tumors.According to the individualized medication approach, the therapeutic response in different patient-derived organoids (PDOs), developed directly from patient tumor tissue is different.The PDO model is used to validate the effectiveness of PMRT5 inhibition as a potential treatment for PDAC[174].EZH2 expression in PC cells is significantly higher than that in normal pancreatic duct cells and fibroblasts.3-Deazaneplanocin A (DZNeP) regulates the expression of EZH2 and H3K27me3, synergically enhancing the anti-proliferative activity of gemcitabine and significantly increasing the apoptosis rate of cells[175].DZNeP is an S-adenosine homocysteine hydrolase inhibitor.DZNeP also enhances the mRNA and protein expression of nucleoside transporter HENT1/HCNT1[175].The combination of DZNeP and DZNeP/gemcitabine significantly reduces the growth volume of PDAC spheres in selective medium[175].
Bromodomain and extra-terminal (BET) inhibitors and EZH2 inhibitors are designed to rescue the dysregulated KMT2C/MLL3-KDM6A/UTX-PRC2 regulatory axis and have achieved preliminary success in preclinical models.The regulatory axis regulates the expression of various downstream tumor suppressor genes[169].Therefore, rebalancing this axis represents a new approach to PDAC therapy.
Defects in KDM6A make sex-specific squamous PC sensitive to bromouracil and BET inhibitors[169].BET inhibitor JQ1 reverses squamous cell differentiation and inhibits tumor growthin vivoby decreasing MYC pathway activity and p63 levels[176].JQ1 affects cancer-associated fibroblast (CAF) activation by acting on the Hedgehog and TGF-β pathways.JQ1 inhibitor converts α-SMA-positive CAFs to α-SMA-negative CAFs, but does not eliminate CAFs[177].
Small molecules containing 8-hydroxyquinoline structure are competitive inhibitors of KDM4 (also known as JMJD2) family, binding active iron to inhibit the activity of KDM4 and regulate demethylation of H3K9 sites[178].KDM4C inhibitor SD70 can inhibit the growth of prostate cancer cells[179].
Many types of inhibitors of KDM1A have been reported, but the inhibitors of this enzyme are mainly targeted at acute myeloid leukemia or small cell lung cancer,etc.And there are few studies on PC.For example, SP2509 is a noncompetitive inhibitor,and is used in current clinical trials for the treatment of acute myeloid leukemia or small cell lung cancer[180].Ory-1001 effectively inactivates LSD1 and is highly selective for FAD-dependent ammonia oxidase[181].The application of histone demethylase inhibitors in the treatment of PC is still limited, so it is necessary to strengthen the exploration of the treatment of PC based on the existing research.
FUTURE DIRECTIONS
Histone writers and erasers do not work independently.In fact, the interactions between writers and erasers include the positive correlativity between EZH2 and KDM2B, and the synergistic effects of EZH2 and KDM6A[182,183].In bladder cancer,the H3K27 demethylase KDM6A gene often has mutations[131,184].This makes cancer tissues that have lost KDM6A more vulnerable to EZH2 attack[185].This accelerates the onset of tumors.The expression of EZH2 and KDM2B in ovarian cancer is positively correlated[183].Therefore, knocking down the KDM2B gene is beneficial to inhibit the migration of ovarian cancer cellsin vitro.
Many problems remain in the research of histone modifications.Research on histone methyltransferases is relatively adequate, but there are few articles about the mechanism of SMYD2, so SMYD2 is not mentioned in our review.Current research on histone arginine residue demethylases has not yet fully achieved results.Therefore,only histone lysine residue demethylases are discussed.However, the effect of KDM7 subfamily demethylases on PC has seldom been proved, so only some guesses about the effect of KDM7A are mentioned.Besides, the study of the interactions between writers and erasers in PC is still in a blank state.The role of the reader domain in PC remains unclear.This review only lists the roles of the PHD domain in the localization of histone modifications and the recruitment of related protein complexes.Reader domain is still a potential research direction in PC.
The study of histone methylation and demethylation has enlightening effects on the diagnosis, treatment, and prognosis of PC.Histone modifications can be used to predict in the prognosis of PC patients[171].Histone methyltransferase and demethylase inhibitors are used clinically to treat PC.The corresponding inhibitors act on the signal regulatory pathway and change the signal expression of downstream target cells, thus regulating the growth and development of cancer cells.At present,the research of histone demethylase inhibitors is inadequate.Therefore, histone demethylase inhibitors need to be further explored.
The effect of histone modifications on PC is interdependent.The interactions between histone modifications and other epigenetic forms can influence the occurrence and progression of cancers such as cervical cancer and breast cancer.The effect of these interactions enlightens the research on PC.DNA methylation and the expression of miRNAs can be regulated by histone methyltransferases and demethylases, thereby causing alternations of developing process of cancer.Histone methyltransferase EZH2 epigenetically silences tumor-suppressor miRNAs, such as miR-139-5p, miR-125b,miR-101, let-7c and miR-200b, thereby promoting cancer cell metastasis[186].Histone demethylase KDM5B targets H3K4 demethylation of miR-let-7e and promotes tumor cell proliferation through epigenetically inhibiting the tumor suppressor miR[187].The combination of EZH2 and promoter region induces the expression of specific target protein H3K27me3, thereby reducing the expression of downstream gene, DNA(cytosine-5)-methyltransferase 3A (DNMT3A)[10].EZH2-H3K27me3-DNMT3A is the key factor of regulating cervical total stimulus molecule Tim-3/galectin-9, which results in immune escape in the process of malignant transformation[10].It is reasonable to speculate that the interaction between histone methylation and other epigenetic modifications may also play a role in PC.This opinion draws some inspiration and reference to future research of PC.
CONCLUSION
This review focuses on the mechanism of histone methylation in PC.Histone methylation is mainly regulated by writer, reader and eraser.Writer refers to histone methyltransferase, eraser refers to histone demethylase and reader refers to the modification domain of histone methyltransferase and demethylase.Reader can be an independent polypeptide or a component of methyltransferase and demethylase.On the one hand, histone methyltransferase can promote the proliferation and invasion of PC cells.On the other hand, histone methyltransferase can inhibit the proliferation of cancer cells.Histone demethylase promotes the occurrence of PC and is related to apoptosis.Reader domain plays a role in guiding related methyltransferases and demethylases to identify corresponding sites during the methylation and demethylation process.
 World Journal of Gastroenterology2021年36期
World Journal of Gastroenterology2021年36期
- World Journal of Gastroenterology的其它文章
- Fluorescent cholangiography: An up-to-date overview twelve years after the first clinical application
- Hepatitis B virus infection and hepatoceIIuIar carcinoma in sub-Saharan Africa: ImpIications for eIimination of viraI hepatitis by 2030?
- Liver disease in the era of COVID-19: Is the worst yet to come?
- Treatment of hepatitis B virus infection in chiIdren and adoIescents
- Basic Study CircRNA_0084927 promotes colorectal cancer progression by regulating miRNA-20b-3p/glutathione S-transferase mu 5 axis
- Basic Study ExosomaI microRNA-588 from M2 poIarized macrophages contributes to cispIatin resistance of gastric cancer ceIIs
