WRKY转录因子在植物干旱响应机制中的作用研究进展
孙晓琛,栗锦鹏,原静静,王惠珍,杜弢
WRKY转录因子在植物干旱响应机制中的作用研究进展
孙晓琛,栗锦鹏,原静静,王惠珍,杜弢
甘肃中医药大学药学院,甘肃 兰州 730000
WRKY转录因子是调控植物发育和应对外界胁迫刺激反应的主要调控因子,在植物抵御干旱过程中的作用至关重要。本文就近年来WRKY转录因子抵御干旱胁迫依赖的信号通路、干旱胁迫下其参与的植物生长发育和生理调控,以及对药用植物次生代谢产物的调控相关研究进行梳理,为进一步探讨植物的抗旱分子机制、培育耐旱品系,探索植物次生代谢产物的药用价值提供参考。
WRKY转录因子;干旱胁迫;信号通路;生长发育;次级代谢产物;综述
植物早期从水生环境迁移到陆地易遭受干旱胁迫、盐害、极端温度等不利因素影响,其中干旱胁迫会对植物造成致命伤害[1],因此,提高植物对干旱胁迫的耐受性是迄今植物保护地区促进可持续发展的有效策略,越来越多的国家和国际组织也在积极开展探索植物抗旱机制的相关研究,以确定提高植物抗旱性的关键基因或工具。在分子水平上,胁迫相关基因的诱导有助于提高植物适应不利环境因子的能力,在该过程中,许多转录调控网络被激活,其中WRKY作为植物中最大的转录因子家族之一,在植物对干旱胁迫反应的调控网络中发挥重要作用[2]。
最初,WRKY被认为是高等植物特有的,后在蕨类植物和蓝氏贾第鞭毛虫与盘状网柄菌中鉴定出WRKY基因[3],表明其为起源于原核生物与真核生物分化之前的转录因子大家族。首个WRKY cDNASPF1是从甘薯[4]中克隆所得,随后ABF1,2在野燕麦[5]中发现并进一步在皱叶欧芹中得到WRKY1、WRKY2和WRKY3,并创造WRKY[6]。WRKY家族成员共同特征是具有高度保守的60个氨基酸区域的结构域,由N末端保守氨基酸序列WRKY GQK和C末端锌指状基序C2H2或C2HC组成[7]。该结构域可形成一种4条链的β-折叠,其稳定性由β-折叠端的锌结合袋所决定,表明N端保守序列可以直接与DNA结合[8]。根据WRKY结构域的数量和锌指状基序的类型,通常将其分为以下3组亚家族,由此反映其不同功能:第一组由2个WRKY结构域组成,分为Ia和Ib亚组,Ia含C2HC锌指,Ib也含有C2HC锌指;第二组只有1个WRKY结构域和1个C2H2锌指,根据其系统发育关系分为5个亚组,即Ia、Ib、Ic、Id和Ie;第三组也只有1个WRKY结构域,但其锌指结构为C2HC[7]。多数WRKY转录因子可通过核定位信号、亮氨酸拉链、丝氨酸/苏氨酸富集区、富含谷氨酰胺的区域、富含脯氨酸的区域、激酶结构域、TIR-NBS-LRr等结构参与植物的多种调节途径[2]。近10年来,WRKY不仅在农作物和果蔬烟草抵御干旱胁迫机制中研究广泛,还在人参、丹参、红豆杉、青蒿等植物次级代谢产物的药用价值中取得进展。兹就相关研究进行综述。
1 WRKY调控植物抗旱反应
WRKY是最大的植物特异性转录因子超家族之一,可通过识别和结合其启动子中的W盒(TTGACC/T)正调控下游基因表达,而这些下游基因又通过多种生理过程赋予植物非生物胁迫和生物胁迫耐受性,使其成为改善干旱胁迫反应中的主要候选靶基因[9]。此外,WRKY家族成员还在调控植株胚胎、茎叶生长,衰老、糖信号传导及毛状体形态中起着重要作用[10]。见表1。
表1 部分响应干旱的WRKY转录因子
基因物种研究方法文献 AtWRKY46,AtWRKY54,AtWRKY70拟南芥Arabidopsis thaliana转录组分析[11] AtWRKY57拟南芥Arabidopsis thaliana获得功能突变体[12] MdWRKY20,MdWRKY22,MdWRKY40,MdWRKY47,MdWRKY53,MdWRKY77,MdWRKY90,MdWRKY91,MdWRKY100,MdWRKY125苹果Malus domestica转录组分析[13] VlWRKY48,VlWRKY30葡萄Vitis vinifera过表达到拟南芥[14-15] HvWRKY38大麦Hordeum vulgare转录组分析[16] TaWRKY1,TaWRKY33小麦Triticum aestivum过表达到拟南芥[17] OsWRKY30,OsWRKY45水稻Oryza satiwa过表达到拟南芥[18-19] ZmWRKY58玉米Zea mays过表达到水稻[20] NtWRKY3,NtWRKY10,NtWRKY12,NtWRKY69烟草Nicotiana tabacum转录组分析[21] GsWRKY20野大豆Glycine soja过表达到苜蓿[22] MtWRKY76蒺藜苜蓿Medicago truncatula过表达[23] CmWRKY10菊花Chrysanthemum过表达[24] ZbWRKY33花椒Zanthoxylum bungeanum过表达[25] PtrWRKY2 枳Poncirus trifoliata RNA印迹分析[26] PbWRKY白梨Pyrus bretschneideri 转录组分析[27] IbWRKY2番薯Ipomoea batatas转录组分析[28] BcWRKY46不结球白菜Brassica campestris过表达到烟草[29] MuWRKY3硬皮豆Macrotyloma uniflorum过表达到花生[30]
1.1 干旱胁迫下WRKY依赖的信号通路
干旱应激信号传导途径包括信号感知、信号传导、应激反应。首先,植物细胞膜上的各种质膜蛋白作为传感器感知胁迫信号,然后由激素和第二信使活性氧自由基、Ca2+、环核苷酸(cAMP和cGMP)等启动相应的信号转导途径,最终影响调节网络并诱导应激反应基因的表达[31]。
1.1.1 自我调节和交叉调节
转录因子可通过与相同蛋白质或同一家族其他成员的物理相互作用分别形成同二聚体或异二聚体,或与其他蛋白质家族的转录因子建立复合物,并在转录调控过程中提供巨大的组合灵活性[31]。因此,该转录因子不仅能实现WRKY-WRKY蛋白作用,如番茄中由干旱诱导的9个WRKY基因,尤其WRKY58,可单独或与其他基因组合进一步作为耐旱转基因的靶标[32];还可以与其他蛋白相互作用,如ZmWRKY25、ZmWRKY47、ZmWRKY80的启动子具有W盒顺式作用元件,这3种蛋白可相互作用,并通过与其他蛋白质相互作用而参与干旱响应[2]。
尽管WRKY具有功能多样性,但几乎所有WRKY蛋白都能识别W盒序列,因此,除识别核心W盒启动子元件外,还需其他机制以实现WRKY转录因子的调控特异性[33]。如单个WRKY蛋白可与多个转录调控辅助因子VQ蛋白的相互作用,赋予WRKY蛋白广泛的生物学功能,干旱能诱导VQ蛋白表达,IbWRKY2通过与IbVQ4和AtVQ4交互作用,充当植物干旱胁迫耐受性的正调节剂[28]。
1.1.2 丝裂原活化蛋白激酶对WRKY的调节
丝裂原活化蛋白激酶(MAPK)级联是植物感受到内部发育和外部生物胁迫和非生物胁迫信号后被激活的中枢信号通路,其中已确定MAPK信号通路参与干旱胁迫响应[34]。WRKY可被MAPK磷酸化,与其启动子区的特定顺式元件相互作用直接调节一系列下游基因表达[35]。如烟草中MAP激酶WIPK和苜蓿中SAMK可被寒冷、干旱、伤害和生物信号激活[36]。Shen等[18]研究表明,MAP激酶激活OsWRKY30的过表达赋予植株耐旱性,其过程为OsWRKY30与OsMPK3、OsMPK4、OsMPK7、OsMPK14、OsMPK20-4及OsMPK20-5相互作用,同时被OsMPK3、OsMPK7和OsMPK14磷酸化。Li等[37]鉴定了1个完整的MAP激酶级联反应,在此反应中其介导GhWRKY转录因子被激活并磷酸化,还阐明了一种由GhMAP3K15-GhMKK4-GhMPK6-GhWRKY59- GhDREB2组成调控模块,该模块参与控制棉花的干旱反应。
1.2 WRKY参与脱落酸介导的信号途径
一般而言,干旱反应途径可分为两类:一类是脱落酸(ABA)依赖途径,另一类则独立于ABA。一方面,ABA作为一种胁迫信号在植物的干旱胁迫反应中充当内源信使,还可在胁迫条件下对植物的生长发育进行微调,控制的生理过程包括生长、气孔孔径和导水率的调节等[38]。9-顺式环氧类胡萝卜素双加氧酶(NCED)是干旱胁迫下ABA合成的关键酶,NCED基因的过度表达可增强ABA积累,增强植物的抗旱能力,Liu等[39]研究表明,PbrWRKY53是一个抗旱积极因子,能直接与PbrNCED1启动子结合并正调控PbrNCED1表达响应干旱。在小麦研究中检测到脱落酸合成基因ABA1和ABA2,意味着ABA生产加速,且ABI5转录丰度增加,表明TaWRKY33通过脱落酸合成和转导途径提高耐旱水平[17]。另一方面,ABA可修饰组成性表达的转录因子,导致早期反应转录激活因子表达,然后激活下游胁迫耐受效应基因[40]。甘油醛-3-磷酸脱氢酶是一种在非生物胁迫和植物发育过程中发挥重要作用的多功能酶。Zhang等[41]研究表明,当植物遭受干旱胁迫时,TaWRKY40与TaGAPC1启动子结合并增强启动子活性,从而增加ABA信号通路中的TaGAPC1基因表达水平,以增强植物抗旱性。Yan等[42]研究显示,在干旱胁迫条件下,野生型棉花ABA诱导基因AREB、DREB、NCED、ERD和LEA的转录水平比转基因植物抑制作用明显,由此得到GhWRKY17通过ABA信号响应干旱的结论。并且,过度表达活性ABA响应元件结合蛋白AREB1和活化的AREB1(AREB1∆QT)的植物能表现出更强的耐旱性[43]。
此外,Fei等[25]通过STRING预测表明WRKY还涉及3种激素信号通路——茉莉酸、水杨酸和乙烯信号通路。WRKY在激素的上下游发挥作用,参与水杨酸和茉莉酸/乙烯的拮抗作用,通过生长素、细胞分裂素和油菜素甾醇类激素(BR)控制发育过程[9],其中ZbWRKY33对干旱胁迫反应强烈,主要依赖于茉莉酸信号通路为中心的调控网络[25]。
1.3 WRKY调控植物发育和生理反应抵御干旱胁迫
1.3.1 根
根系是植物吸收土壤水分和养分、感知和传递土壤水分亏缺信号的重要器官,植物通过更深的根系从更深的土壤剖面中提取水分的能力是干旱胁迫下直接影响产量的最相关特征之一[44]。BR除调节根系生长发育还具有保护植物免受各种环境胁迫的能力[45],Chen等[11]发现,一组WRKY转录因子WRKY46、WRKY54、WRKY70,其中WRKY54与BES1协同介导BR调节的干旱反应,分别通过与W盒和G盒区域结合抑制干旱诱导基因GLYI7转录,而ABA会调节BIN2糖原合成酶激酶GSK-3抑制BR信号转导,导致干旱胁迫下BIN2水平升高、WRKY54和BES1蛋白失稳,最终导致干旱诱导基因的表达增加以抗旱生存。此外,WRKY转录因子可以和干旱诱导转录因子相互作用调节根系发育。Wang等[46]研究表明,GmWRKY27可与GmMYB174相互作用,通过抑制GmNAC29表达提高转基因大豆毛状根抗旱能力。而ZmWRKY40过表达激活干旱应激反应基因STZ、DREB和RD29A的表达促进干旱胁迫下转基因拟南芥根系生长,降低水分流失率,提高了转基因拟南芥的耐旱性[47]。
1.3.2 叶
叶片对干旱胁迫的形态和生理响应是减少水分流失、提高水分利用效率的基础[48]。在热休克蛋白101启动子控制下,OsWRKY11过表达导致叶片萎蔫率降低,维持叶绿素和叶面积稳定,这些因素有助于提高作物的耐旱性[49]。叶片中气孔介导植物与大气之间交换,其周围保卫细胞膨胀压力的变化控制着气孔开闭。
保卫细胞转录组富含转录因子编码基因,其中WRKY46已被证明参与调节光依赖性气孔开放并调节干旱胁迫的反应[50]。Sun等[51]研究表明,激活AtWRKY53表达可通过促进淀粉代谢,促进气孔开放、调节气孔运动,减少保卫细胞中的H2O2含量,从而负调节干旱耐受性。但GmWRKY54可直接激活并结合PYL8、SRK2A、CIPK11及CPK3等基因,通过ABA和Ca2+信号通路促进气孔关闭,减少水分流失以赋予植株耐旱性[52]。
1.3.3 渗透调节和抗氧化反应
转录因子本身在转录水平上受到其他上游成分调控,在转录后水平上经过泛素化等修饰后,形成一个复杂的调控网络,调节应激反应基因表达、各种生理和代谢过程。耐旱植物可通过积累蔗糖、海藻糖及棉子糖系列寡糖增强其抗旱性[53],肌醇半乳糖苷合成酶(GolS)是棉子糖代谢过程中第一个限速步骤的关键酶。
Wang等[54]研究表明,BhGolS1赋予转基因烟草脱水耐受性,未受胁迫的烟草中未检测到半乳糖醇内部积累,当处于干旱胁迫环境时,WRKY转录因子与BhGolS1启动子的W盒结合导致转基因烟草耐旱性提高。同样,OsWRKY11能激活一些棉子糖合成相关基因表达,促进植株体内棉子糖的积累以抵御干旱胁迫[49]。此外,植物遭受干旱胁迫后,会伴随次生胁迫损伤,如活性氧损伤,VlWRKY48在干旱胁迫下能提高过氧化氢酶、过氧化物酶、超氧化物歧化酶等抗氧化酶活性,清除活性氧(ROS)以提高葡萄抗旱性[14]。GhWRKY41/ SpWRKY1则调节气孔导度和诱导抗氧化基因表达、降低丙二醛含量和ROS水平,提高转基因烟草的耐旱性[55-56]。PbrWRKY53还能通过抗坏血酸途径将H2O2转化为H2O清除,减轻干旱造成的氧化损伤[39]。
2 WRKY参与植物次级代谢产物合成
根据化学成分,次生代谢产物大致分为两类,即含氮分子(生物碱)和缺氮分子(萜类和酚类)。植物中生物碱、萜类、黄酮等化合物与植物逆境反应有关,且干旱胁迫等逆境信号会诱导大量的转录因子形成调控网络,其中WRKY转录因子可介导次生代谢产物在植物胁迫反应中的作用[57]。见图1。
许多植物合成的生物碱具有很高药用价值,已被用于治疗多种晚期疾病[58]。长春花中大量萜类吲哚生物碱(TIA)是天然或半合成抗癌药的重要来源,从其幼苗分离出的CrWRKY1被证明在TIA生物合成中起关键作用[59]。酚类化合物通常有一个带有羟基的芳香环,其生物合成依赖于2条途径(莽草酸途径和丙二酸途径),木质素是该类群的重要成员[57]。已有研究表明,干旱会影响细胞壁木质素的生物合成,拟南芥WRKY13通过直接结合NST2 18的启动子积极调节茎中木质素生物合成[60];此外,WRKY还控制黄酮醇和单宁化合物生成,WRKY23以生长素诱导方式调节黄酮醇产生,实现根系正常生长和发育[61]。
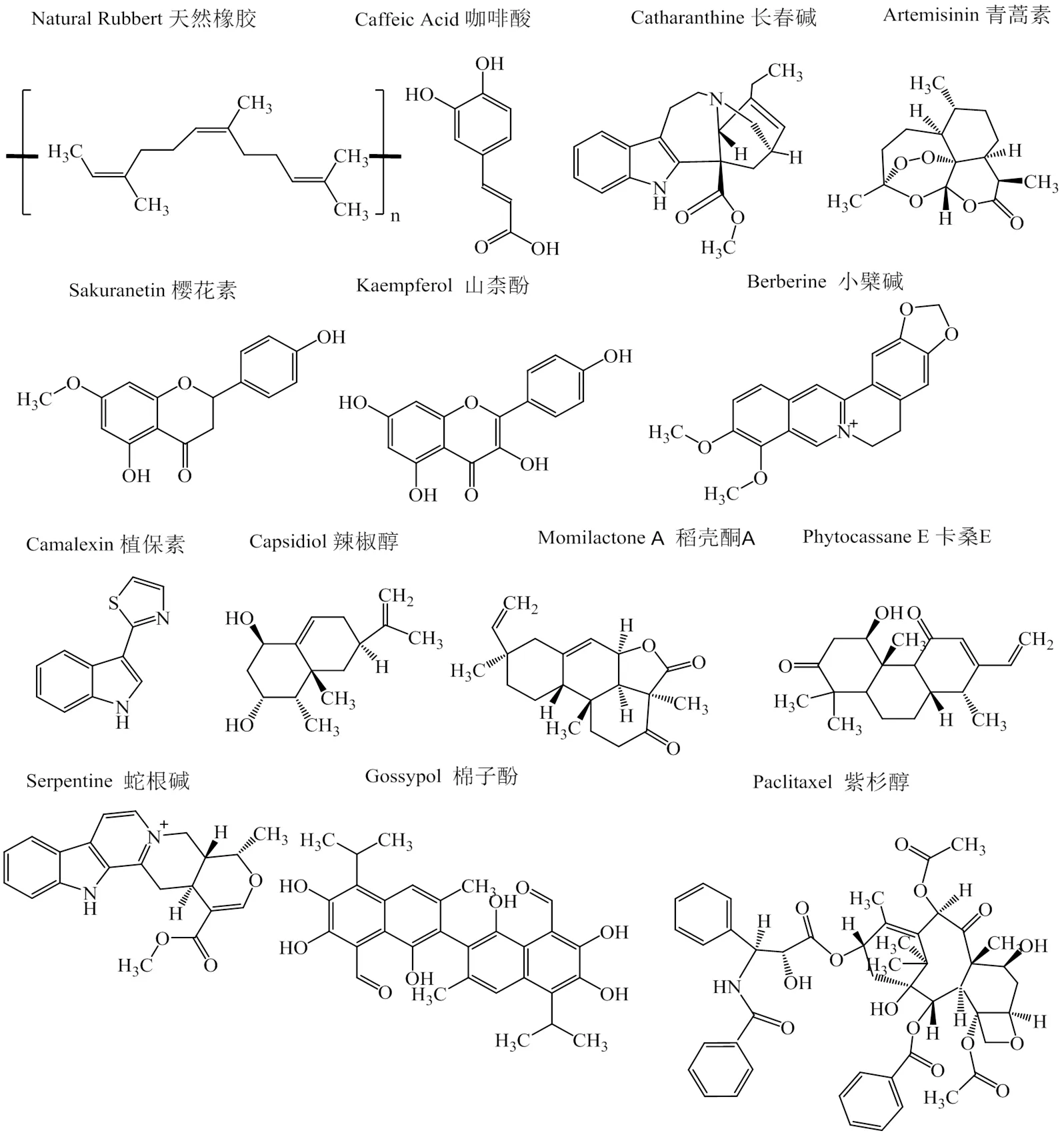
图1 WRKY转录因子调控代谢途径合成的植物化学物质
萜类化合物具有多种生物学功能,丹参酮是一类具有生物活性的二萜类化合物,广泛应用于治疗心血管疾病。Cao等[62]发现,SmWRKY1过度表达显著提高MEP途径中编码酶基因转录本,尤其1-脱氧-D-木糖-5-磷酸合成酶和1-脱氧-D-木酮糖-5-磷酸还原酶,使转基因系中丹参酮产量提高5倍以上。并且,青蒿素中紫穗槐-4,11-二烯合酶(ADS)可催化法尼基焦磷酸转化为紫穗槐-4,11-二烯1-羟化酶(CYP71AV1)。Ma等[63]研究表明,ADS是AaWRKY1靶基因,表明AaWRKY1参与了青蒿素生物合成调控,实现抗疟药青蒿素生物合成的第一步,且进一步研究发现过表达的AaWRKY1激活了CYP71AV1转录,上调的CYP71AV1可促进青蒿素生物合成。此外,过表达TcWRKY8和TcWRKY47能显著提高红豆杉中紫杉醇生物合成相关基因的表达水平,该成分是目前治疗癌症最有效的化疗药物[64]。总之,WRKY可正调控植物次级代谢产物合成与积累,有助于增强植株抗旱能力并发挥药用价值。
3 小结
干旱胁迫会造成植物生长不良和作物产量受限,是大部分植物生长过程面临的一种严重威胁因素,因此,必须通过一系列生理生化和分子应激性反应抵御干旱胁迫才能更好生存。转录调控对植物抗旱反应起着至关重要作用,转录因子能调节干旱胁迫反应途径中一整套基因,近年来应用RT-PCR、荧光定量PCR、基因芯片、酵母双杂交等方法分析植物中WRKY家族在干旱胁迫下基因表达网络的研究已成为热点。尽管WRKY转录因子作为现代基因组工具对干旱胁迫耐受性的作用研究已取得重大进展,但单个转录因子的完整调节机制,包括上下游的共同调节因子及其相互作用仍不清楚,未来可通过转录调控因子调控和活性,对驯化作物品种的遗传改良,特别是抗逆性和防御反应发挥作用;另外,WRKY能参与植物次级代谢产物调控,今后有必要深入研究干旱胁迫下WRKY调控植物次级代谢产物合成与积累机制,探索植物的药用价值。总之,研究干旱胁迫下WRKY的响应机制,进一步阐明WRKY转录因子在植物抗旱中的作用,为后期应用于植物的抗旱工程、培育抗旱新种质,以及探索植物次生代谢产物药用价值具有重要意义。
[1] ASHRAF M. Inducing drought tolerance in plants:recent advances[J]. Biotechnol Adv,2010,28(1):169-183.
[2] ZHANG T, TAN D, ZHANG L, et al. Phylogenetic analysis and drought-responsive expression profiles of the WRKY transcription factor family in maize[J]. Agri Gene,2017,3:99-108.
[3] ZHANG Y, WANG L. The WRKY transcription factor superfamily:its origin in eukaryotes and expansion in plants[J]. BMC Evol Biol,2005,5(1):1.
[4] ISHIGURO S, NAKAMURA K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and β- amylase from sweet potato[J]. Mol Gen Genet,1994,244(6):563-571.
[5] RUSHTON P J, MACDONALD H, HUTTLY A K, et al. Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of α-Amy2 genes[J]. Plant Mol Bio,1995,29(4):691-702.
[6] RUSHTON P J , TORRES J T, PARNISKE M, et al. Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes[J]. EMBO J, 1996,15(20):5690-5700.
[7] EULGEM T, RUSHTON P J, ROBATZEK S, et al. The WRKY superfamily of plant transcription factors[J]. Trends in Plant Science,2000, 5(5):199-206.
[8] YAMASAKI K, KIGAWA T, INOUE M, et al. Solution structure of an Arabidopsis WRKY DNA binding domain[J]. Plant Cell,2005,17(3):944-956.
[9] TRIPATHI P, RABARA R C, RUSHTON P J. A systems biology perspective on the role of WRKY transcription factors in drought responses in plants[J]. Planta,2014,239(2):255-266.
[10] BAKSHI M, OELMÜLLER R. WRKY transcription factors:Jack of many trades in plants[J]. Plant Signal Behav,2014,9(2):e27700.
[11] CHEN J, NOLAN T M, YE H, et al. Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid- regulated plant growth and drought responses[J]. The Plant Cell, 2017,29(6):1425-1439.
[12] JIANG Y, LIANG G, YU D. Activated expression of WRKY57 confers drought tolerance in Arabidopsis[J]. Mol Plant,2012,5(6):1375- 1388.
[13] MENG D, LI Y, BAI Y, et al. Genome-wide identification and characterization of WRKY transcriptional factor family in apple and analysis of their responses to waterlogging and drought stress[J]. Plant Physi Biochem,2016,103:71-83.
[14] ZHAO J, ZHANG X, GUO R, et al. Over-expression of a grape WRKY transcription factor gene, VlWRKY48, in Arabidopsis thaliana increases disease resistance and drought stress tolerance[J]. Plant Cell, Tissue and Organ Culture,2018,132(2):359-370.
[15] ZHU D, CHE Y, XIAO P, et al. Functional analysis of a grape WRKY30 gene in drought resistance[J]. Plant Cell, Tissue and Organ Culture,2018,132(3):449-459.
[16] RUSHTON P J, SOMSSICH I E, RINGLER P, et al. WRKY transcription factors[J]. Trends Plant Sci,2010,15(5):247-258.
[17] HE G H, XU J Y, WANG Y X, et al. Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis[J]. BMC Plant Biol,2016,16(1):116.
[18] SHEN H, LIU C, ZHANG Y, et al. OsWRKY30 is activated by MAP kinases to confer drought tolerance in rice[J]. Plant Mol Biol, 2012,80(3):241-253.
[19] TAO Z, KOU Y, LIU H, et al. OsWRKY45 alleles play different roles in abscisic acid signalling and salt stress tolerance but similar roles in drought and cold tolerance in rice[J]. J Exp Bot,2011, 62(14):4863-4874.
[20] CAI R H, ZHAO Y, WANG Y F, et al. Overexpression of a maize WRKY58 gene enhances drought and salt tolerance in transgenic rice[J]. Plant Cell, Tissue and Organ Culture,2014,119(3):565-577.
[21] RABARA R C, TRIPATHI P, REESE R N, et al. Tobacco drought stress responses reveal new targets for Solanaceae crop improvement[J]. BMC Genomics,2015,16(1):484.
[22] TANG L, CAI H, ZHAI H, et al. Overexpression of Glycine soja WRKY20 enhances both drought and salt tolerance in transgenic alfalfa (L.)[J]. Plant Cell, Tissue and Organ Culture,2014,118(1):77-86.
[23] LIU L, ZHANG Z, DONG J, et al. Overexpression of MtWRKY76 increases both salt and drought tolerance in Medicago truncatula[J]. Environmental & Experimental Botany,2016,123:50-58.
[24] JAFFAR M A, SONG A, FAHEEM M, et al. Involvement of CmWRKY10 in drought tolerance of chrysanthemum through the ABA-signaling pathway[J]. Int J Mol Sci,2016,17(5):693.
[25] FEI X, HOU L, SHI J, et al. Patterns of drought response of 38 WRKY transcription factors of zanthoxylum bungeanum maxim[J]. Int J Mol Sci,2019,20(1):68.
[26] ŞAHIN-ÇEVIK M. A WRKY transcription factor gene isolated fromshows differential responses to cold and drought stresses[J]. Plant Omics,2012,5(5):438-445.
[27] HUANG X, LI K, XU X, et al. Genome-wide analysis of WRKY transcription factors in white pear () reveals evolution and patterns under drought stress[J]. BMC Genomics,2015,16:1104.
[28] ZHU H, ZHOU Y, ZHAI H, et al. A novel sweetpotato WRKY transcription factor, IbWRKY2, positively regulates drought and salt tolerance in transgenic Arabidopsis[J]. Biomolecules,2020, 10(4):506.
[29] WANG F, HOU X, TANG J, et al. A novel cold-inducible gene from Pak-choi (ssp. chinensis), BcWRKY46, enhances the cold, salt and dehydration stress tolerance in transgenic tobacco[J]. Mol Biol Rep,2012,39(4):4553-4564.
[30] KIRANMAI K, LOKANADHA R G, PANDURANGAIAH M, et al. A novel WRKY transcription factor, MuWRKY3 (Lam. Verdc.) enhances drought stress tolerance in transgenic groundnut (L.) plants[J]. Front Plant Sci,2018,9:346-346.
[31] KHAN S A, LI M Z, WANG S M, et al. Revisiting the role of plant transcription factors in the battle against abiotic stress[J]. Int J Mol Sci,2018,19(6):1634.
[32] KARKUTE S G, GUJJAR R S, RAI A, et al. Genome wide expression analysis of WRKY genes in tomato () under drought stress[J]. Plant Gene,2018,13:8-17.
[33] ZHOU Y, YANG Y, ZHOU X, et al. Structural and functional characterization of the VQ protein family and VQ protein variants from soybean[J]. Sci Rep,2016,6(1):34663.
[34] BIGEARD J, HIRT H. Nuclear signaling of plant MAPKs[J]. Front Plant Sci,2018,9:469.
[35] SHEIKH A H, ESCHEN-LIPPOLD L, PECHER P, et al. Regulation of WRKY46 transcription factor function by mitogen-activated protein kinases in Arabidopsis thaliana[J]. Front Plant Sci, 2016,7:61.
[36] BARTELS D, SUNKAR R. Drought and salt tolerance in plants[J]. Critical Reviews in Plant Sciences,2005,24(1):23-58.
[37] LI F, LI M, WANG P, et al. Regulation of cotton (Gossypium hirsutum) drought responses by mitogenactivated protein (MAP) kinase cascade-mediated phosphorylation of Gh WRKY 59[J]. New Phytologist,2017,215(4):1462-1475.
[38] ULLAH A, MANGHWAR H, SHABAN M, et al. Phytohormones enhanced drought tolerance in plants:a coping strategy[J]. Environ Sci Pollut Res Int,2018,25(33):33103-33118.
[39] LIU Y, YANG T, LIN Z. et al. A WRKY transcription factor PbrWRKY53 from pyrus betulaefolia is involved in drought tolerance and AsA accumulation[J]. Plant Biotechnology Journal, 2019,17(9):1770-1787.
[40] UMEZAWA T, OKAMOTO M, KUSHIRO T, et al. CYP707A3, a major ABA 8'-hydroxylase involved in dehydration and rehydration response in Arabidopsis thaliana[J]. Plant J,2006,46(2):171-182.
[41] ZHANG L, XU Z, JI H, et al. TaWRKY40 transcription factor positively regulate the expression of TaGAPC1 to enhance drought tolerance[J]. BMC Genomics,2019,20(1):795.
[42] YAN H, JIA H, CHEN X, et al. The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic Nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species production[J]. Plant Cell Physiol, 2014,55(12):2060-2076.
[43] LEITE J P, BARBOSA E G, MARIN S R, et al. Overexpression of the activated form of the AtAREB1 gene (AtAREB1ΔQT) improves soybean responses to water deficit[J]. Genet Mol Res,2014,13(3):6272-6286.
[44] MOUMENI A, SATOH K, KONDOH H, et al. Comparative analysis of root transcriptome profiles of two pairs of drought-tolerant and susceptible rice near-isogenic lines under different drought stress[J]. BMC Plant Biol,2011,11(1):174.
[45] ANWAR A, LIU Y, DONG R, et al. The physiological and molecular mechanism of brassinosteroid in response to stress:a review[J]. Biological Res,2018,51(1):46.
[46] WANG F, CHEN H W, LI Q T, et al. Gm WRKY 27 interacts with Gm MYB 174 to reduce expression of Gm NAC 29 for stress tolerance in soybean plants[J]. Plant J,2015,83(2):224-236.
[47] WANG C T, RU J N, LIU Y W, et al. The maize WRKY transcription factor ZmWRKY40 confers drought resistance in transgenic Arabidopsis[J]. Int J Mol Sci,2018,19(9):2580.
[48] BALDONI E, GENGA A, COMINELLI E. Plant MYB transcription factors:their role in drought response mechanisms[J]. Int J Mol Sci,2015,16(7):15811-15851.
[49] WU X, SHIROTO Y, KISHITANI S, et al. Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter[J]. Plant Cell Rep,2009, 28(1):21-30.
[50] DING Z J, YAN J Y, XU X Y, et al. Transcription factor WRKY 46 regulates osmotic stress responses and stomatal movement independently in Arabidopsis[J]. Plant J,2014,79(1):13-27.
[51] SUN Y, YU D. Activated expression of AtWRKY53 negatively regulates drought tolerance by mediating stomatal movement[J]. Plant Cell Rep,2015,34(8):1295-1306.
[52] WEI W, LIANG D W, BIAN X H, et al. GmWRKY54 improves drought tolerance through activating genes in abscisic acid and Ca2+signaling pathways in transgenic soybean[J]. Plant J,2019, 100(2):384-398.
[53] ALBINI F M, MURELLI C, FINZI P V, et al.Galactinol in the leaves of the resurrection plant Boea hygroscopica[J]. Phytochemistry, 1999,51(4):499-505.
[54] WANG Z, ZHU Y, WANG L, et al. A WRKY transcription factor participates in dehydration tolerance in Boea hygrometrica by binding to the W-box elements of the galactinol synthase (BhGolS1) promoter[J]. Planta,2009,230(6):1155-1166.
[55] CHU X, WANG C, CHEN X, et al. The cotton WRKY gene GhWRKY41 positively regulates salt and drought stress tolerance in transgenic Nicotiana benthamiana[J]. PLoS One,2015,10(11):e0143022.
[56] LI J B, LUAN Y S, LIU Z. Overexpression of SpWRKY1 promotes resistance to Phytophthora nicotianae and tolerance to salt and drought stress in transgenic tobacco[J]. Physiol Plant,2015, 155(3):248-266.
[57] MERAJ T A, FU J, RAZA M A, et al. Transcriptional factors regulate plant stress responses through mediating secondary metabolism[J]. Genes (Basel),2020,11(4):346.
[58] PATRA B, SCHLUTTENHOFER C, WU Y. et al. Transcriptional regulation of secondary metabolite biosynthesis in plants[J]. Biochim Biophys Acta,2013,1829(11):1236-1247.
[59] SUTTIPANTA N, PATTANAIK S, KULSHRESTHA M, et al. The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus[J]. Plant Physiol,2011,157(4):2081-2093.
[60] LI W, TIAN Z, YU D. WRKY13 acts in stem development in Arabidopsis thaliana[J]. Plant Sci,2015,236:205-213.
[61] GRUNEWALD W, DE SMET I, LEWIS D R, et al. Transcription factor WRKY23 assists auxin distribution patterns during Arabidopsis root development through local control on flavonol biosynthesis[J]. Proc Natl Acad Sci USA,2012,109(5):1554-1559.
[62] CAO W, WANG Y, SHI M, et al. Transcription factor SmWRKY1 positively promotes the biosynthesis of tanshinones in salvia miltiorrhiza[J]. Front Plant Sci,2018,9:554.
[63] MA D, PU G, LEI C, et al. Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the amorpha-4,11-diene synthase gene, a key gene of artemisinin biosynthesis[J]. Plant Cell Physiol,2009,50(12):2146-2161.
[64] ZHANG M, CHEN Y, NIE L, et al. Transcriptome-wide identification and screening of WRKY factors involved in the regulation of taxol biosynthesis in Taxus chinensis[J]. Sci Rep,2018,8(1):5197.
Research Progress in the Role of WRKY Transcription Factor inPlant Drought Response Mechanism
SUN Xiaochen, LI Jinpeng, YUAN Jingjing, WANG Huizhen, DU Tao
WRKY transcription factor is the main regulator that regulates plant development and responds to external stress stimuli, and plays an important role in the process of plant resistance to drought. This article summarized the research on the signal pathways that WRKY transcription factors rely on to resist drought stress, the plant growth and physiological regulation under drought stress, and the regulation of secondary metabolites of medicinal plants, in order to provide references for further exploring the molecular mechanism of drought resistance in plants, cultivating drought-tolerant strains, and exploring the medicinal value of plant secondary metabolites.
WRKY transcription factor; drought stress; signal pathway; growth and development; secondary metabolites; review
R2-05;R282.5
A
1005-5304(2021)06-0138-07
10.19879/j.cnki.1005-5304.202007627
国家自然科学基金(81760683);现代农业产业体系建设专项(CARS-21)
王惠珍,E-mail:whz1974828@163.com
(2020-07-31)
(2020-11-09;编辑:梅智胜)

