A new snake species of the genus Gonyosoma Wagler,1828 (Serpentes:Colubridae) from Hainan Island,China
A new species of the genusGonyosomaWagler,1828 is described herein based on six specimens from the Diaoluoshan Mountains,Hainan Island,Hainan Province,China.The new species,Gonyosoma hainanensesp.nov.,is most similar to its continental sister species,Gonyosoma boulengeri(Mocquard,1897).Both taxa have a scaled protrusion on the anterior portion of the rostrum,distinct from other congeners.However,Gonyosoma hainanensesp.nov.can be distinguished fromG.boulengeriby two significant morphological characters:(1) black orbital stripe absent in adults (vs.present inG.boulengeri);and (2) two loreals (vs.one loreal inG.boulengeri).The new species is also genetically divergent and forms a unique clade from its sister species and all other congeners based on sequences of the mitochondrial gene cytochromeb(cytb).
Gonyosoma boulengeriwas first described by Mocquard(1897) based on six specimens from the Gulf of Tonkin in Vietnam.Its common name,rhinoceros snake,is derived from the distinctive scaled protrusion at the distal end of its rostrum.In adults,the body is green and abdomen is yellowish green.In contrast,neonate and juvenile color patterns are gray,but gradually change to green as they mature (Orlov et al.,1999).
Formerly,the rhinoceros snake was considered a single species,i.e.,G.boulengeri,and was reported from Hainan,Guangxi,Guangdong,and Yunnan provinces in China,as well as Vietnam (Fan,1931;Hecht et al.,2013;Luu et al.,2013;Mocquard,1897;Nguyen et al.,2020;Peng et al.,2017;Yang et al.,2018;Zhao,2006).From 2010 to 2019,we collected six rhinoceros snake specimens from the Diaoluoshan Mountains in Hainan Island,China.Based on morphological and molecular comparisons,the Hainan Island rhinoceros snake population differs from the mainlandG. boulengeripopulations.Thus,we treat the Hainan Island population as a new species.
In total,16 specimens,including six specimens from Hainan Island,nineG.boulengerispecimens from Yunnan (3) and Guangdong (3),China,and Lào Cai (3),Vietnam,and oneG.frenatumspecimen from Huangshan,Anhui Province,China,were sampled and used in the present study.Fresh liver and muscle tissues were maintained in 90% ethanol and stored at−80 °C.Identification of the sex of the four adults from Hainan Island was determined via gonadal examination and the presence of a hemipenis.The specimens examined in this study were preserved in 75% alcohol and deposited at the Anhui Normal University Museum (ANU) and Hainan Normal University Museum (HNU).
Genomic DNA was extracted from macerated liver or muscle tissues using an Ezup Column Animal Genomic DNA Purification Kit (Map Biotech,China),according to the protocols of the manufacturer.The sequences of the mtDNA fragment of cytbwere obtained and amplified by polymerase chain reaction (PCR) using primers L14910/H16064 (Burbrink et al.,2000).The PCR products were sequenced using Sanger sequencing by a commercial company (Map Biotech,China).Sequences were assembled manually using SeqMan in Lasergene v15.1 (DNASTAR,USA).The six Hainan Island rhinoceros snakes shared four haplotypes,while the nine continental rhinoceros snakes (G.boulengeri) shared three haplotypes.All newly generated sequences (1 116 bp),including that of the singleG.frenatumspecimen,were deposited in GenBank (accession Nos.:MW495258–MW495263).
Homologous sequences of all congeners (accession Nos.in Figure 1A,from Alencar et al.,2016;Burbrink & Lawson,2007;Chen et al.,2014a,2014b;Lawson et al.,2005;Li et al.,2020) and outgroup taxa (Boiga kraepelini,MN962360,Weinell et al.,2020) were retrieved from GenBank.The sequences were aligned in MEGA X using the ClustalW algorithm with default parameters (Kumar et al.,2018),and checked by eye for ambiguous alignments.
Bayesian inference (BI) and maximum-likelihood (ML)approaches were used to reconstruct the phylogenetic bifurcating trees. Prior to analyses,ModelFinder(Kalyaanamoorthy et al.,2017) was used to select the best-fit model (HKY+F+G4) under Bayesian information criterion.MrBayes v3.2.6 (Ronquist et al.,2012) was used for BI analyses.All searches consisted of four independent runs,each initiated with a random tree.Each run consisted of four Markov chains (three heated chains and a single cold chain)estimated for 4×106generations and sampled every 1 000 generations,with the initial 25% of sampled data discarded as burn-in.Convergence was assessed based on effective sample size (ESS>200) and likelihood plots against generation time using Tracer v1.7 (Rambaut et al.,2018),as well as similarity among runs.After confirming that the four analyses reached stationarity at a similar likelihood score and topologies were similar,the resulting trees were combined to calculate posterior probabilities (PP) for each node in a 50%majority-rule consensus tree.The ML tree was reconstructed using RAxML v7.2.6 (Stamatakis,2006) in the GTRGAMMA model with 1 000 ultrafast (Minh et al.,2013) bootstrap (BS)replicates. Uncorrected pairwise distances (P-distances)among ingroup taxa were calculated using the neighborjoining method (Tamura et al.,2004) in MEGA X (Kumar et al.,2018).
Morphological data,including 11 meristic characters,were examined as follows:loreal (Lor),preocular (PrO),postocular counts (PtO),supralabial (SL),infralabial (IL),temporal counts(Tem),dorsal scale row numbers (DSR,counting at one head length behind head,at midbody,and at one head length before vent,respectively),ventral counts (VEN,according to Dowling (1951)),subcaudal counts (SC,terminal scute excluded from number of subcaudals),preanal plate (PreAn),and infralabial contact with first pair of chin shields (IL-CS).Nine measurements were also recorded as follows:total length (TL,distance from tip of snout to tip of tail),snout-vent length (SVL,distance from tip of snout to posterior margin of cloaca),tail length (TaL,distance from posterior margin of cloaca to tail tip),head length (HL,posterior edge of jaw to tip of snout),head width (HW,maximum head width),eye length(EL,maximum vertical eye length),eye width (EW,maximum horizontal eye width),head height (HH),and rostral width(RW).We also analyzed the TaL/TL and HW/HL ratios.Here,TL,SVL,and TaL were measured with a ruler to the closest 1 mm,all other measurements were obtained using a digital caliper to 0.1 mm.
The topologies of the BI and ML bifurcating trees were identical.The six Hainan Island rhinoceros snakes formed a monophyletic clade with high support (1.00 PP and 93 BS,Figure 1A),and were sister toG.boulengeri.TheGonyosomaspecies relationships were similar to those reported in Pyron et al.(2013) and Chen et al.(2014b),i.e.,(G.oxycephalum,G.jansenii)+((G.margaritatum,G.prasinum)+(G.frenatum+(G.boulengeri,Hainan Island rhinoceros snakes))).
Supplementary Table S1 shows the intraspecific and interspecific uncorrectedP-distances. The intraspecific distances among the six Hainan Island rhinoceros snakes ranged from 0.0% to 0.6%,whereas the distances between the Hainan rhinoceros snake and continental sister species,G.boulengeri,ranged from 2.5% to 3.0%.Thus,combined with morphological data,the specimens from Diaoluoshan,Hainan,China,are considered a new species and described below.
Taxonomic account
Rhynchophis boulengeriF.Mocquard,1897,Bull.Mus.Hist.Nat.,Paris,[ser.1],3:215.Type locality:Baie d’Along,Iles Norway,Gulf of Tonkin,Vietnam.
Proboscidophis versicolorT.-H.Fan,1931,Bull.Dept.Biol.Coll.Sun Yatsen Univ.,Canton,11:114.Type locality:Kutchen (=Guchen),Yaoshan (=Dayao Shan),Guangxi Zhuang Autonomous Region,China.
Gonyosoma boulengeriX.Chen et al.,2014,Zootaxa,3 881(6):532–548.
Gonyosoma hainanense sp.nov.L.Peng,Y.Zhang,S.Huang,F.T.Burbrink,and J.Wang,2021
(Figure 1B–F;Supplementary Figure S1A–F;Supplementary Table S2)
Suggested English name:Hainan rhinoceros snake.
Suggested Chinese name:海南尖喙蛇 (Bopomofo:hǎi nán jiān huì shé).
Holotype:ANU20190002 (collection number:HSR2010074),adult female (Figure 1B–F),captured by H.Yang and M.Hou in the summer of 2010 from Diaoluoshan (N18°39′58.57″,E109°54′50.48″;elevation of 204 m a.s.l.),Hainan,China.
Paratypes (five specimens):One subadult (ANU20190003,collection number:HSR2012105,Supplementary Figure S1D,E),sampled by Y.W.Zhu and L.F.Peng from Diaoluoshan,Hainan,China,in the summer of 2012.One male and one subadult (HNU20190001–2,dead on road),collected by Y.Zhang and J.C.Wang on 28 April 2019 from the same site as the holotype. One adult female and one male(HNU20190003–4),collected by J.C.Wang in the summer of 2010 from Diaoluoshan,Hainan,China,elevation~900 m a.s.l.
Diagnosis:The new species,Gonyosoma hainanensesp.nov.,can be distinguished by the following characters:(1)Long pointed rostral appendage covered with small smooth scales;(2) SVL 652–927 mm in adults;(3) Dorsal scales slightly keeled,19-19-15(13);(4) Ventral and subcaudal scales strongly angulated laterally; (5) Ventral scales 216–221;(6) Subcaudals paired,122–135;(7) Anal plate divided;(8) One preocular,two postoculars;(9) Supralabials 9/9,fourth to sixth in contact with eye;(10) Infralabials 10–12,first five in contact with first pair of chin shields;(11)Temporals 2+2+3 (rarely 2+3+3);(12) Black orbital stripe absent in adults;(13) Two loreals oriented longitudinally.
Description of holotype:Adult female;SVL 927 mm;TaL 302 mm;TL 1 229 mm;TaL/TL 0.246;HW/HL 0.425;PrO 1;PtO 2;Tem 2-3-3;SL 9/9,fourth to sixth in contact with eye;CS two pairs;IL 11/12,first pair in contact behind mental,first five in contact with first pair of chin shields (Figure 1C);Loreals 2 (Figure 1F),posterior loreal partially or completely extending below preocular;DSR 19-19-13,dorsal scales slightly keeled;VEN 216;SC 122;anal plate divided.
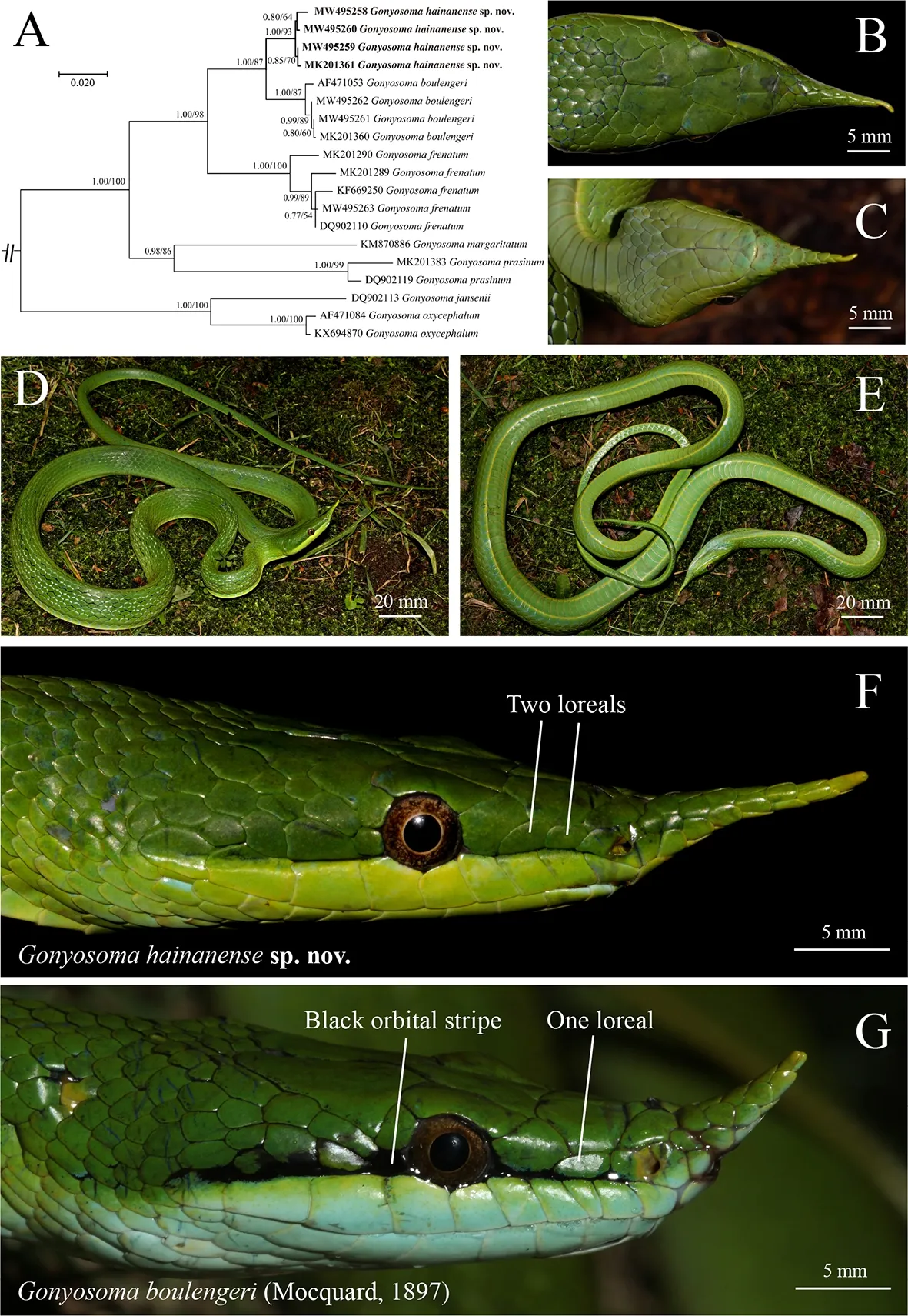
Figure 1 Phylogenetic tree,holotype of Gonyosoma hainanense sp.nov.,and comparisons with Gonyosoma boulengeri (Mocquard,1897)
Holotype possesses prominent,distinctive,scaled protrusion on front of snout,~10 mm long.Head triangular,distinct from neck.Eye large with round pupil.Prefrontal nearly twice as long as internasal;frontal very broad and flat in front,sharp at back,inverted triangle in shape,slightly shorter than parietals (Figure 1B).
Coloration in life:Dorsal ground color uniform green,changing to pale green on sides of body.Mid and upper flanks of body with black interstitial skin,one or two short white stripes on anterior lateral margin of most dorsal scales exposed and visible when body inflated.Ventral surface pale green,yellow line along each lateroventral edge(Figure 1D,E).
Coloration in preservation:Dorsal ground color blue,ventral ground color gray,off-white line along each lateroventral edge.
Morphological variation:Measurement and pholidosis details for holotype and paratypes are listed in Supplementary Table S2.
Comparisons:Gonyosoma hainanensesp.nov.is most similar toG.boulengeri,with both distinguished from all other species ofGonyosomaby a distinctive scaled protrusion on front of snout.
The new species can be distinguished fromG.boulengeriby black orbital stripe in adults absent (vs.present inG.boulengeri) and two loreals (vs.one loreal inG.boulengeri)(Figure 1F,G).
Etymology:The specific name refers to the province of its type locality,i.e.,Hainan Island,Hainan Province,China.
Distribution and natural history:Gonyosoma hainanensesp.nov.is currently known only from the Diaoluoshan Mountains (type locality) and the Jianfengling Mountains of Hainan Island,Hainan Province,China.This species inhabits subtropical rainforests (Supplementary Figure S1G) at elevations between~200 and 900 m a.s.l.,particularly in valleys with streams.It is generally arboreal and nocturnal(Zhao et al.,1998;Zhao,2006).Furthermore,it is oviparous with a clutch size of six (white) eggs and an incubation period of 62 days (20 April–21 June 2019).Neonates and juveniles are gray,with black orbital stripes.Captive individuals(ANU20190003) consume mice.Wild individuals are preyed on byLycodon rosozonatus(Supplementary Figure S1A–F).Coloration gradually changes to green as the snake matures,and the black orbital stripes gradually fade.
Comments
The generaGonyophis,Rhadinophis,andRhynchophishave been synonymized within the single genusGonyosomabased on molecular and morphological similarities,as well as their comparable habitats throughout Southeast Asia (Chen et al.,2014b).The genusGonyosomacurrently contains six species:i.e.,G.oxycephalum(Boie,1827),G.frenatum(Gray,1853),G.prasinum(Blyth,1854),G.jansenii(Bleeker,1859),G.margaritatum(Peters,1871),andG.boulengeri(Mocquard,1897). In the present study,we describeGonyosoma hainanensesp.nov.as a new species,which increases the number of species withinGonyosomato seven.
Hainan Island is located in southern China,facing the Leizhou Peninsula of Guangdong,across the Qiongzhou Strait in the north,Guangxi and Vietnam across the Beibuwan Gulf in the west,and the South China Sea in the south.It is the second largest island in China,covering an area of more than 30 000 km2,and harbors many endemic species (Peng et al.,2018).Six specimens of the new species were obtained from the Diaoluoshan Mountains,and one juvenile and one female with six neonates (Supplementary Figure S1A,C) were observed and released in the Jianfengling Mountains.These two distribution sites are approximately 200 km apart,and span much of the southern part of Hainan Island.Thus,we believe the new species is likely to be distributed in other mountainous areas of Hainan Island.The adults of the rhinoceros snake previously recorded in Hainan also have no black orbital stripe (Shi et al.,2011;Zhao et al.,1998;Zhao,2006).The rhinoceros snakes distributed in Hainan should be only the new speciesGonyosoma hainanensesp.nov.rather thanG.boulengeri.
The updated List of Key Protected Wild Animals in China was approved and implemented by the State Council of the People’s Republic of China on 4 January 2021(http://www.forestry. gov.cn).Gonyosoma boulengerihas been upgraded to the Second Class of Nationally Protected Animals.We suggest thatGonyosoma hainanensesp.nov.be given the same attention,listing,and protection.
NOMENCLATURAL ACTS REGISTRATION
The electronic version of this article in portable document format represents a published work according to the International Commission on Zoological Nomenclature (ICZN),and hence the new names contained in the electronic version are effectively published under that Code from the electronic edition alone (see Articles 8.5–8.6 of the Code).This published work and the nomenclatural acts it contains have been registered in ZooBank,the online registration system for the ICZN.The ZooBank LSIDs (Life Science Identifiers) can be resolved and the associated information can be viewed through any standard web browser by appending the LSID to the prefixhttp://zoobank.org/.
Publication LSID:
urn:lsid:zoobank.org:pub:3D2E9839-A584-407B-9B43-693D851 42FE2
Gonyosoma hainanenseLSID:
urn:lsid:zoobank.org:act:B326744E-972D-4C39-97DC-19D122 69C8E1
SCIENTIFIC FIELD SURVEY PERMISSION INFORMATION
All sampling and procedures involving live snakes were performed in accordance with the Wild Animals Protection Law of the People’s Republic of China and approved by the Animal Ethics Committees at Anhui Normal University and Hainan Normal University.Permission for field surveys in Hainan Province was granted by the Management Office of the Diaoluoshan Scenic Area,Hainan Province,China.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
S.H.,J.C.W.,L.F.P.,and F.T.B.conceived and designed the study.L.F.P.,Y.Z.,and J.M.C.performed the experiments,data analyses,and manuscript preparation.M.H.,H.Y.,S.H.,Y.W.Z.,J.C.W.,L.F.P.,and Y.Z.collected materials.S.H.,J.C.W.,and F.T.B.revised the manuscript.All authors read and approved the final version of the manuscript.
ACKNOWLEDGEMENTS
We thank the Key Laboratory of Tropical Animal and Plant Ecology of Hainan Province,Diaoluoshan National Nature Reserve,Yinggeling National Nature Reserve,and Jianfengling National Nature Reserve for grant support.We thank Tong-Liang Wang,Xiao-Fei Zhai,Rong-Ping Bu,and Chen-Xu Wang (HNU) for their help with field work,and Mei-Yan Ke and Yu-Zhe Wang (HNU) for their help with morphometrics.We thank Nikolay A.Poyarkov (Lomonosov Moscow State University),Darrel R.Frost (American Museum of Natural History),Gernot Vogel (Society for Asian Herpetology),Patrick David (Muséum National d'Histoire Naturelle) and Nguyen Van Tan (Save Vietnam's Wildlife) for correcting the Latin name of the new species.
- Zoological Research的其它文章
- 3DPhenoFish:Application for two-and threedimensional fish morphological phenotype extraction from point cloud analysis
- PINK1 gene mutation by pair truncated sgRNA/Cas9-D10A in cynomolgus monkeys
- A bright future for the tree shrew in neuroscience research:Summary from the inaugural Tree Shrew Users Meeting
- Inhibition of mTOR signaling by rapamycin protects photoreceptors from degeneration in rd1 mice
- Clonal spread of Escherichia coli O101:H9-ST10 and O101:H9-ST167 strains carrying fosA3 and blaCTX-M-14 among diarrheal calves in a Chinese farm,with Australian Chroicocephalus as the possible origin of E.coli O101:H9-ST10
- Genome and population evolution and environmental adaptation of Glyptosternon maculatum on theQinghai-Tibet Plateau

