The Streptococcus agalactiae Ribose Binding Protein B (RbsB) Mediates Quorum Sensing Signal Uptake via Interaction with Autoinducer-2 Signals
FAN Bolin, PAN Lixia, WANG ZhongliangWANGKAHART Eakapol, HUANG Yuchong3, YANG Dengfeng, JIAN Jichang3, HUANG Yu3, and WANG Bei3, *
TheRibose Binding Protein B (RbsB) Mediates Quorum Sensing Signal UptakeInteraction with Autoinducer-2 Signals
FAN Bolin1),#, PAN Lixia2),#, WANG Zhongliang1), WANGKAHART Eakapol4), HUANG Yuchong1), 3), YANG Dengfeng5), JIAN Jichang1), 3), HUANG Yu1), 3), and WANG Bei1), 3), *
1) College of Fishery, Guangdong Ocean University, Guangdong Provincial Key Laboratory of Pathogenic Biology and Epidemiology for Aquatic Economic Animal, Key Laboratory of Control for Disease of Aquatic Animals of Guangdong Higher Education Institutes, Zhanjiang 524088, China 2) National Engineering Research Center for Non-Food Biorefinery, State Key Laboratory of Non-Food Biomass and Enzyme Technology, Guangxi Academy of Sciences, Nanning 536000,China 3) Key Laboratory of Experimental Marine Biology, Institute of Oceanology, Chinese Academy of Sciences, Qingdao 266071,China; Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Techno-logy, Qingdao 266237, China 4) Research Unit of Excellence for Tropical Fisheries and Technology, Division of Fisheries, Department of Agricultural Technology, Faculty of Technology, Mahasarakham University, Kantarawichai, Mahasarakham, Thailand 5) Guangxi Key Laboratory of Marine Natural Products and Combinatorial Biosynthesis Chemistry, Guangxi Beibu Gulf Marine Research Center, Guangxi Academy of Sciences, Nanning 536000, China
Understanding aquatic pathogen in sediments or aquacultural water is crucial to protect public health from soilborne and waterborne diseases. Quorum sensing (QS) was increasingly reported in biological wastewater treatment processes because of their inherent roles in biofilm development, bacterial aggregation and so on. The widely QS signals was Antoinducer-2 (AI-2), primarily involved to allow the possibility of interspecies communication. However, the cellular components that mediate the response ofto AI-2 have not been fully characterized. Analysis of the complete genome sequence ofindicated that its RbsB protein has similarity toLsrB andRbsB proteins that bind AI-2We hypothesized that RbsB protein mediates quorum sensing signal uptakeinteraction with AI-2. To evaluate the regulatory effect of RbsB on QS system, the recombinant plasmid pGEX-6p-1-RbsB was constructed and RbsB protein was purified with GST-tag. To further elucidate the role of RbsB protein binding to DPD (AI-2 precursor dihydroxypentanedione), the systematically throughput circular dichroism (CD) spectroscopy, isothermal titration calorimetry200(ITC200) and molecular docking methods were employed. The high expression of soluble RbsB protein with molecular weight of 33kDa was obtained. The thermodynamics results (Δ<0, Δ<0, Δ<0) with ITC determination indicated that the binding process between DPD and RbsB was exothermic and spontaneous, with hydrogen bonds and van der Waals forces as the main binding forces. Obviously, DPD can be more easily combined with RbsB in a dose-dependent manner, suggesting that RbsB was changed in the microenvironment of DPD when the DPD concentration was between 0.8–1.0mmolL−1and reaching the maximum binding amount. According to molecular docking, 3 hydrophobic residues involved in DPD and RbsB protein stable binding were be found, and also hydrogen bonding plays a key role in the formation of the new complex. RbsB efficiently inhibitedbioluminescence induced by bothAI-2 andAI-2 in a dose-dependent manner. However, our results suggest that RbsB may play a role in the response ofto AI-2.
;RbsB protein; circular dichroism (CD) spectroscopy; isothermal titration calorimetry200(ITC200); molecular docking
1 Introduction
In humans,or group B strep-tococcus (GBS) is a frequent colonizer of the rectovaginal tract, and a major cause of neonatal and non-pregnant adults infectious diseases. It alsocauses invasive disease in fish, compromising food security and posing a zoonotic hazard (Delannoy, 2013). However, it has been reported thatcauses menigoencephalitis in fish leading to abnormal swimming behaviour, popeye, haemorrhages, and rapid severe mortalities (Hu, 2017). Unlike terrestrial animals, aquatic animals are directly exposed to water, where bacterial population in aquaculture system play crucial roles in nutrient absorption, water quality control, pathogen defense, antibiotic resistance and maintaining host heath (Blancheton, 2013; Carbone and Faggio, 2016). In recent times, it has been recognized that the large amounts of fecal materials that were applied to agricultural soils as beneficial fertilizers or soil amendments introduced various pathogens in soil environments (Tyrrel and Quinton, 2003). This bacterial strain,was widely detected in sediments and aquacultural wastewater and thus, attracted considerable public and scientific interests (Al-Harbi and Uddin, 2005). However, the(ST491) strain isolated from fish shared a 3-set genotype identical to that of(ST283) clone associated with invasive disease of adult humans in Asian (Delannoy., 2013). The apparent association of the aquatic pathogen in sediments or aquacultural water suggests that some strains of aquaticmay present a zoonotic or anthro- ponotic hazard. Therefore, a thorough understanding of the pathogenicity ofhas important implications in protecting public health from diseases.
In recent years, quorum sensing (QS) has been widely quoted among a variety of bacteria species, depending on the function of coordinating communal behavior, for instance, biofilm formation, bacterial virulence and motility, which are essential for establishing a symbiotic or pathogenic relationship with their respective eukaryotic hosts successfully (Ohtani, 2002; Marketon, 2003; Quiñones, 2005; Rice.,2005; Suntharalingam and Cvitkovitch, 2005; Shrout and Nerenberg, 2012; Song, 2014; Cao, 2020). Furthermore, previous investigations have equally showed that QS embodied in some bacterial species, might amend their behavior by cell-to-cell communication which was conductedchemical signal molecules, and this can serve as a population density assessment parameter. Consequently, when the detected quantity reaches a certain value, specific genes expression is induced and a relevant community- level behaviors are activated (Davies., 1998; Han- cock and Perego, 2004; Suntharalingam and Cvitkovitch, 2005; Parsek and Greenberg, 2013a; Kristich.,2018). Recent research has revealed that various genes are adjusted by many bacteria species, thus, presenting different types of QS signaling mechanisms. It is well known that one QS signal called autoinducer-2 (AI-2) is shared by both Gram-positive and Gram-negative bacteria. AI-2 is also discovered in numerous bacterial species and universally is responsible for interspecies communication. Previous study has disclosedas the crucial gene for AI-2 signal synthesis (Balestrino, 2005; Ramsey, 2009).is a key regulatory gene for the AI-2- mediated quorum sensing system, and its product catalyzes the conversion of S-ribosylhomocysteine (SRH) to homocy- steine and 4,5-dihydroxy-2,3-pentanedione (DPD) (Mar- keton., 2003; Balestrino., 2005), with spontaneous generation of AI-2 by DPD (Fig.1) (Han and Lu, 2009; Gu, 2018).

Fig.1 The AI-2 synthesis pathway. SAH is hydrolyzed by the nucleosidase Pfs to S-ribosylhomocysteine (SRH), which is subsequently converted to 4,5-dihydroxy-2,3-pentanedione (DPD) and homocysteine by LuxS. DPD can spontaneously cyclize and hydrate to AI-2.
Moreover, since AI-2 molecule is a hydrophilic molecule with low binding force with lipids, bacteria must rely on AI-2 transporter protein and AI-2 receptor to complete the physiological process when exporting and internalizing the molecule (Kamaraju, 2011; Pereira, 2013b). An AI-2 signal molecule receptor, Furanoyl borate diester receptor plays an important role in the process of bacterial recognition and perception of environmental changes and adaptive responses (bacterial density sensing system and quorum sensing) (Bassler and Losick, 2006; Pereira., 2013b). So far, researchers have identified two AI-2-specific surface binding receptors (Chen; Miller, 2004) at the genetic, biochemistry and bioin- formatics levels as LuxP and Lsrb proteins (Pereira, 2013a). In, sensing of AI-2 occurs through a two-component system involving LuxP and LuxQ proteins (Bassler, 1994). LuxP is similar to periplasmic ribose binding proteins and binds AI-2, while LuxQ contains sensor kinase and response regulator domains to propagate the signal (Henke and Bassler, 2004).,,andutilize the Lsr (LuxS regulated) ABC transporter, an AI-2 transport system similar to the ribose ABC transporter (Miller., 2004). In addition to LsrB, it was determined inthat the ribose binding protein RbsB also interacts with AI-2 (James, 2006; Shao, 2007).
ZQ0910 (SAZQ0910) does not possess a homologue of LuxPQ transport system. However, the SAZQ0910 genome contains an ABC transporter annotated as a ribose transport system that incorporates a binding protein. The SAZQ0910gene product has 76% identity toLsrB, and 86% identity toRbsB that was found to bind AI-2. The predicted RbsB proteins fromZQ0919 andare virtually identical in length (298. 293 amino acids respectively). Based on these observations and the similarity between the Lsr-AI- 2 transport system and ribose transporters, it was hypothesized that SAZA0910 may utilize RbsB for internalization of and response to AI-2.
However, with regard to RbsB role in AI-2 system, there is no experimental evidence to confirm these hypotheses. The present study was undertaken to investigate whether RbsB is indeed linked with the DPD, as is the case for LsrB and LuxP receptor or their equivalent in AI-2 system; if so, we would expect to see interactions between the two molecules. Therefore, we utilised CD spectroscopy, ITC200and molecular docking to determine whether DPD specifically interacts with purified RbsB, which would be consistent with either or both of the above roles. We confirm the binding of DPD to RbsB and further show that RbsB-DPD ligand binding results in stabilisation of RbsB structural conformation. Thus, the DPD binding stabilised and exerted changes in both the secondary and tertiary structural conformations of RbsB. These results confirm the role of RbsB in cell growth, toxicity, EPS production (Yu, 2019), and that the microflora exist in the form of flocs, biofilms or particles in biological wastewater treatments (Song., 2014).
2 Materials and Methods
2.1 Molecular Cloning
strain ZQ0910 was isolated, stored in our laboratory and deposited at China microbial culture preservation center (storage number: CGMCC 7.264). Genomic DNA fromZQ0910 was prepared by the following the manufacturer’s protocol (TIANGEN, China). The extracted DNA was used as a template for PCR amplification using upstream (5’-CGCATGAAATTTGGAAAAAAACTT-3’) and downstream (5’-CCGTTACCAATTGTATTTTTCAACATTG-3’) primers with the restriction sitesand(underlined) respectively. Amplification was done at 95℃ for 5min and then at 95℃ for 30s, 60℃ for 30s, and 72℃ for 60s for a total of 35 cycles, followed by 72℃ for 10min. All positive clones were confirmed by restriction enzyme digestion and DNA sequence analysis (Sangon Biotech, Guangzhou).
2.2 Construction of Expression Vectors for the Fused Protein of RbsB to GST in E. coli Cells
To express RbsB as a soluble form incells, RbsB was fused to the GST (217 aa) using the pGEX-6P- 1 expression vector. The amplified product was double digested with the restriction enzymes BamH I and XhoI and ligated with BamHI-XhoI site of pGEX-6P-1 to give pGex-RbsB. Subsequently, it was transformed into competent DH5α cells and selection of positive clones was done on ampicillin-supplemented agar plates. The colonies obtained were screened using colony PCR and double enzyme digestion followed by gene sequencing (Sangon Biotech, Guangzhou).
2.3 Expression of RbsB
A seed culture ofstrain BL21 carrying the pGex-RbsB expression vector was grown in 20mL of Luria-Bertani (LB) medium containing ampicillin (100μg mL−1) at 37℃ with rapid shaking at 220rmin−1overnight and was then transferred into 500 mL of LB-broth containing 100μgmL−1ampicillin in a 2-L Sakaguchi flask and cultured with reciprocal shaking (220rmin−1) until the OD600reached 0.6–0.8. RbsB was expressed at 37℃ for an additional 4h after induction with 0.2mmolL−1isopropyl β-D-thiogalactopyranoside (IPTG). The cells (weight 5.4g) were harvested by centrifugation at 4000for 15min at 4℃, and then stored at −80℃.
2.4 Purification of RbsB
For chromatographic purification of RbsB, 5.4g of cell pellet was re-suspended in 40mL equilibration buffer (50mmolL−1Tris-HCl (pH 8.0), 150mmolL−1NaCl, 10% glycerol) in a 50mL falcon tube, and ultrasonicated for 15 min on ice with an amplitude of 30% and a repeated pulse rate of 5s pulse ON and 10s OFF using a Branson 450 Digital Sonifier. The supernatant was collected after centrifugation at 12000for 40min at 4℃. The crude cell lysate was applied to Glutathione Sepharose 4B (GE Healthcare) beads for affinity purification. The column was equilibrated with approx. 5 column volumes of equi- libration buffer (50mmolL−1Tris-HClpH 8.0, 150mmolL−1NaCl, 10% glycerol, pH 8.0). The supernatant containing RbsB was loaded on the column and was washed with approx. 10 column volumes of the equilibration buffer. One column volume of GST elution buffer (50mmolL−1Tris-HCl PH 8.0, 150mmolL−1NaCl and 15 mmolL−1reduced glutathione, pH 8.0) was used to recover the target protein. The sample was then concentrated with an AmiconUltra-15 centrifugal filters (3000 MWCO) at 4℃, followed by re-suspension with HRV 3C reaction buffer (Thermo scientific). Thereafter, 100μL of HRV 3C protease was added into the lysate and the mixture incubated overnight at 4℃ for complete cleavage. The target protein was purified by Glutathione Sepharose 4B equilibrated against HRV 3C reaction buffer. Further- more, the target protein was purified by passing it through a SuperdexTM75 column.
2.5 SDS-PAGE Assay of the Proteins of Interest
SDS-PAGE was performed on 15% running gel, and after electrophoresis, proteins were stained with Coomas- sie Brilliant Blue R-250. Total protein concentrations of supernatant were measured by TaKaRa Bradford Protein Assay Kit with bovine serum albumin as the standard.
2.6 Circular Dichroism (CD) Spectra
The circular dichroism (CD) spectra of DPD in mixtures with RbsB in equilibration buffer (pH 8.0) were run with a Chirascan CD spectrometer (Applied Photophysics, Surrey, UK). The sample was scanned three times in the region from 200 to 300nm to obtain an average spectrum. The data was obtained at interval of 1nm and at scan speed of 96nmmin−1. The concentration of the RbsB was kept constant (2.0×10−5molL−1) and the molar ratios of RbsB to DPD series 1–6 were varied as follows: 1:0, 1:5, 1:10, 1:30, 1:40 and 1:50. In CD spectra measurements, the CD spectra of these compounds were corrected by subtracting the CD spectrum of equilibration buffer alone to have a background correction.
2.7 Thermodynamic Measurements
The thermodynamic measurements of RbsB and its complex with DPD were performed in a MicroCalTMiTC200(GE Healthcare Life Sciences, USA) at 25℃. RbsB solution (0.02mmolL−1, 300μL) was placed into the sample cell and the DPD solution (0.8mmolL−1) was respectively loaded into the 300μL injection syringes. All the samples were prepared in equilibration buffer. After it reached equilibrium, 2.0μL of DPD injection volumes were in dropwise titrated into the sample cell with a sequence of 19 injections. Each injection lasted 4s and the time delay between successive injections was 150s. The RbsB-DPD solutions in the titration cell were stirred at a constant speed of 1000rmin−1throughout the experiments. A blank experiment was performed by titrating DPD into equilibration buffer. The change in enthalpy (Δ) and in entropy (Δ) were determined using the software package supplied by ITC instruments.

Δ= Δ−Δ. (2)
2.8 Molecular Docking
Molecular docking is a widely used computational te- chnique for improving the understanding of ligand-protein interactions and ultimately elucidating interaction mechanisms. In order to elucidate the mechanism of RbsB and DPD, the docking studies were carried out against RbsB and DPD. The structures of DPD were constructed using ChemOffice 2004 software (Cambridge Soft Co., Cambridge, MA, USA). 3D structures of the RbsB were builted by using SWISS-MODEL (https:// swissmodel.expasy.org/). The docking was performed using the AutoDock Vina program. Docking calculations were performed based on the Lamarckian genetic algorithm (LGA). The best ranked docking pose of RbsB with DPD were obtained according to the binding-energy values.
2.9 Partial Purification of V. barveyi and S. agalactiae AI-2
Induction ofbioluminescence was carried out essentially as described by Surette and Bassler (Su- rette and Bassler, 1998). Preparations ofAI-2 were obtained from conditioned medium of overnightcultures grown in AI broth (Surette and Bassler, 1998). After cells were harvested, the medium was filtered through a 0.22-µm-pore-size filter and used immediately or stored at −70℃.
AI-2 fromwas partially purified by chromatography of conditioned medium from an exponential-phase culture on a Sep-Pak C18column (Waters) as described by Sperandio. (Sperandio, 2003). Briefly, cells were harvested from 4mL of culture and the supernatant was filtered through a 0.22-μm-pore-size filter and subsequently through a Centricon YM-3 1-kDa exclusion filter (Millipore Co.). The filtered solution was lyophilized and concentrated by re-suspension in 200μL of cold 5mmolL−1sodium phosphate, pH 6.2. Chromatography on a C18Sep-Pak column was carried out according to the instructions of the manufacturer. AI-2 activity was present in the column flow-through fraction.
2.10 Determination of V. harveyi Bioluminescence
For the determination ofbioluminescence, an aliquot of an overnightculture was diluted 1:5000 into fresh sterile AB medium and 80μL of cells was added to wells on a 96-well microtiter dish. The positive control wells received cell-free conditioned medium fromor partially purifiedAI-2 (20μL) While the negative-control wells received 20μL sterile AB medium. The experimental wells received 20μL oforAI-2 that was pre-incu- bated for 30min at 30℃ with various concentrations of purified RbsB. Samples were then added to wells containing the dilutedcells to give a final concentration of 0.1 to 5000ngmL−1RbsB. The microtiter plate was shaken on a rotary shaker at 500rmin−1at 30℃, and bioluminescence was measured at half-hour or hourly intervals by the use of a Wallac Victor31420 MultiLabel counter (PerkinElmer). Bioluminescence of each well was initially calculated as induction (n-fold) of light relative to that for the negative control reaction (BB170 cells incubated in sterile AI medium). Bioluminescence of the positive control reaction was subsequently normalized to a value of 1.0, and experimental data were calculated as percentage of the positive-control value.
3 Results
3.1 Preparation of Construct, Expression, Purification and Confirmation of the Recombinant Protein
Thegene was PCR amplified and fused with pGEX-6P-1 to form the construct pGEX-RbsB plasmid. The pGEX-6P-1 had a GST fusion tag, which was sparingly soluble in, and also participates in the process of disulfide bond reduction in the system. It made the purification and identification of subsequent proteins much easier. The protein expression was observed in BL21 (DE3). On comparing the expression profile at different temperature conditions, it was observed that expression was best at 37℃ (Fig.2). The pGEX-RbsB plasmid was transformed intoBL21 (DE3) competent cells, after induction under optimized conditions (0.2 mmolL−1IPTG, 4h, 37℃) and the eluate was collected according to the HUXI UV detector (wavelength of A280 nm). The eluate was identified by SDS-PAGE. The results showed that a higher content of the 59kDa fusion protein (10.464mgmL−1) was obtained after purification (Fig.3).

Fig.2 SDS-PAGE analysis of expression of the RbsB fusion protein at different temperatures. MW, Molecular weight marker; Ø, Non-induced bacteria culture (negative control). *Lysis with in native buffer (50mmolL−1 Tris-HCl, 150mmolL−1 NaCl, 10% glycerol, pH 8.0). *Clarification (centrifugation C1): The supernatant is the native protein extract (NPE). *Solubilization of the pellet from C1 with a denaturing buffer (Urea 8molL−1). *Clarification (centrifugation): The supernatant is the denatured protein extract (DPE).

Fig.3 Overexpression of GST-RbsB in E. coli BL21(DE3) cells. The expression level was tested on a 20% SDS- PAGE gel. MW, molecular weight marker; 1, purified GST-RbsB fusion protein (59kDa); 2, supernatant after cell lysis; 3, GST-RbsB (59kDa) after induction with 0.2 mmolL−1 IPTG for 4h at 37℃.

Fig.4 Fusion protein was digested with PierceTM HRV 3C protease overnight (1µL PierceTM HRV 3C protease: 200 ug fusion protein). The Cleavage test result was tested on a 20% SDS-PAGE gel. MW, molecular weight marker; 1, before cleavage sample; 2, after cleavage sample.
The eluate was concentrated with ultra-filtration tube, and after purification by glutathione agarose gel 4B, the eluent was collected and reacted with HRV 3C protease for overnight. The results were validated by SDS-PAGE(Fig.4). The target protein was purified further after passing through a SuperdexTM75 column using an ÄKTA pure system (GE Healthcare).According to the chromatographs obtained, 8 tubes corresponding to the desired peaks were selected for SDS-PAGE identification (Fig.5). The results showed that the molecular weights of RbsB (33kDa) was correct (Fig.6). Table 1 summarizes the amounts and percent purity of RbsB after each step of the purification process.
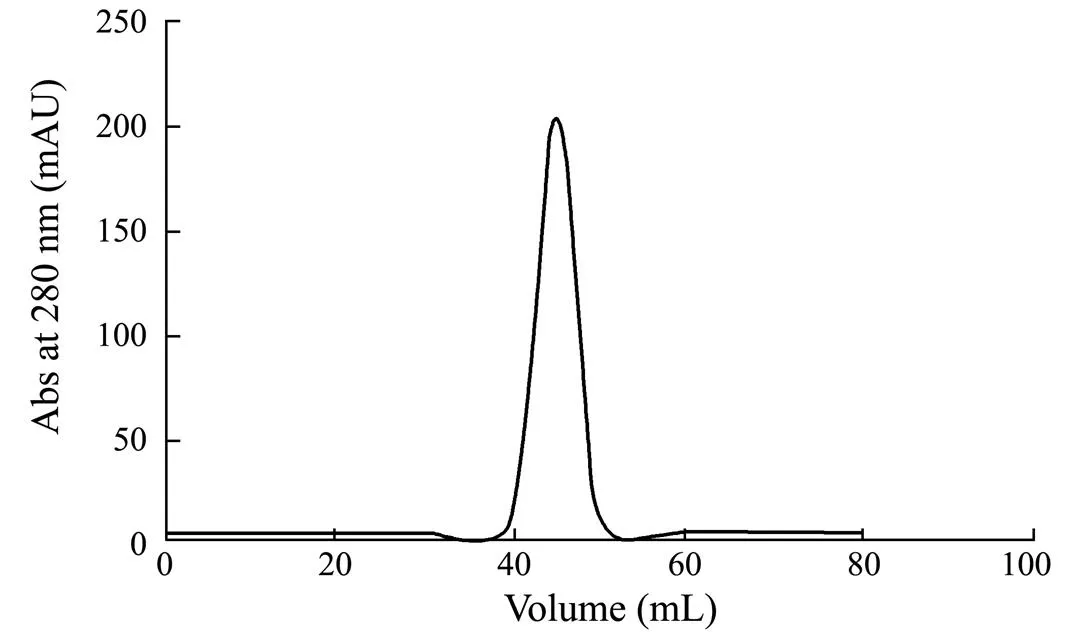
Fig.5 Gel filtration purification. RbsB (33kDa) was eluted as a peak at 45mL retention volume from a Superdex 75 (16/600, GE Healthcare) column at a flow rate of 1.0mL min−1 in the buffer (50mmolL−1 Tris-HCl, 150mmolL−1 NaCl, 10% glycerol, pH 8.0). ÄKTA pure system (GE Healthcare) was used.

Fig.6 Purity of the affinity peak was analyzed by a 20% SDS-PAGE gel. MW, molecular weight marker; 1–8, different fractions from the affinity peak of AKTATM purifier.
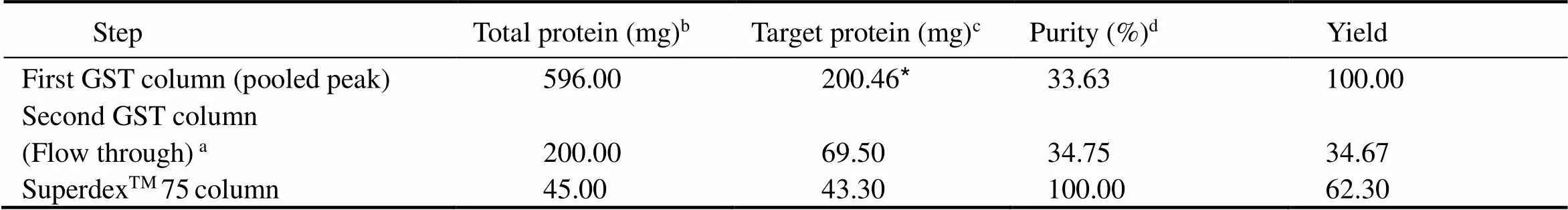
Table 1 Summary of the RbsB purification
Notes:a: second GST column purification step, after tag cleavage (flow through collected);b: protein concentration determined by Bradford assay using BSA as a standard protein;c: determined from total protein concentration and purity;d: purity determined by densitometric assessment of SDS-PAGE; *: recombinant protein.
3.2 Circular Dichroism (CD) Spectra Analysis
The CD spectrum was used to verify the interaction of RbsB with AI-2 precursor 4,5-dihydroxy-2,3-pentanedione (DPD) (Fig.1). The curve of RbsB has two ne- gative peaks, at 208 and 222 nm, which are typically characteristic of the a-helix structure of proteins, caused by n→π* transfer for the peptide bond of α-helix. As shown in Fig.7, this banding pattern is characteristic of the α-helical structure of proteins. The interaction between DPD and RbsB causes a slight increase in band intensity at all wavelengths of the far UC CD without any significant shift of peaks. The α-helix structure of RbsB decreased by the additions of DPD from 53.8% in free RbsB to 43.0% bound RbsB (Table 2). With the continuous increase of DPD concentration, the content of α-helix structure of RbsB was continuously reduced; DPD can be more easily combined with RbsB in a dose-dependent manner, suggesting that RbsB was changed in the microenvironment of DPD when the DPD concentration was between 0.8–1.0mmolL−1and reaching the maximum binding amount. This is an indication that the RbsB-DPD system has taken effect, resulting in the RbsB structural change.

Fig.7 CD spectra of RbsB in the absence and presence of DPD. RbsB concentration was 0.02mmolL−1 and DPD concentration were 0.1, 0.2, 0.6, 0.8 and 1.0mmolL−1, respectively.

Table 2 Predicted and calculated secondary structures of RbsB
3.3 Thermodynamic Measurements
Isothermal calorimetric titration (ITC) can directly be obtained from the thermodynamic information of the interaction between substances. The ITC curve of DPD acting on RbsB is shown in Fig.8, and it is evident that each drop of DPD added to RbsB solution produced a negative absorption sharp peak. This indicates that the binding process of RbsB and DPD is exothermic, and the ITC peak gradually decreased with the reaction, because the free DPD molecules in the sample pool gradually decreased. As the titration gradually occupies the binding site on the protein, the peak exotherm decreases and eventually saturates. The complex of DPD and RbsB gradually reached equilibrium. The integrated curve of ITC were obtained by single point bond and model fitting, and the related thermodynamic data Ka, Δ, Δ, Δof DPD combined with RbsB were calculated, as shown in Fig.8.

Fig.8 Titration with 0.02mmolL−1 RbsB and 0.8mmolL−1 DPD. N is stoichiometry, K is the association constant, ΔH is enthalpy change, and ΔS is entropy change.
It can be seen from Fig.8 that Δ<0 in the binding process of DPD and RbsB, was spontaneous. According to the relationship between the thermal effect and the acting force when Δ<0 and Δ<0, the acting force between Rbsb and DPD was van der Waals force and hydrogen bonding. The binding constant Kaof DPD and Rbsb is more than 105order of magnitude, which indicates that the combination between DPD and RbsB is of medium strength. With the increase in polymerization degree of DPD, the binding constant increases, which makes it easier to combine with RbsB, thus, prolongs the drug release time, and increases the efficacy time.
3.4 Molecular Docking Analysis
After several simulations of docking, the lowest energy docking results obtained are shown in the (Fig.9), the energy where the compounds entered the cavity of each protein molecule. The binding energy of the most favorable orientation of DPD bound with RbsB was −3.48kcalmol−1.The hydrophobic residues that interacted with DPD were ASN150, ASP185 and VAL151. The existence of these hydrophobic residues indicates that the binding between DPD and RbsB protein involved hydrophobic interactions. In addition, it can be seen from the Fig.9 that a hydrogen bond was formed between the DPD and the RbsB, which indicates that the hydrogen bond also played a role in the formation of a new complex. This is consistent with the results obtained from the analysis of thermodynamic data, RbsB stably binds to the AI-2 signaling molecule DPD at these sites.
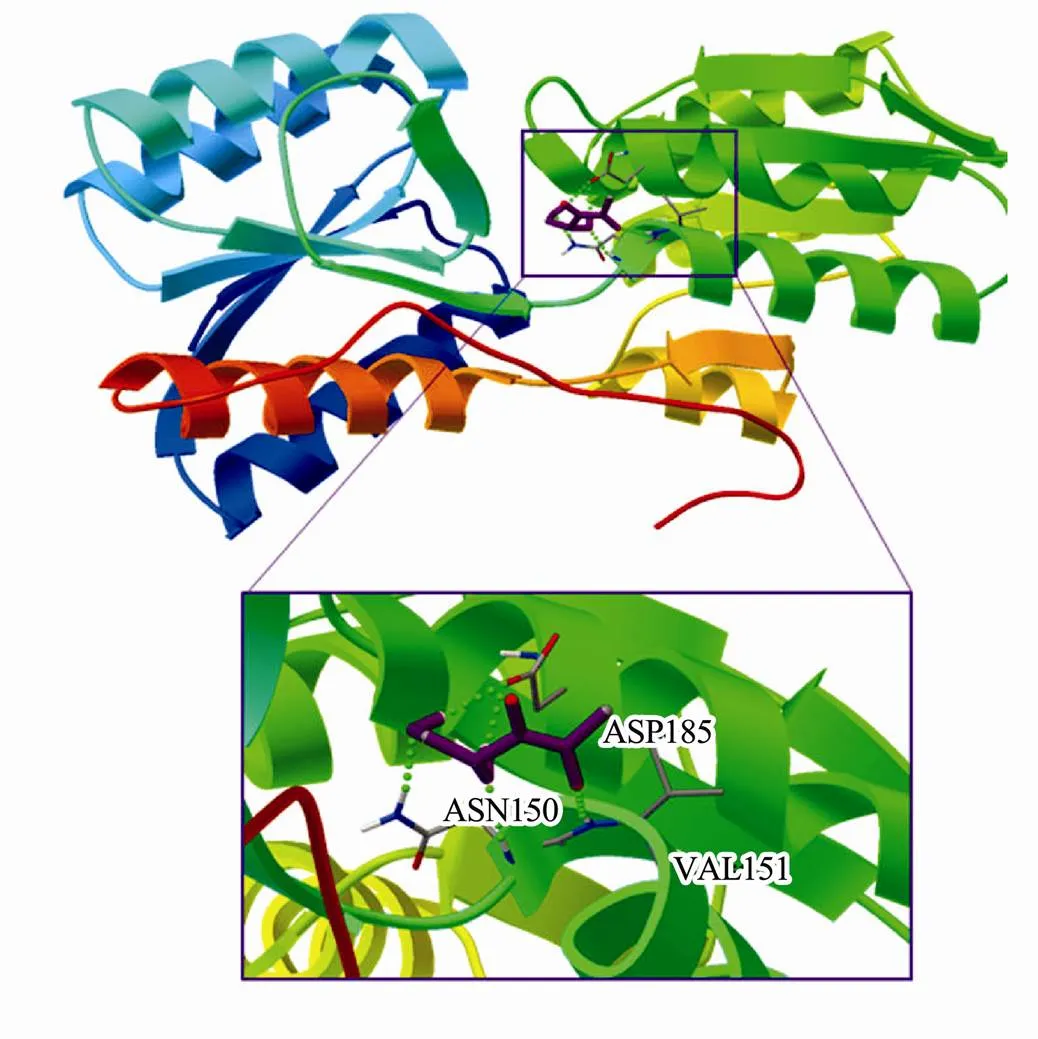
Fig.9 Molecular simulation analysis of interactions between DPD and RbsB. Detailed illustration of the amino acid residues lining the binding site in RbsB protein. RbsB and ligands (DPD) are displayed in stick format. The green dotted line represents the hydrogen bond.
3.5 S. agalactiae RbsB Inhibits AI-2-Dependent Induction of V. harveyi Bioluminescence
Incubation of purified RbsB withBB170 andAI-2 potently inhibited the induction ofbioluminescence in a dose-dependent manner (Fig.10A). The level ofbioluminescence that was induced in the positive-control reaction was approximately 1000-fold greater than light production byBB170 incubated with sterile AB medium. The concentration of RbsB required to inhibitAI-2-mediated induction ofbioluminescence by 50% was approximately 0.3ngmL−1. In addition, RbsB was also an effective inhibitor of bioluminescence induced by AI-2 from(Fig.10B), with 50% inhibition of light induction occurring in the presence of approximately 40ng per mL of RbsB. The induction ofbioluminescence caused by the positive control in these reactions was approximately 500-fold greater than that achieved by thenegative control. These results suggest that exogenously addedRbsB effectively competes withLuxP for AI-2 and that RbsB is capable of interacting not only with its cognate signal but also with a heterologous signal produced by an unrelated organism.
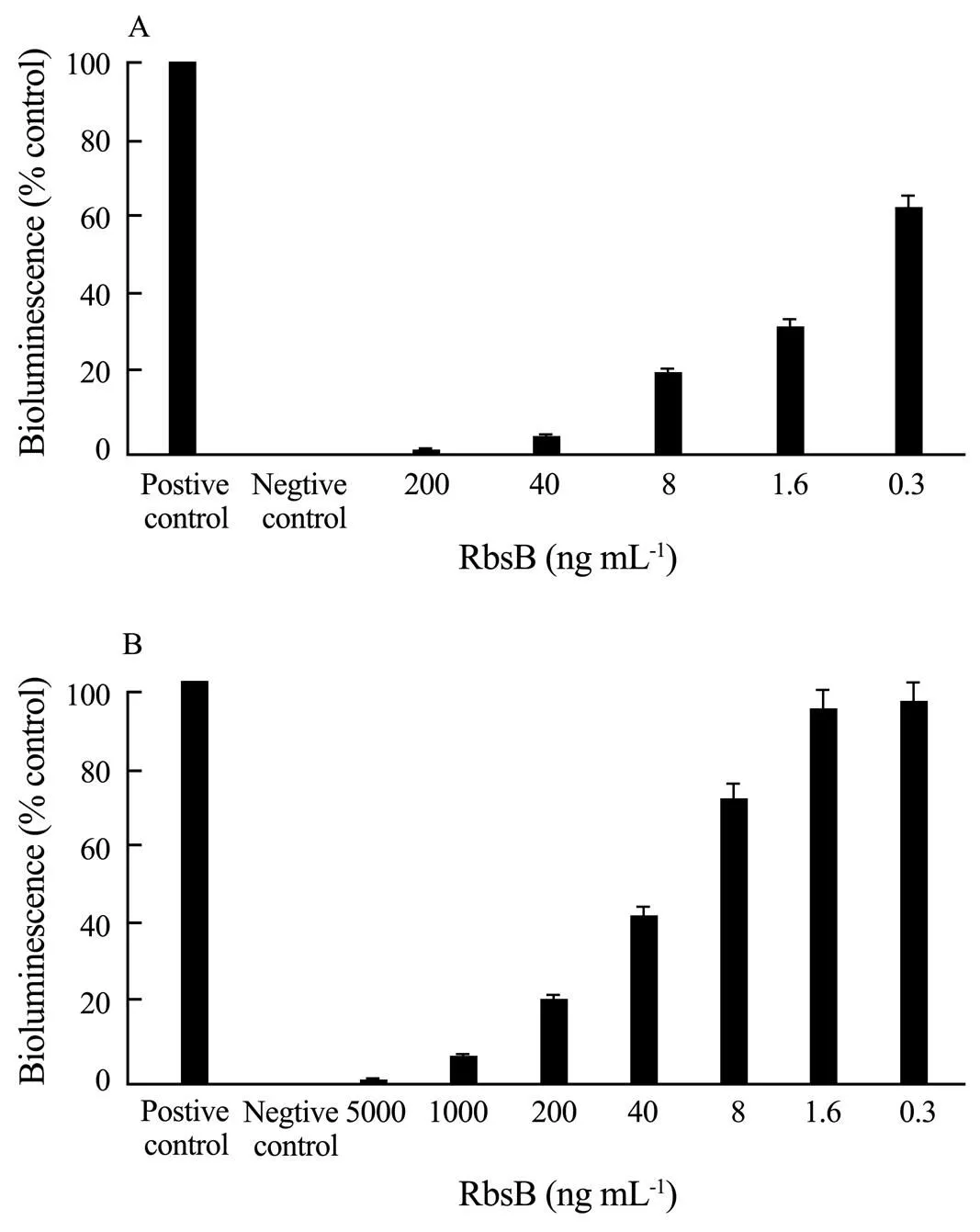
Fig.10 RbsB inhibits V. harveyi BB170 bioluminescence induced by S. agalactiae AI-2 (A) and V. harveyi AI-2 (B) in a dose-dependent manner.
4 Discussion
Quorum sensing can globally control many traits and behaviors of bacteria, allowing the bacteria to respond to the changes in the number of cells present in a community (Cao., 2020). In addition, QS signaling have a great potential for enhancement of community cooperation during the wastewater treatment process. The impacts of QS signaling were investigated in the wastewater and sludgeexploring the signaling species and receptor, physicochemical property and microflora (Huang, 2019). Some enzymes related to QS activity were applied as exogenous additives to the wastewater and sludge system and the corresponding signaling reaction to enzymes was effective. The QS signaling AI-2 of QS-related organisms in the community was not clear due to the complex microbial community structure in wastewater and sludge system (Maddela, 2019). The detailed mechanism of QS function and the effective application of QS signaling need to be elucidated by continuous investigation. In this study, we confirmed thatZQ0910 possesses a functional RbsB protein and produces AI-2. To evaluate the function of RbsB/AI-2 quorum sensing, we used these spectroscopic techniques: CD, ITC, and molecular docking to identify RbsB as a requirement for the uptake of and response to AI-2 byZQ0910, and equally synthesized the signal molecule AI-2. Our current studies showed thatRbsB interacted with AI-2 and played a role in the cellular response to AI-2, suggesting that RbsB may also function as AI-2 transporter.
Our previous work revealed that when RbsB was expressed using pET28 vector, most of the His-tagged products were inclusion bodies, and the purified protein was relatively unstable in solution. Although, RbsB complex could be obtained by a tandem expression method, we were unable to get high quality protein from this sample because of the fusion of the His-tag fused onto the RbsB. This was improved by overexpressing the GST- tagged RbsB into obtain a purer RbsB. However, this made it easier for the exploration of its function, collection of diffraction data and crystallization in future. After expression, the protein was purified to homogeneity by utilizing chromatographic techniques which included GST affinity purification by using at first a Glutathione Sepharose column (GE Healthcare), the target protein was purified by Glutathione Sepharose 4B equilibrated against HRV 3C reaction buffer and finally by using a Superdex 75 10/300 GL gel filtration column (GE Health- care). GST affinity chromatography could remove most of impure proteins. Remarkably, the RbsB elution gave a perfect single peak during gel chromatography and the purity of the protein as estimated by SDS-PAGE was >95%. Our work provided a particular method of protein expression and purification and laid the foundation for function and structure determination of RbsB and more similar enzymes.
Circular dichroism (CD) is widely used to analyze the interactions, secondary structure and folding of proteins (Fasman., 1970; Adler, 1973; Bertucci, 2011). It is often desirable to study the protein structure and interactions at the concentrations, when the proteins are highly potent and hence administered at low concentrations or are formulated at high concentrations for parenteral injections (Bowen, 2012; Wang.,2015; Baek and Zydney, 2018; Beate., 2018). The effect of AI-2 precursor DPD on the secondary structures of RbsB was further confirmed using CD spectroscopy. It is evident from Fig.7 that RbsB showed two characteristic valleys at 208 and 220nm indicating the high content of α helixl. The presence of DPD reduced the ellipticityof the CD spectra of RbsB indicating the reduction in the secondary structure of RbsB. Furthermore, CD Pro software was employed to decipher the changes occurring in the α-helical content of RbsB. From the above data, it was evident that the interactions of DPD with RbsB caused a slight conformational change of the protein, which leads to a decrease of helical content. In addition, upon interacting with DPD, a decrease in the ellipticity of RbsB was observed in the CD spectrum, indicating a partial unfolding of the helical structure. The data confirmed that the binding of DPD perturbs the helical content in RbsB.
The isothermal titration calorimetry (ITC) assay is a convenient and widely used approach to directly measure the amount of heat released or absorbed during association processes of biomolecules (such as protein-protein, protein-DNA, or protein-small molecules) in solution and to quantitatively estimate the interaction affinity (Lin and Wu, 2019). By fitting the interaction pattern of two reactants by the Origin software, change in enthalpy, Δ, the binding stoichiometry, the binding capacity,, the dissociation constant,d, and the change of entropy, Δcan be obtained. When the binding stoichiometry () and the concentration of the reactants in the sample cell are fixed, the greater the, the stronger the binding capacity between the two reactants will be. Whenis too small, correct, Δ, anda cannot be obtained because the curve is too flat. In this research,=392.7,value of 5–500 is generally considered as a good range to get the above parameters.=(7.22±1.74)×10−6 M, as a rule of thumb, Kdon the micromolar (μmolL−1or 10−6M) level describes a medium-strength association.
Computational methods have played pivotal role in drug discovery efforts for many years. Their principles and implementations have evolved together with the concepts of molecular recognition on protein surface. In particular, the historic ‘lock and key’ mechanism that served as a textbook explanation of substrate recognition at the enzyme active site has gradually developed into ‘hand and glove’ concept to account for protein flexibility and mutual adaptive ability of receptor and ligand (Sledz and Caflisch, 2018). Regarding the molecular docking simulation, The binding energy of the most favorable orientation of DPD bound with RbsB was −3.48kcalmol−1. The selected residues, including ASN150, ASP185, VAL151 were observed to form hydrogen bonds with DPD. These interactions have been confirmed in the CD spectrometry and thermodynamic data.
Many gram-positive and gram-negative bacteria species express LuxS and secrete and respond in various ways to a signal that is related to autoinducer-2 of(Sperandio., 2003). However, few of these organisms possess the dedicated two-component circuit that mediates the cell density-dependent response ofto AI-2 (James., 2006). So, in other species of bacteria, what acts at the receptor for the AI-2 signal molecule? Analysis of the complete genome sequence ofZQ0910 indicates that this organism lacks direct homologs of the dedicated sensor kinase/phosphatase (LuxQ) and the phospho transfer polypeptide (LuxU) that exist inspp. Inand, a soluble cell periplasmic protein family member called Ribose binding protein B (RbsB) was found, which is part of an ABC transporter involved in the uptake of AI-2. TheRbsB is similar to theandperiplasmic ribose binding proteins (76% sequence identity to each) encoded by theoperon (), which function to transport ribose into the cell. The RbsB ofhas recently been shown to bind to AI-2 and function as part of a transport system encoded by theoperon, which internalizes AI-2 (Taga, 2001, 2003; James., 2006). Since AI-2 is derived from the ribose moiety of S-ribosylhomocysteine, we hypothesized that theoperon (gene) may also function in the transport of AI-2 in. In our study, RbsB that was purified fromZQ0910 inhibitedbioluminescence induced by bothAI-2 andin a dose-dependent manner, suggesting that RbsB not only competes with LuxP (the periplasmic receptor for AI-2 in) for AI-2, but also appeared to interact with theAI-2. Of concern is that RbsPB was a more effective competitive inhibitor ofbioluminescence that was induced byAI-2 than of that induced byAI-2. Fifty percent inhibition of light production induced byAI-2 occurred with the addition of 0.3ng mL−1of RbsB, whereas approximately 40ngmL−1RbsB was required for 50% inhibition of bioluminescence produced byAI-2. The result is similar toRbsB, which was also more effective competitive inhibitor ofbioluminescence (James., 2006). These results suggest that, there is a difference in binding affinity forAI-2 andAI-2 by RbsB, possibly arising from structural variation of theandAI-2 signals. This observation also showed that there exists difference between known isoforms of AI-2 in their structural similarity. Indeed, multiple forms of AI-2 are known to exist. LsrB inserovar Typhimurium produces a form of AI-2 that differs in stereochemistry from and lacks the borate diester that is present in AI-2 produced by(Miller., 2004). Thus, it is possible thatmay utilize different means for uptake of quorum signal or that it may be responding to AI-2 signaling in a different manner in special environments. Further investigation into the affinity with the AI-2 isoform in terrestrial and aquatic environments, which provides way for future wastewater treatment, relies on QS-based strategies.
5 Conclusions
The focus of this study was to determine if the periplasmic ribose binding protein RbsB interacts with AI-2 and represents a potential receptor for AI-2 in. Here, we utilised CD spectroscopy, ITC200and molecular docking to determine if AI-2 (4,5-dihydroxy-2, 3-pentanedione, DPD) specifically interacts with purified RbsPB. In addition, we showed that purified RbsB inhibits AI-2-mediated induction ofbioluminescence. Collectively, these results suggest that RbsB may play a role in the response ofto AI-2. Our studies provide the first evidence that theoperon may be involved in the recognition of AI-2 byThe next stage of works to be investigated include: the physiologic role of AI-2, determination of the structure of theAI-2 and clarification of the molecular pathway that governs the response of the organism to AI-2. Additional studies are required to determine the downstream events that occur within the bacterium after internalization of signal.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (Nos. 31702386, 31660251, 31860245 and 31960203), the International Cooperation Science & Technology Planning Project of Guangdong Province of China (No. 2017A050501037), the Natural Science Foundation of Guangxi Province (Nos. 2018 GXNSFAA281019, 2017GXNSFAA198010), and the Central Government Directs Special Funds for Local Science and Technology Development Projects (No. ZY1949 015). E. Wangkahart was supported financially by the Ministry of Science and Technology of Thailand and Mahasarakham University.
Adler, A. J., Greenfield, N. J., and Fasman, G. D., 1973. Circular dichroism and optical rotatory dispersion of proteins and polypeptides. In:. Academic Press, 675-735.
Al-Harbi, A. H., and Uddin, N., 2005. Bacterial diversity of tilapia () cultured in brackish water in Saudi Arabia., 250: 566-572.
Baek, Y., and Zydney, A. L., 2018. Intermolecular interactions in highly concentrated formulations of recombinant thera- peutic proteins., 53: 59-64.
Balestrino, D., Haagensen, J. J., Rich, C., and Forestier, C., 2005. Characterization of type 2 quorum sensing inand relationship with biofilm formation., 187: 2870-2880.
Bassler, B. L., and Losick, R., 2006. Bacterially speaking., 125: 237-246.
Bassler, B. L., Wright, M., and Silverman, M. R., 1994. Multi- ple signalling systems controlling expression of luminescence in: Sequence and function of genes encoding a second sensory pathway., 13: 273- 286.
Beate, B., Wolfgang, R., and Johannes, S., 2018. Subcutaneous administration of biotherapeutics: An overview of current challenges and opportunities., 32: 425-440.
Bertucci, C., Pistolozzi, M., and De Simone, A., 2011. Structural characterization of recombinant therapeutic proteins by circular dichroism., 12 (10): 1508-1516.
Blancheton, J. P., Attramadal, K. J. K., Michaud, L., D’orbcastel, E. R., and Vadstein, O., 2013. Insight into bacterial popula- tion in aquaculture systems and its implication., 53: 30-39.
Bowen, M., Armstrong, N., and Maa, Y. F., 2012. Investigating high-concentration monoclonal antibody powder suspension in nonaqueous suspension vehicles for subcutaneous injection., 101 (12): 4433-4443.
Cao, Q., Ma, K., Nie, M., Dong, Y., Lu, C., and Liu, Y., 2020. Role ofin immune evasion and pathogenicity of piscineis not dependent on autoinducer-2., 99: 274-283.
Carbone, D., and Faggio, C., 2016. Importance of prebiotics in aquaculture as immunostimulants. Effects on immune system ofand., 54:172-178.
Chen, X., Schauder, S., Potier, N., Van Dorsselaer, A., Pelczer, I., Bassler, B. L., and Hughson, F. M., 2013. Structural identi- fication of a bacterial quorum-sensing signal containing bo- ron., 415: 545-549.
Davies, D. G., Parsek, M. R., Pearson, J. P., Iglewski, B. H., Costerton, J. W., and Greenberg, E. P., 1998. The involve- ment of cell-to-cell signals in the development of a bacterial biofilm., 280 (53): 295-298.
Delannoy, C. M. J., Crumlish, M., Fontaine, M. C., Pollock, J., Foster, G., Dagleish, M. P.,., 2013. Humanstrains in aquatic mammals and fish., 13:41.
Fasman, G. D., Hoving, H., and Timasheff, S. N., 1970. Circular dichroism of polypeptide and protein conformations. Film studies., 9 (17): 3316-3324.
Gu, Y., Li, B., Tian, J., Wu, R., and He, Y., 2018. The response of LuxS/AI-2 quorum sensing in2-1 to changes in environmental growth conditions., 68:287-294.
Han, X. G., and Lu, C. P., 2009.biosynthesis of autoinducer 2 ofSerotype 2 using recombi- nant LuxS and Pfs., 44 (1): 40-45.
Hancock, L. E., and Perego, M., 2004. Thetwo-component system controls biofilm develop- ment through production of gelatinase., 186 (17): 5629-5639.
Henke, J. M., and Bassler, B. L., 2004. Three parallel quorum-sensing systems regulate gene expression in, 186 (20): 6902-6914.
Hu, W. T., Guo, W. L., Meng, A. Y., Sun, Y., Wang, S. F., Xie, Z. Y.,., 2017. A metabolomic investigation into the effects of temperature onfrom Nile tilapia () based on UPLC-MS/MS., 210: 174-182.
Huang, J., Yi, K., Zeng, G., Shi, Y., Gu, Y., Shi, L.,., 2019. The role of quorum sensing in granular sludge: Impact and future application: A review., 236: 124310.
James, D., Shao, H., Lamont, R. J., and Demuth, D. R., 2006. Theribose binding protein RbsB interacts with cognate and heterologous auto- inducer 2 signals., 74 (7):4021-4029.
Kamaraju, K., Smith, J., Wang, J., Roy, V., Sintim, H. O., Bentley, W. E.,., 2011. Effects on membrane lateral pre- ssure suggest permeation mechanisms for bacterial quorum signaling molecules., 50 (32): 6983-6993.
Kristich, C. J., Li, Y. H., Cvitkovitch, D. G., and Dunny, G. M., 2018. Esp-independent biofilm formation by., 186 (1): 154-163.
Lin, K., and Wu, G., 2019. Isothermal titration calorimetry assays to measure binding affinities., 189: 257-272.
Maddela, N. R., Sheng, B., Yuan, S., Zhou, Z., Villamar-Torres, R., and Meng, F., 2019. Roles of quorum sensing in biolo- gical wastewater treatment: A critical review., 221: 616-629.
Marketon, M. M., Glenn, S. A., Eberhard, A., and González, J. E., 2003. Quorum sensing controls exopolysaccharide pro- duction in., 185 (1): 325-331.
Miller, S. T., Xavier, K. B., Campagna, S. R., Taga, M. E., Semmelhack, M. F., Bassler, B. L.,., 2004.recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2., 15 (5): 677-687.
Ohtani, K., Hayashi, H., and Shimizu, T., 2002. Thegene is involved in cell-cell signalling for toxin production in., 44 (1): 171- 179.
Parsek, M. R., and Greenberg, E. P., 2013. Sociomicrobiology: The connections between quorum sensing and biofilms., 13 (1): 27-33.
Pereira, C. S., Thompson, J. A., and Xavier, K. B., 2013. AI-2- mediated signalling in bacteria., 37 (2): 156-181.
Quiñones, B., Dulla, G., and Lindow, S. E., 2005. Quorum sen- sing regulates exopolysaccharide production, motility, and virulence in., 18 (7): 682-693.
Ramsey, M. M., Korgaonkar, A. K., and Whiteley, M., 2009. Quorum-sensing in bacteria. In:3rd edition, Schaechter, M., ed., Oxford Academic Press, Oxford, 357-374.
Rice, S. A., Koh, K. S., Queck, S. Y., Labbate, M., Lam, K. W., and Kjelleberg, S., 2005. Biofilm formation and sloughing inare controlled by quorum sensing and nutrient cues., 187 (10):3477-3485.
Shao, H., James, D., Lamont, R. J., and Demuth, D. R., 2007. Differential interaction of()LsrB and RbsB proteins with auto- inducer 2., 189 (15): 5559-5565.
Shrout, J. D., and Nerenberg, R., 2012. Monitoring bacterial twitter: Does quorum sensing determine the behavior of water and wastewater treatment biofilms?, 46 (4): 1995-2005.
Sledz, P., and Caflisch, A., 2018. Protein structure-based drug design: From docking to molecular dynamics., 48: 93-102.
Song, X. N., Cheng, Y. Y., Li, W. W., Li, B. B., Sheng, G. P., Fang, C. Y.,., 2014. Quorum quenching is responsible for the underestimated quorum sensing effects in biological wastewater treatment reactors., 171: 472-476.
Sperandio, V., Torres, A. G., Jarvis, B., Nataro, J. P., and Kaper, J. B., 2003. Bacteria-host communication: The language of hormones., 100 (15):8951-8956.
Suntharalingam, P., and Cvitkovitch, D. G., 2005. Quorum sen- sing in streptococcal biofilm formation., 13 (1): 3-6.
Surette, M. G., and Bassler, B. L., 1998. Quorum sensing inand,95 (12): 7046-7050.
Taga, M. E., Miller, S. T., and Bassler, B. L., 2003. Lsr-mediat- ed transport and processing of AI-2 in.,50 (4): 1411-1427.
Taga, M. E., Semmelhack, J. L., and Bassler, B. L., 2001. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in., 42 (3):777- 793.
Tyrrel, S. F., and Quinton, J. N., 2003. Overland flow transport of pathogens from agricultural land receiving faecal wastes., 94 (s1): 87-93.
Wang, S., Zhang, N., Hu, T., Dai, W., Feng, X., Zhang, X.,., 2015. Viscosity-lowering effect of amino acids and salts on highly concentrated solutions of two IgG1 monoclonal anti- bodies., 12 (12): 4478-4487.
Yu, Z., Hu, L., and Lo, I. M. C., 2019. Transport of the arsenic (As)-loaded nano zero-valent iron in groundwater-saturated sand columns: Roles of surface modification and as loading., 216: 428-436.
#The two authors contributed equally to this work.
. E-mail: wong19820204@126.com
October 20, 2020;
March 10, 2021;
March 31, 2021
© Ocean University of China, Science Press and Springer-Verlag GmbH Germany 2021
(Edited by Ji Dechun)
 Journal of Ocean University of China2021年5期
Journal of Ocean University of China2021年5期
- Journal of Ocean University of China的其它文章
- Precipitation Distribution of the Extended Global Spring–Autumn Monsoon and Its Possible Formation Mechanism
- Observation of Internal Tides in the Qiongzhou Strait by Coastal Acoustic Tomography
- Sedimentary Changes of a Sand Layer in Liquefied Silts
- Optimizing Winding Angles of Reinforced Thermoplastic Pipes Based on Progressive Failure Criterion
- Seismic Images of Shallow Waters over the Shatsky Rise in the Northwest Pacific Ocean
- Distribution and Sources of Particulate Organic Matter in the Northern South China Sea: Implications of Human Activity
