Clonal spread of Escherichia coli O101:H9-ST10 and O101:H9-ST167 strains carrying fosA3 and blaCTX-M-14 among diarrheal calves in a Chinese farm,with Australian Chroicocephalus as the possible origin of E.coli O101:H9-ST10
Wan-Yun He ,Xing-Xing Zhang ,Guo-Long Gao ,Ming-Yi Gao ,Fa-Gang Zhong ,Lu-Chao Lv ,Zhong-Peng Cai,Xing-Feng Si,Jun Yang,*,Jian-Hua Liu,*
1 College of Veterinary Medicine, South China Agricultural University, Guangzhou, Guangdong 510642, China
2 Guangdong Laboratory for Lingnan Modern Agriculture, Guangzhou, Guangdong 510642, China
3 State Key Laboratory for Sheep Genetic Improvement and Healthy Production, Institute of Animal Husbandry and Veterinary, Xinjiang Academy of Agricultural and Reclamation Science, Shihezi, Xinjiang 832000, China
4 Zhejiang Tiantong Forest Ecosystem National Observation and Research Station, School of Ecological and Environmental Sciences,East China Normal University, Shanghai 200241, China
ABSTRACT During a 2018 antimicrobial resistance surveillance of Escherichia coli isolates from diarrheal calves in Xinjiang Province,China,an unexpectedly high prevalence (48.5%) of fosfomycin resistance was observed. This study aimed to reveal the determinants of fosfomycin resistance and the underlying transmission mechanism. Polymerase chain reaction (PCR) screening showed that all fosfomycin-resistant E.coli carried the fosA3 gene.Pulsed-field gel electrophoresis (PFGE) and southern blot hybridization revealed that the 16 fosA3-positive isolates belonged to four different PFGE patterns (i.e.,A,B,C,D).The fosA3 genes of 11 clonally related strains (pattern D) were located on the chromosome,while others were carried by plasmids.Whole-genome and long-read sequencing indicated that the pattern D strains were E.coli O101:H9-ST10,and the pattern C,B,and A strains were O101:H9-ST167,O8:H30-ST1431,and O101:H9 with unknown ST,respectively.Among the pattern C strains,the blaCTX-M-14 gene was colocalized with the fosA3 gene on the F18:A-:B1 plasmids.Interestingly,phylogenetic analysis based on core genome single nucleotide polymorphisms(cgSNPs) showed that the O101:H9-ST10 strains were closely related to a Australian-isolated Chroicocephalus-origin E.coli O101:H9-ST10 strain producing CTX-M-14 and FosA3,with a difference of only 11 SNPs.These results indicate possible international dissemination of the high-risk E.coli clone O101:H9-ST10 by migratory birds.
Keywords: Clonal spread;Bovine;fosA3;blaCTX-M-14; O101:H9-ST10; Chroicocephalus
INTRODUCTION
Bacterial infections in domesticated bovines continue to increase year by year (Ruegg,2017).Diarrhea in calves,which is partly caused by pathogenicEscherichia coli,is one of the three major bovine diseases causing economic loss to cattle producers (Wieler et al.,2007).Antimicrobials are often used to treat calf diarrhea caused by pathogenicE.coli(Constable,2004).However,due to the abuse and misuse of antimicrobials,antimicrobial resistance (AMR) among bovineE.colihas become an important issue,especially for bacterial disease treatment and public health.Interestingly,in many cases,antibiotic resistance in cattle-originE.coliis lower than in that originating from pigs or chickens (Ho et al.,2011;Li et al.,2019).
As an old antibiotic used in the treatment of uncomplicated urinary tract infections,fosfomycin has been reintroduced with other antimicrobials for the clinical treatment of multidrugresistant (MDR) bacteria due to its excellent antimicrobial activity (Bassetti et al.,2019;Falagas et al.,2016).Although fosfomycin is not approved for animal use in China,fosfomycin resistance is widely reported among food animals nationwide.In addition,the plasmid-mediatedfosA3gene is reported to be a major determinant of fosfomycin resistance and is often co-localized with CTX-M β-lactamase genes (He et al.,2013,2017;Huang et al.,2020).Consequently,thefosA3gene can be co-selected under the use of β-lactam antibiotics.During AMR surveillance ofE.colifrom a cattle farm in Xinjiang Province,China,an unexpectedly high prevalence (48.5%) of fosfomycin resistance was observed,which was significantly higher than previously reported rates in bovines (Chan et al.,2014;Wang et al.,2017b).Hence,this study aimed to uncover the determinants of fosfomycin resistance and the underlying transmission mechanism in diarrheal calf-derivedE.coliisolates.
MATERIALS AND METHODS
Bacterial strain
A total of 51 fecal samples were collected from diarrheal calves aged less than one month from a farm located in Yili,Xinjiang,China,in May 2018.These calves had been treated with enrofloxacin,ceftiofur,gentamycin,ampicillin,penicillin,florfenicol,colistin,and tulathromycin.The collected samples were enriched in Luria-Bertani (LB) broth at 37 ℃ for 16–18 h.The overnight culture was then incubated on a MacConkey agar plate.One isolate showingE.colimorphology from each sample was further identified using matrix-assisted laser desorption ionization-time of flight-mass spectrometry (MALDITOF MS) (Shimadzu-Biotech Corp.,Japan).
Antimicrobial susceptibility testing and detection of resistance genes
According to the Clinical Laboratory Standard Institute (CLSI)guidelines (M07-A11),the minimum inhibitory concentrations(MICs) of allE.coliisolates against fosfomycin with 25 mg/L glucose-6-phosphate,beta-lactams (ampicillin,cefoxitin,ceftazidime,cefquinome,cefotaxime,and imipenem),aminoglycosides (amikacin,streptomycin,apramycin,gentamicin,and neomycin),tetracyclines (tetracycline and doxycycline),florfenicol,trimethoprim-sulfamethoxazole,and ciprofloxacin were determined using agar dilution or broth microdilution methods (colistin and tigecycline).TheE.coliATCC 25 922 strain was used for quality control.The MICs were interpreted according to the criteria of the CLSI (M100-S30) (for fosfomycin,ampicillin,cefoxitin,ceftazidime,cefotaxime,imipenem,gentamicin,amikacin,tetracycline,doxycycline,trimethoprim-sulfamethoxazole,and ciprofloxacin),EUCAST (http://www.eucast.org) (for colistin and tigecycline),US Food and Drug Administration (FDA) for streptomycin (S,≤32 mg/L;R,≥64 mg/L),US National Antimicrobial Resistance Monitoring System (NARMS) for apramycin (S,≤8 mg/L;R,≥64 mg/L),and veterinary CLSI(VET06-S1) (for cefquinome,neomycin,and florfenicol).
Polymerase chain reaction (PCR) amplification was used to screen the fosfomycin resistance genefosA3and other important antimicrobial resistance genes (ARGs),including the extended-spectrum beta-lactamase geneblaCTX-M-1G/9G,AmpC beta-lactamase geneblaCMY-2,16S rRNA methyltransferase genesarmAandrmtB,florfenicol resistance genefloR,and colistin resistance genemcr-1,using previously described primers (Supplementary Table S1) (Cao et al.,2020;Chen et al.,2007;Yan et al.,2004).PCR mapping was used to determine the genetic background offosA3with known primers (Supplementary Table S1) (Hou et al.,2012).The PCR products were subjected to Sanger sequencing (TsingKe Biological Technology,Beijing,China),and the obtained sequences were ascertained without mutation by NCBIBLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Pulsed-field gel electrophoresis (PFGE),S1-PFGE,and southern blot hybridization
The clonal relationship offosA3-positiveE.coliisolates was assessed based on a rapid PFGE protocol (Gautom,1997).Total DNA was digested by theXbaI enzyme (TaKaRa,Japan),embedded in low-melting-point agarose (Bio-Rad,USA),and subjected to PFGE using the CHEF-MAPPER System (Bio-Rad,USA).The electrophoretic conditions were:initial switch time,2.16 s;final switch time,63.8 s;running time,19 h;angle,120°;gradient,6.0 V/cm;temperature,14 °C;ramping factor,linear.TheSalmonella entericaserotype Braenderup H9812 was used as a molecular size marker.The gel was dyed with ethidium bromide,visualized using a gel imaging system (Bio-Rad,USA),and analyzed with BioNumerics v6.6 (Applied Maths,Belgium).DNA patterns were interpreted based on proposed criteria (Tenover et al.,1995).The S1-PFGE protocol was the same as that of PFGE,except that total DNA was digested by S1 nuclease (TaKaRa,Japan).The products were subsequently used to perform southern blot hybridization with a digoxigenin-labelledfosA3DNA probe (Roche,Germany).
Conjugation experiment
Horizontal transmission ability was determined for allfosA3genes,and sodium azide-resistantE.coliJ53 was used as the recipient for conjugation.The transconjugant was selected on a MacConkey agar plate supplemented with 150 mg/L sodium azide,128 mg/L fosfomycin,and 25 mg/L glucose-6-phosphate.The transconjugant underwent PCR amplification and Sanger sequencing to confirm the transfer of thefosA3gene.
Whole-genome sequencing analysis
Total genomic DNA,extracted using a Hipure Bacterial DNA Kit (Magen,China),was subjected to whole-genome sequencing by Novogene (Beijing Novogene Bioinformatics Co.,Ltd.,China) using Illumina platform Novo-PE150 technology and to long-read sequencing using Oxford Nanopore MinION (Oxford Nanopore Technologies,UK).SPAdes v3.8.7 (Bankevich et al.,2012) was used forde novoassembly.Unicycler v0.4.7 (Wick et al.,2017) was used to obtain the assembled genome.Whole-genome sequencing data were analyzedin silicousing MLST v2.11 (https://github.com/tseemann/mlst) for multi-locus sequence typing,ABRicate v0.8 (https://github.com/tseemann/abricate) for screening ARGs,plasmid types,and virulence factors,and SeroTypeFinder v2.0 for serotyping (Joensen et al.,2015).Sequence alignment was performed by Easyfig v2.1 (Sullivan et al.,2011).A phylogenetic tree based on core genome single nucleotide polymorphisms (cgSNPs) was constructed using Parsnp v1.5.4 (https://github.com/marbl/parsnp).Snippy v4.6.0 (https://github.com/tseemann/snippy) was used to calculate total SNP quantity.
Nucleotide sequence accession number
The assembled genomes of theE.coliisolates (XJW9B263 and XJW9B277) based on long-read sequencing were submitted to GenBank under accession Nos.CP067399–CP067401 and CP068041–CP068045,respectively.The raw reads (Illumina) of theE.coliisolates(XJW9B298,XJW9B290,XJW9B274,XJW9B277,XJW9B263,and XJW9B285) were deposited in the Genome Sequence Archive (GSA) under accession No.CRA004296.
RESULTS
Overall resistance phenotypes of E.coli isolates
A total of 33 non-duplicateE.colistrains were obtained.The antimicrobial susceptibility results showed that of the 33E.coliisolates,29 (87.9%) exhibited resistance to five or more antimicrobials,and most were resistant to critically important antimicrobials (CIAs),including third and fourth generation cephalosporins (n=24) and ciprofloxacin (n=24) (Figure 1).In particular,16 (48.5%) isolates showed resistance to fosfomycin,as well as cephalosporins and ciprofloxacin.
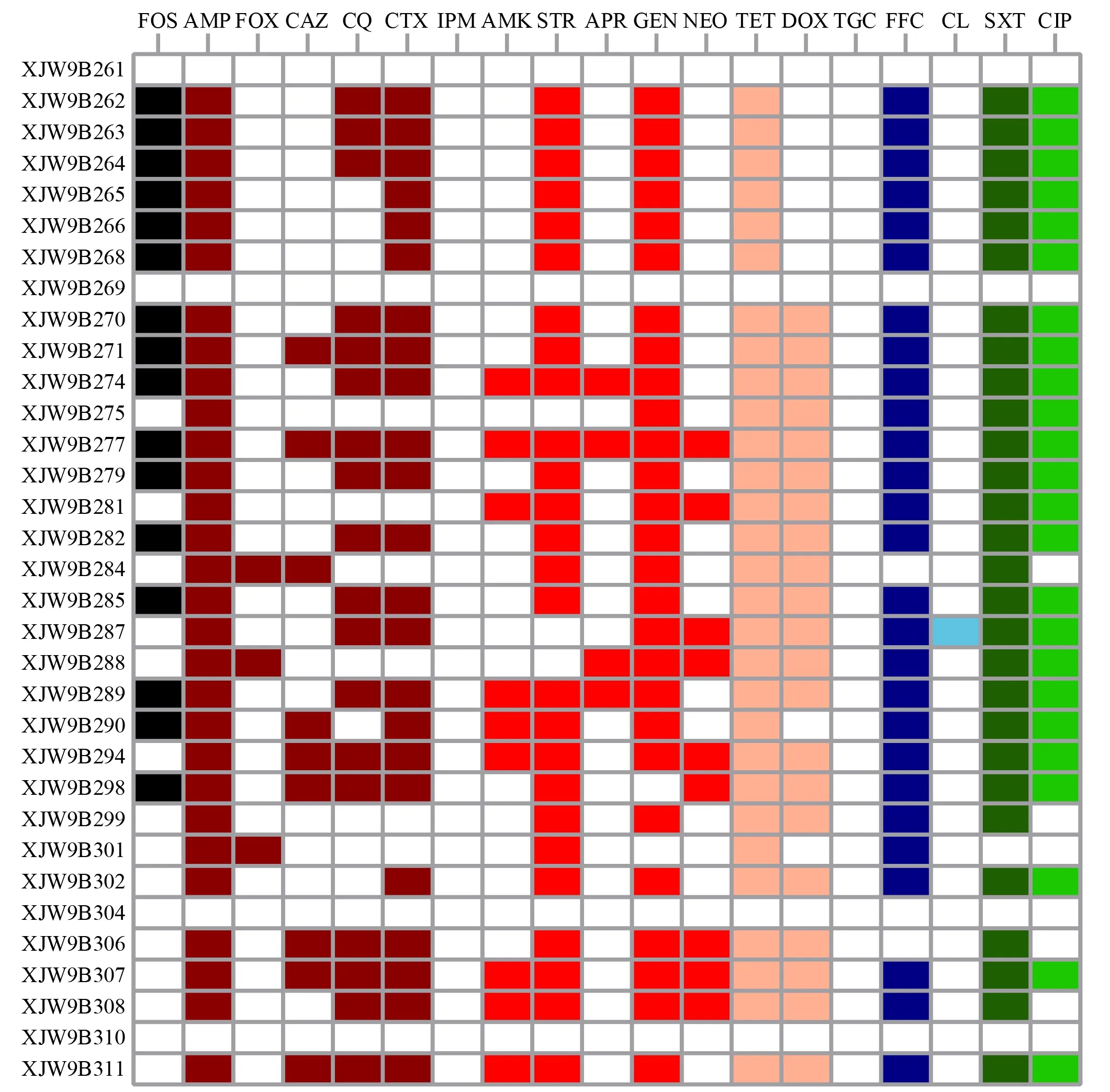
Figure 1 Antimicrobial resistance phenotypes of all E. coli isolates
Molecular characterization of fosA3-positive E.coli
PCR screening confirmed that all fosfomycin-resistant isolates were positive forfosA3.In total,93.8% (n=15),93.8% (n=15),and 25.0% (n=4) offosA3-positive isolates co-harboredblaCTX-M,floR,andrmtB,respectively,withblaCMY-2,armA,andmcr-1not detected.
PFGE was successfully performed for allfosA3-carrying isolates,and four differentXbaI PFGE patterns (A to D) were observed (Figure 2).Pattern D,which included 11 (68.8%)fosA3-carrying isolates,was dominant,followed by pattern C(n=3),suggesting pandemic pattern DfosA3-carryingE.coliisolates in this cattle farm.
The conjugation results indicated that fivefosA3genes in A-,B-,and C-pattern isolates were successfully transferred toE.coliJ53.The S1-PFGE results confirmed that thefosA3genes in the A-and C-pattern isolates were located on single plasmids of the same size (~138.9 kb),and thefosA3gene in the B-pattern isolate was located on a single plasmid(~78.2 kb).Southern blot hybridization revealed that the other 11fosA3genes that failed in the conjugation experiment were located on the same-size band (~1 135 kb) in all D-pattern isolates,indicating a chromosomal location for thefosA3gene.
The PCR mapping results demonstrated that allfosA3genes were flanked by IS26.In total,three common types of IS26composite transposons were found,including IS26-ΔISEcp1-blaCTX-M-14-ΔIS903-fosA3-orf1-Δorf2-IS26(n=4),IS26-fosA3-orf1-orf2-Δorf3-IS26(n=1),and IS26-fosA3-orf1-Δorf2-IS26(n=11) (Supplementary Figure S1).
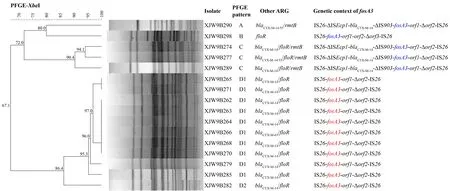
Figure 2 PFGE profiles,antimicrobial resistance genes,and genetic structure of fosA3-positive E.coli
Genomic analysis of fosA3-positive E.coli
Whole-genome sequencing was performed on sixfosA3-harbouringE.coliisolates with four different PFGE patterns,and the obtained data were analyzedin silico.The sequence type (ST) of the dominant D-patternfosA3-harbouringE.coliisolates (XJW9B263 and XJW9B285) was ST10.The other isolates were ST167 (XJW9B274 and XJW9B277),ST1431(XJW9B298),and unknown ST (XJW9B290).The O101:H9 serotype was obviously dominant among the six wholegenome sequencing isolates,except that the serotype ofE.coliXJW9B298 (ST1431) was O8:H30.In addition,multiple ARGs were detected among all whole-genome sequencing isolates,three of which carried the F18:A-:B1 plasmid(Supplementary Table S2).
The D-pattern isolate XJW9B263 and C-pattern isolate XJW9B277 were further subjected to long-read sequencing to obtain assembled genomes.Sequence analysis confirmed thatfosA3andblaCTX-M-14were co-located on the chromosome of XJW9B263 (GenBank:CP067399) and on the 133 299 bp long F18:A-:B1 plasmid pHNXJB277 of XJW9B277 (GenBank accession No.:CP068043).
NCBI-BLAST analysis revealed that pHNXJB277 showed genomic sequence identity with thefosA3-positive F18:A-:B1 plasmid pT28-2R of pet dog origin in Henan Province,China(GenBank accession No.:CP049355.1) (Figure 3A).The backbones of the two F18:A-:B1 plasmids were highly similar,while the multi-resistance region (MRR) varied.Similarly,fosA3genes co-localized withblaCTX-M-14on these two plasmids were surrounded by IS26.
Core genome SNP calling of E.coli ST10 and ST167
To explore the origin of pandemicE.coliST10 and ST167,the assembled contigs of 88 164E.coliisolates from GenBank were collected to identify STs.A total of 3 444E.coliisolates of ST10 and 336E.coliisolates of ST167 were detected and further subjected to cgSNP-based phylogenetic analysis with the pandemicE. coliST10 (isolates XJW9B263 and XJW9B285) andE.coliST167 (isolates XJW9B274 and XJW9B277) identified in this cattle farm.Based on our results,allE.coliST167 isolates from GenBank showed >390 cgSNP differences from XJW9B277,whereas isolates XJW9B274 and XJW9B277 were clonal with only one cgSNP distance.We also calculated the number of cgSNPs in 45E.coliST10 isolates that were related to the XJW9B263 clone against the XJW9B263 reference isolate. Isolates XJW9B285 and XJW9B263 from the cattle farm were clonal with no cgSNP difference.However,most of theE.coliST10 isolates from GenBank showed >100 cgSNP differences from XJW9B263,including isolates originating from humans and animals in China;only seven isolates derived from humans and animals outside China had <100 cgSNP differences from XJW9B263.Of note,anE.coliST10 strain (GenBank:GCA_014156895.1)isolated from a rectal swab of an Australian silver gull(Chroicocephalus) in 2017 differed from isolate XJW9B263 by just 11 cgSNPs (Figure 4),suggesting a close relationship according to the recommended ≤10 SNP threshold ofE.coli(Schürch et al.,2018).
Serotypes,ARGs,virulence genes,and plasmids were screened using whole-genome sequences of the original Australian silver gullE.coliisolate (GCA_014156895.1),and further compared with isolate XJW9B263.Results showed that both wereE.coliO101:H9-ST10 and carried identical resistance genes (blaCTX-M-14,blaTEM-1B,aph(3'')-Ib,aph(6)-Id,aac(3)-IId,tet(A),cmlA1,floR,mdf(A),mph(A),fosA3,sul2,dfrA14) and plasmids (IncFIB and IncY) (Table 1).XJW9B263 was distinguished from GCA_014156895.1 by only four virulence-associated genes (ecpD,entD,espL4,andespX4),and GCA_014156895.1 additionally carried Col-like replicon Col(MG828).Moreover,the contigs of GCA_014156895.1 successfully matched a partial chromosome of XJW9B263 containingfosA3,blaCTX-M-14,and other ARGs (Figure 3B)
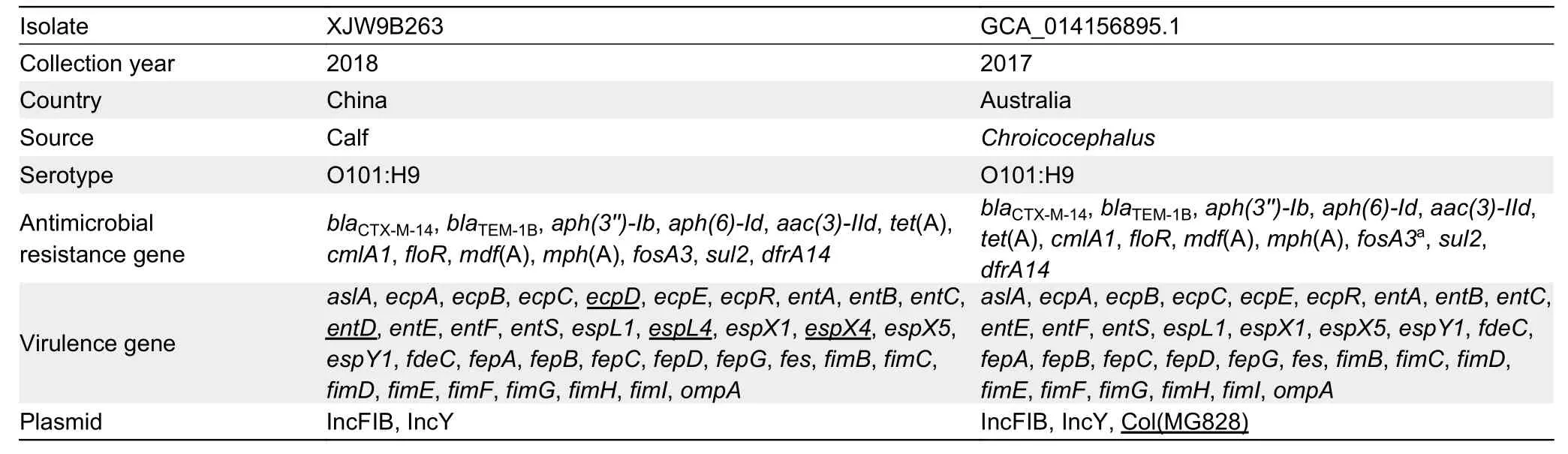
Table 1 Genomic analysis of clonal E.coli O101:H9-ST10
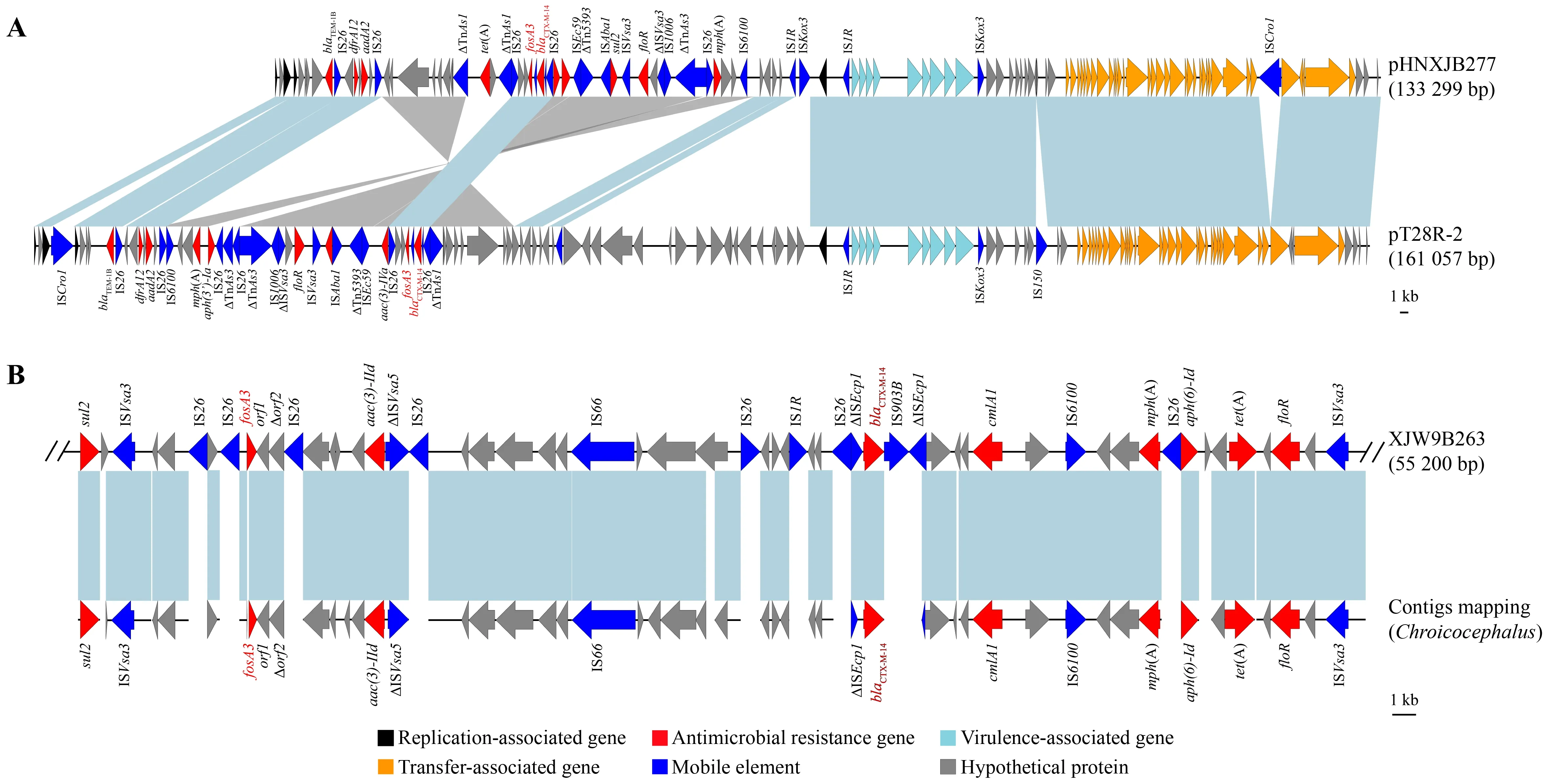
Figure 3 Genetic environment of fosA3 and blaCTX-M-14
DISCUSSION
With the widespread use of antimicrobials among humans and animals,MDR bacteria have emerged (Nikaido,2009;Schürch et al.,2018),challenging the clinical treatment of bacterial infections.During routine surveillance of antimicrobial resistance in a cattle farm located in Xinjiang,a high prevalence of MDRE.coliisolates of diarrheal calf origin was observed. In particular,a surprisingly high fosfomycin resistance rate (48.5%) was noted,much higher than that of other food animals.For example,fosfomycin resistance rates in chicken-originE.coliisolates from Guangdong and Northeast China are reported at 27.9% (He et al.,2017) and 27.4% (Jiang et al.,2017),respectively.PCR screening showed thatfosA3genes were present in all fosfomycinresistant isolates,and thus may mediate fosfomycin resistance.All FosA3-producers also showed resistance to cephalosporins and ciprofloxacin,and almost allfosA3-producers co-harboredblaCTX-M,as commonly described nation-and worldwide (Cunha et al.,2017;Hou et al.,2012;Lupo et al.,2018;Lv et al.,2020;Yang et al.,2014).Regarding this,co-selection by long-term use of cephalosporins and enrofloxacin in this cattle farm may account for the prevalence offosA3,as reported in a Chinese broiler farm,in which co-selection was considered the hypothetical driving force for the prevalence of plasmidmediated colistin resistance genemcr-1(Cao et al.,2020).
Previous studies have reported that thefosA3gene inE.coliis not generally spread by clonal transmission,but rather by plasmid-mediated horizontal transmission.Here,however,we found that mostfosA3-positive isolates shared similar PFGE profiles,belonging toE.coliO101:H9-ST10 (n=2) and O101:H9-ST167 (n=2),indicating that the spread offosA3genes in this cattle farm was likely mediated by vertical clonal transmission.
Serotype O101,which is associated with animal and human diseases,is frequently detected among pathogenicE.coli(Chirila et al.,2017;Mandal et al.,2001;Tan et al.,2012).To the best of our knowledge,however,serotype O101:H9 has only been reported in Shiga toxin-producingE.coli(STEC)from humans with diarrheal disease in Germany (Beutin et al.,2008),in enterotoxigenicE.coli(ETEC) from diarrheal calves in Europe (Contrepois et al.,1998) and children with diarrheal disease in New Caledonia (Begaud et al.,1993),and inE.coliisolated from humans with acute suppurative cholangitis(Sung et al.,1994).In view of the limited number of reports of serotype O101:H9 in China,core genomes of the O101:H9-ST10 and O101:H9-ST167 clones were compared with those of allE.coliisolates submitted in GenBank to explore the origin of theE.coliclones in this cattle farm.Results demonstrated that all GenBankE.coliST167 isolates were clonally unrelated to the O101:H9-ST167 clone (isolates XJW9B274 and XJW9B277) detected in this study,while core genomes of 45E.coliST10 isolates from GenBank were relatively similar to those of the O101:H9-ST10 clone (isolates XJW9B263 and XJW9B285).Further analysis confirmed that most showed >100 cgSNP differences from the O101:H9-ST10 clone;nevertheless,six isolates,all isolated from humans outside of China,including Canada and several European countries (UK,Germany,France,and Estonia),differed from the O101:H9-ST10 clone by <100 cgSNPs.Surprisingly,the core genome sequence of an AustralianChroicocephalus-derivedE.coliisolate showed high similarity to the O101:H9-ST10 clone,with just 11 cgSNP differences,and the ARG profiles and plasmid types of both were very similar,indicating a significant clonal relationship.We note that wild birds forage for food at this cattle farm throughout the year,and that cattle feed is often contaminated by bird droppings (Supplementary Figure S2).Considering that the core genome of this clone exhibits greater similarity to foreign isolates,we suspect that theE.coliO101:H9-ST10 clone spreading in this cattle farm originated from foreign wild birds,i.e.,theChroicocephalus-bearingE.coliO101:H9-ST10.In accordance with a migration map of waterbirds worldwide(https://www.eaaflyway.net/),gulls and terns annually traverse the East Asian-Australasian Flyway (EAAF) covering East Asian countries and Australia. Furthermore,AustralianChroicocephalusoften mix with the great crested terns(Thalasseus bergii cristatus) that fly to Australia in the south and to Ryukyu Islands and southeastern China in the north(https://birdsoftheworld.org/bow/home). Therefore,although the AustralianChroicocephalusdoes not migrate to Xinjiang,wild birds foraging for food at the cattle farm may mix with the great crested terns that show an overlapping distribution with the AustralianChroicocephalus,from where they acquire theE.coliO101:H9-ST10 clone (Figure 5).
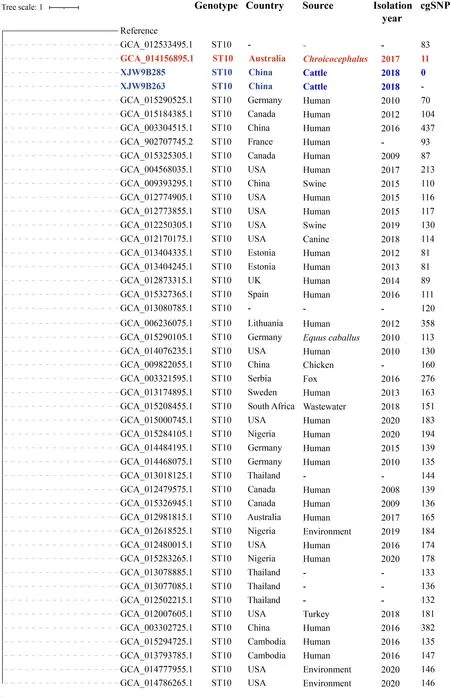
Figure 4 Core genome SNP-based phylogenetic tree of E.coli ST10 strains
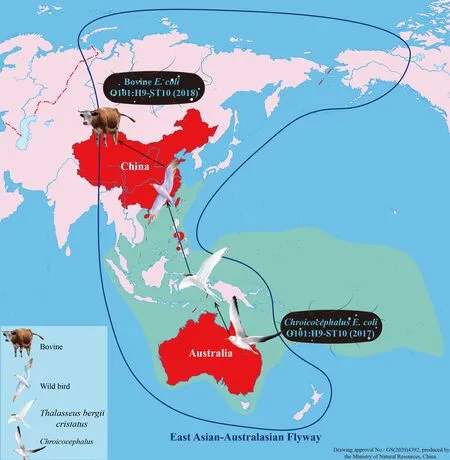
Figure 5 Schematic of possible global dissemination of E.coli O101:H9-ST10 from Australian Chroicocephalus to Chinese cattle
Furthermore,though the estimated number of core genome SNPs per year forE.coliis unclear,a cutoff of ≤21 SNPs per genome per year forKlebsiella pneumoniaeand a ≤23 SNP threshold forEnterobacterialesof local transmission have been reported (David et al.,2019;Sherry et al.,2019).Therefore,the 11 cgSNP distance between the twoE.coliO101:H9-ST10 strains derived fromChroicocephalusand cattle suggests short-term clonal transmission of the high-riskE.coliST10 from migratoryChroicocephalusbirds to the calves.
Although wild birds (gulls) are not exposed to antimicrobials directly,their coastal habitats result in high-level human contact.As such,these birds have been described as reservoirs and vectors of MDR bacteria for the global diffusion of ARGs mediating resistance to CIAs (Mukerji et al.,2019,2020;Villa et al.,2015;Wang et al.,2017a).To date,Chroicocephalusbirds have not been reported in Yili in Xinjiang;however,indirect transmission of MDR bacteria fromChroicocephalusto wild birds that visit this cattle farm is possible.Due to their outdoor breeding,calves may potentially acquire MDR bacteria spread by wild birds that forage or fly over the farm.Thus,greater attention should be paid to wild birds visiting farms and sanitation should be strengthened to slow the potential risk of migratory bird dissemination of MDR bacteria.
CONCLUSIONS
This study described the clonal spread of FosA3-and CTX-MproducingE.coliO101:H9-ST10 among diarrheal calves from a cattle farm in Xinjiang,China.We speculate that the clones originated from migratory (foreign) birds and were transmitted by wild birds foraging on the farm.This is the first direct evidence of migratory birds disseminating bacteria resistant to CIAs across land and countries.These results highlight theneed to pay greater attention to the risk of migratory birds spreading MDR microorganisms on a global scale.Moreover,biosafety prevention and control should not only focus on terrestrial pathogen contact,but also pathogens disseminated by birds and/or insects during aerial flight.
DATA AVAILABILITY
The datasets in this study can be found in GenBank under accession Nos. CP067399-CP067401 (XJW9B263) and CP068041-CP068045 (XJW9B277),and in the GSA databank under accession No.CRA004296 (XJW9B298,XJW9B290,XJW9B274,XJW9B277,XJW9B263,and XJW9B285).
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
J.H.L.and J.Y.conceived the research.W.Y.H.,X.X.Z.,J.Y.,G.L.G.,M.Y.G.,Z.P.C.,and L.C.L.collected the data.J.H.L.,W.Y.H.,X.X.Z.,J.Y.,L.C.L.,F.G.Z.,and X.F.S.analyzed and interpreted the data.W.Y.H.drafted the manuscript,J.H.L.,J.Y.,and X.F.S.revised the report.All authors read and approved the final version of the manuscript.
ACKNOWLEDGEMENTS
We are grateful to Xiao-Jun Yang from the Kunming Institute of Zoology,Chinese Academy of Sciences,for helpful comments on this study.
- Zoological Research的其它文章
- 3DPhenoFish:Application for two-and threedimensional fish morphological phenotype extraction from point cloud analysis
- A bright future for the tree shrew in neuroscience research:Summary from the inaugural Tree Shrew Users Meeting
- Inhibition of mTOR signaling by rapamycin protects photoreceptors from degeneration in rd1 mice
- A new snake species of the genus Gonyosoma Wagler,1828 (Serpentes:Colubridae) from Hainan Island,China
- PINK1 gene mutation by pair truncated sgRNA/Cas9-D10A in cynomolgus monkeys
- Genome and population evolution and environmental adaptation of Glyptosternon maculatum on theQinghai-Tibet Plateau

