Leelamine suppresses cMyc expression in prostate cancer cells in vitro and inhibits prostatecarcinogenesis in vivo
Krishna B.Singh, Eun-Ryeong Hahm, Shivendra V.Singh,2
1Department of Pharmacology & Chemical Biology, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania, PA 15261, USA.
2UPMC Hillman Cancer Center, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania, PA 15213, USA.
Abstract Aim: Leelamine (LLM) inhibits the growth of human prostate cancer cells but the underlying mechanism is not fully understood.The present study was undertaken to determine the effect of LLM on cMyc, which is overexpressed in a subset of human prostate cancers.
Keywords: Prostate cancer, cMyc, leelamine, chemoprevention
INTRODUCTION
Prostate cancer is a leading cause of cancer-related deaths in the United States[1].Several oncogenic drivers of prostate cancer have been identified, including androgen receptor (AR) and cMyc transcription factors[2].Studies using transgenic mice and human specimens indicate an oncogenic role for cMyc in prostate cancer[3-7].For example, transgenic expression of cMyc in prostate epithelial cells resulted in transformation to prostatic intraepithelial neoplasia (PIN) at 2-4 weeks of age that progressed to adenocarcinoma by 6 months[3].About 67% of human prostate adenocarcinoma (n= 9) exhibited highcMyclevels when compared to benign hypertrophied prostate tissue (n= 19)[4].An androgen-responsive cell line (LNCaP)derived from lymph node metastasis of a human prostate cancer patient also showed gene amplification,rearrangement, and overexpression of cMyc[5].The nuclear level of cMyc protein was shown to be an early event in the pathogenesis of human prostate cancer[7].cMyc overexpression was reported in about 70% of early-stage prostate cancer and up to 37% of metastatic prostate cancer patients[8-10].The 8q gain leading to cMyc overexpression was associated with poor survival in prostate cancer patients[11].Moreover, cMyc overexpression is suggested to be an important mechanism in the resistance of prostate cancers to antiandrogen therapy[12].cMyc overexpression also causes increased cell proliferation and metabolic deregulation, a hallmark of different cancers including prostate cancer, including increased glycolysis andde novosynthesis of fatty acids[13-15].It is not surprising that considerable efforts have been devoted to the therapeutic targeting of cMyc in prostate cancer[16].
Chemoprevention using natural products or synthetic chemicals is attractive for blunting the death and suffering from prostate cancer, but a clinically effective chemopreventive intervention for this malignancy is still lacking[17-22].Leelamine (LLM) is one such phytochemical whose effectiveness has been tested against prostate and other cancers[23-25].The antitumor effect of LLM was initially reported in melanoma but our laboratory was the first to document its effectiveness against prostate cancer cells[24,25].In the melanoma model, the antitumor effect of LLM was attributed to the suppression of multiple oncogenic signalling pathways, including phosphatidylinositol 3-kinase, mitogen-activated protein kinase, and signal transducer and activation of transcription 3 (STAT3)[24].In prostate cancer cells, LLM was shown to inhibit cellular growth by suppressing the expression and activity of AR[25].Intraperitoneal administration of about 9 mg LLM/kg body weight (5 times/week) to 22Rv1 xenograft bearing mice resulted in a significant decrease in Ki-67 expression, mitotic fraction, expression of full-length AR and AR-V7 splice variants, and prostatespecific antigen[25].
In the present study, we investigated the effect of LLM treatment on cMyc expression and activity using human prostate cancer cell lines and the Hi-Myc transgenic mouse model.The foundation for this study was based on the following observations: (1) LLM suppresses AR activity that is a ligand-independent regulator of cMyc expression in prostate cancer[25,26]; and (2) cMyc promotes fatty acid synthesis in prostate cancer that is inhibited by LLM treatmentin vitroandin vivo[15,27].
METHODS
Reagents
LLM was purchased from Cayman Chemical Company (Ann Arbor, MI).The reagents for cell culture were purchased from Life Technologies-Thermo Fisher Scientific (Waltham, MA).Antibodies against cMyc,hexokinase II (HKII), pyruvate kinase M2 (PKM2), and lactate dehydrogenase A (LDH-A) were purchased from Cell Signaling Technology (Danvers, MA).The antibodies against glucose transporter 1 (Glut1) and Cyclin D1 were purchased from Abcam (Cambridge, MA) and Santa Cruz Biotechnology (Dallas, TX),respectively.An antibody against glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was purchased from GeneTex (Irvine, CA).The anti-β-Actin antibody was purchased from Sigma-Aldrich (St.Louis, MO).Alexa Fluor 488-conjugated goat anti-rabbit antibody was from Life Technologies.A kit for colorimetric measurement of lactate was purchased from BioVision (Milpitas, CA).
Cell lines
The human prostate cancer cell lines, LNCaP, 22Rv1, and PC-3, were purchased from the American Type Culture Collection (ATCC; Manassas, VA) and cultured by following the supplier’s recommendations.The University of Pittsburgh has a formal material transfer agreement with ATCC.These cell lines were last authenticated by us in March of 2017.The Myc-CaP cell line derived from the prostate tumor of a Hi-Myc mouse was a kind gift from Dr.Charles L.Sawyers (Memorial Sloan Kettering Cancer Center, New York,NY, USA).The Myc-CaP cells were cultured in Dulbecco’s modified essential medium supplemented with 4.5 g/L glucose, L-glutamine, sodium pyruvate, 10% fetal bovine serum, and antibiotic mixture.The PC-3 and 22Rv1 cells were stably transfected with an empty pcDNA3 vector or the same vector encoding cMyc(Addgene, #16011), as described previously[28].
Western blotting
The prostate tumors from control and LLM-treated mice from our previously published study were used for western blotting[25].Details of cell lysate preparation and western blotting have been described previously[29,30].
Immunocytochemistry
LNCaP (2 × 104), 22Rv1 (3 × 104), and Myc-Cap (3 × 104) cells were plated on coverslips in 24-well plates.After overnight incubation, the cells were treated with ethanol or LLM (2.5 or 5 µM) for 24 h.The cells were then fixed and permeabilized with 2% paraformaldehyde and 0.5% Triton X-100, respectively.After blocking with phosphate-buffered saline (PBS) containing 0.5% bovine serum albumin and 0.15% glycine,the cells were treated overnight at 4 °C with the cMyc primary antibody followed by Alexa Fluor 488-conjugated goat anti-rabbit antibody (1:2000 dilution) for 1 h and then counterstained with DRAQ5(nuclear stain) for 5 min at room temperature in the dark.
Quantitative reverse-transcriptase polymerase chain reaction
Total RNA was extracted using RNeasy Mini Kit from Qiagen (Germantown, MD) and by following the manufacturer’s instructions.Complementary DNA was synthesized and reverse transcribed using oligo(dT)20primer and SuperScript™ III reverse transcriptase.Real-time polymerase chain reaction (PCR) was done from 1:10 diluted complementary DNA using DyNAmo HS SYBR Green qPCR kits (Thermo Fisher Scientific) on ABI StepOnePlus PCR Systems (Applied Biosystems-Life Technologies).Primers for humancMycandGAPDHwere as follows: Forward (cMyc): 5’-GCCACGTCTCCACACATCAG-3’; reverse: (cMyc)5’-TGGTGCATTTTCGGTTGTTG-3’; forward (GAPDH): 5’-GGACCTGACCTGCCGTCTAGAA-3’;reverse (GAPDH): 5’-GGTGTCGCTGTTGAAGTCAGAG-3’.The PCR conditions were as follows: 95 °C for 10 min followed by 40 cycles at 95 °C for 15 s, 60 °C for 1 min, and 72 °C for 30 s.Gene expression change was calculated using the method of Livak and Schmittgen[31].
Luciferase reporter assay
The LNCaP or 22Rv1 cells were co-transfected with 2 µg pBV-Luc wt MBS1-4 plasmid and 0.2 µg of pCMV-RL using FuGENE6.Twenty-four hours after co-transfection, the cells were treated with ethanol or the indicated doses of LLM for 12 h.Luciferase activity was determined using Dual-Luciferase® Reporter Assay kit from Promega, and by following the manufacturer’s instructions.The values were corrected for protein concentration and renilla luciferase.
Clonogenic assay
Empty vector (hereafter abbreviated as EV) transfected control cells and cMyc overexpressing 22Rv1 (1000 cells) were seeded in 6-well plates in triplicate and allowed to incubate overnight.The cells were then treated with ethanol or desired doses of LLM.The medium containing ethanol or LLM was replaced every third day.After 10 days, the cells were fixed with 100% methanol for 5 min at room temperature and stained with 0.5% crystal violet solution in 20% methanol for 30 min at room temperature.The colonies from control and LLM treatment groups were counted under an inverted microscope.
Cell viability and cell migration assay
Trypan blue dye exclusion assay was performed to determine the effect of LLM on cell viability as described previously[32].Cell migration assay was performed as described previously[33].
Determination of lactate
The lactate level of plasma and tumors archived from our previous study[25]was measured using a colorimetric assay kit.Plasma specimens and tumor lysates were filtered using a 10 kDa filter.Assays were performed according to the manufacturer’s instructions.
The in vivo study using Hi-Myc mice
Female transgenic FVB-Tg (ARR2/Pbsn-MYC) mice were crossed with male wild-type FVB/N mice, both purchased from the NCI Mouse Repository, to establish a colony.Following transgene verification, 5-weekold male transgenic Hi-Myc mice were divided into two groups.Mice of group 1 were treated with 100 µL vehicle, while group 2 mice received 10 mg LLM/kg body weight by intraperitoneal injection 5 times/week(Monday through Friday).The vehicle consisted of 10% ethanol, 10% dimethyl sulfoxide, 30% kolliphor EL(Sigma-Aldrich, St.Louis, MO), and 50% PBS.After 5 weeks of treatment, the animals were euthanized by CO2overdose (suppliedviacompressed gas cylinder), and blood and prostate tissue were collected.
Statistical tests
The Student’st-test was used for two-group comparisons.One-way analysis of variance (ANOVA) followed by Bonferroni’s test was used for multiple comparisons.ANOVA followed by Dunnett’s test was used for dose-response comparisons.Statistical significance for the effect of LLM on the incidence of hyperplasia(HYP), low-grade PIN (LG-PIN), high-grade PIN (HG-PIN), adenocarcinoma (AC)in situ, and microinvasion (Mic-inv) was determined by Fisher’s test.GraphPad Prism (version 8.0.0) was used to perform statistical analyses.
RESULTS
Effect of LLM on the expression of cMyc
Pharmacokinetics of LLM has been investigated using male ICR mice after a single oral administration with 10 mg/kg body weight dose[33].The peak plasma concentration of LLM was shown to be about 2.8 µM[33].Thus, LLM concentrations of 2.5 and 5 μM were used to determine its effect on the expression of cMyc.As can be seen in Figure 1A, the expression of cMyc protein was decreased in a concentration-dependent manner.The LLM-mediated downregulation of the cMyc protein was confirmed by confocal microscopy[Figure 1B].LLM treatment also inhibited mRNA levels ofcMyc[Figure 1C] and suppressed its transcriptional activity [Figure 1D].Collectively, these results indicated suppression of cMyc expression and activity following LLM treatment in LNCaP and 22Rv1 human prostate cancer cell lines.
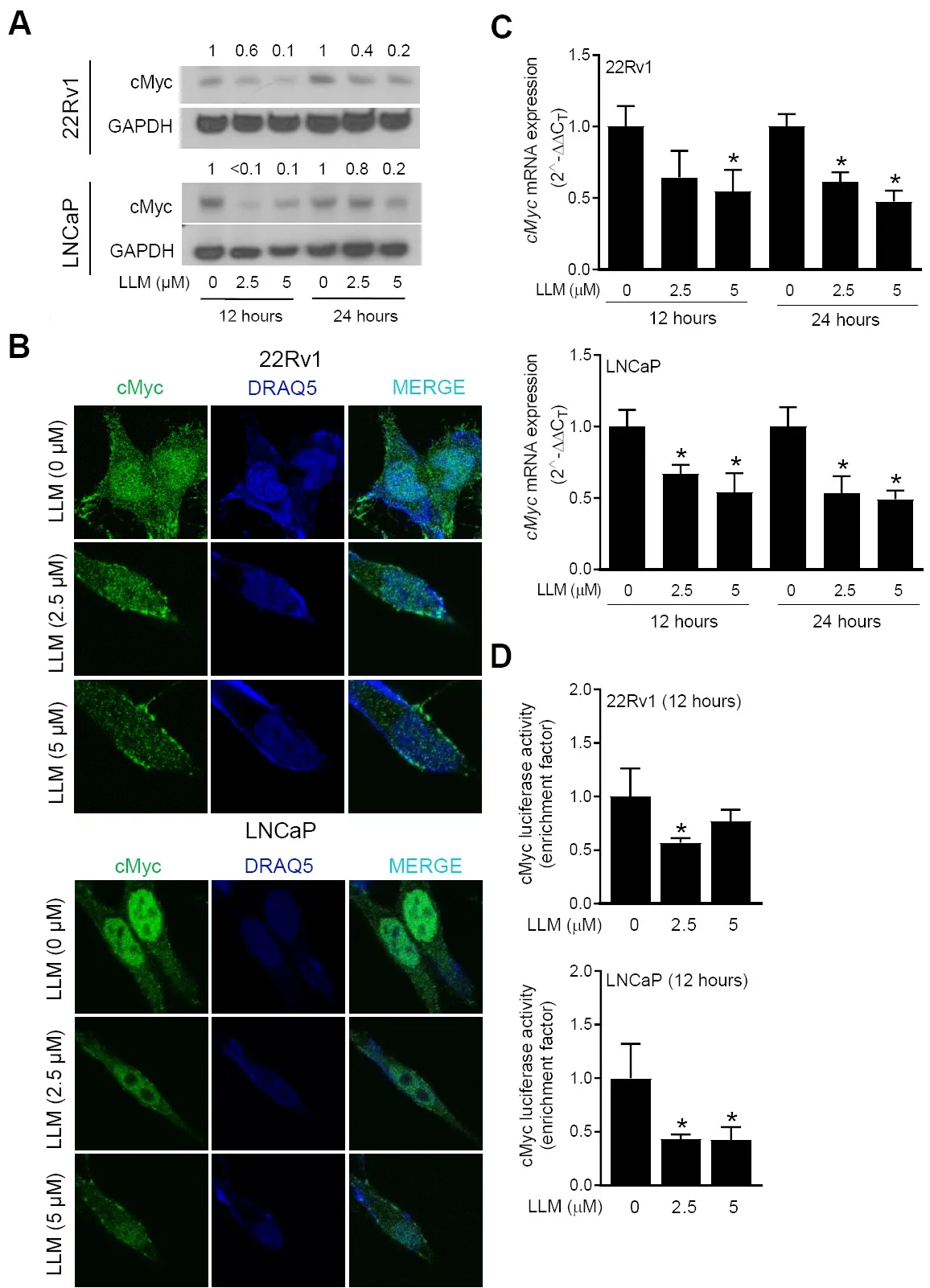
Figure 1.Effect of leelamine (LLM) treatment on expression and activity of cMyc in human prostate cancer cells.The effect of LLM treatment on the protein level of cMyc in 22Rv1 and LNCaP cells as determined by western blotting.The blots were probed with an anti-GAPDH antibody to correct for differences in protein loading.The numbers above bands represent quantitation of the cMyc protein level change relative to corresponding ethanol-treated control (A).Confocal microscopic images of cMyc protein (green fluorescence)in vehicle-treated control and LLM-treated 22Rv1 and LNCaP cells.DRAQ5 (blue fluorescence) was used for nuclear staining (B).The effect of LLM treatment on mRNA level of cMyc.The results shown are mean ± SD (n = 3).*Significantly different (P < 0.05) compared with corresponding ethanol-treated control by one-way ANOVA followed by Dunnett’s test (C).The effect of LLM treatment on the activity of cMyc as determined by luciferase reporter assay.The results shown are mean ± SD (n = 3).*Significantly different (P < 0.05)compared with ethanol-treated control by one-way ANOVA followed by Dunnett’s test (D).Each experiment was performed twice, and the results were generally consistent.
Effect of cMyc overexpression on anticancer effects of LLM
Figure 2A shows overexpression of cMyc protein in 22Rv1 and PC-3 cells stably transfected with the cMyc plasmid (hereafter abbreviated as cMyc) in comparison with EV control cells.The clonogenicity of EV cells was inhibited by LLM treatment in a dose-dependent manner [Figure 2A].The inhibition of clonogenicity resulting from LLM exposure was partly attenuated by cMyc overexpression [Figure 2A].Similarly, thecMyc overexpression conferred partial protection against LLM-mediated inhibition of cell viability[Figure 2B] and cell migration [Figure 2C] in 22Rv1 and/or PC-3 cells.These results indicated the functional significance of cMyc suppression in anticancer effects of LLM.
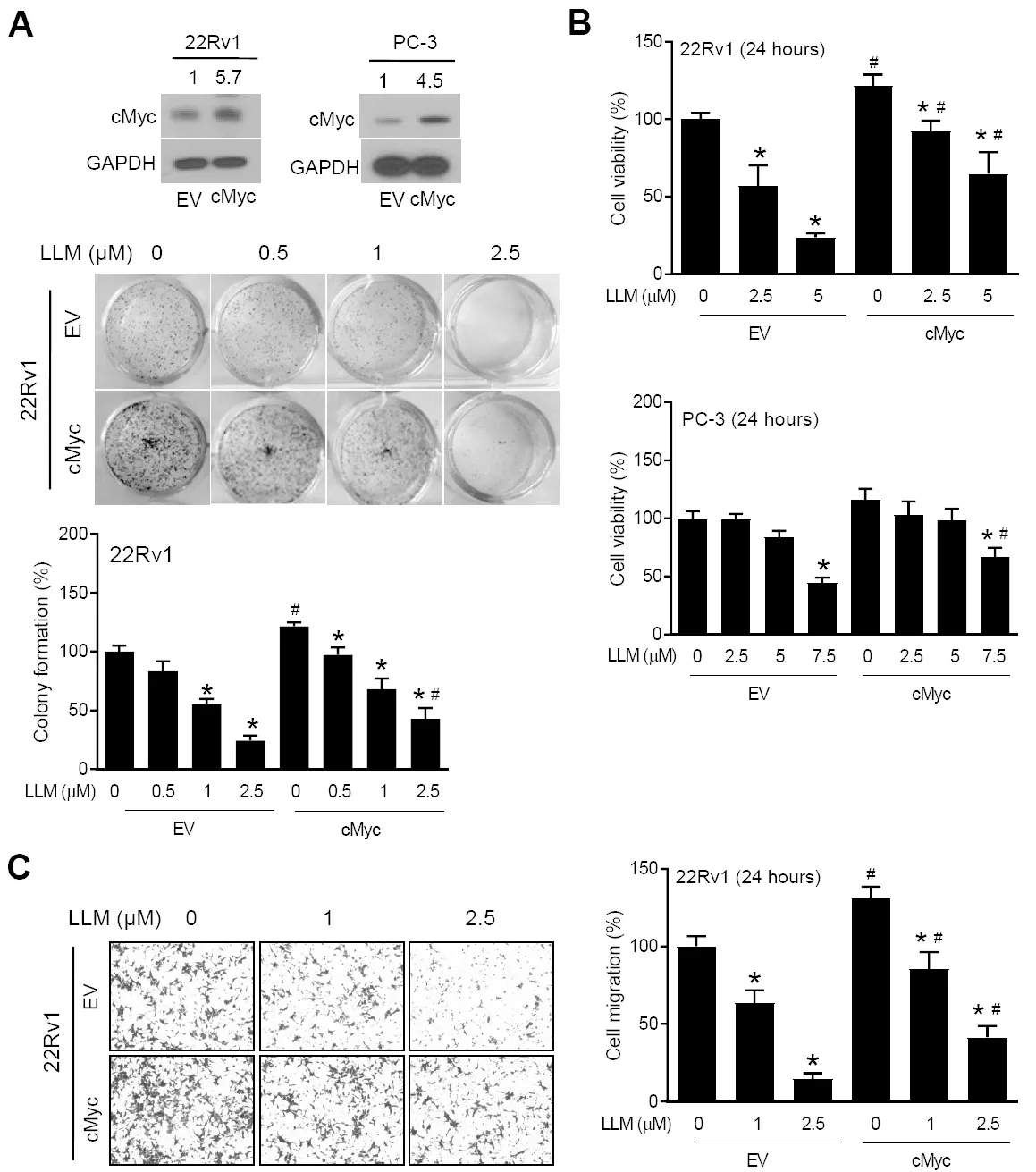
Figure 2.Effect of cMyc overexpression on the anticancer effects of leelamine (LLM).upper: Western blots showing overexpression of the cMyc protein in stable cMyc overexpressing cells (cMyc) compared to empty vector (EV) transfected control cells.The numbers above bands represent quantitation of the cMyc protein level relative to corresponding EV cells.middle and bottom: Clonogenic assay using EV and cMyc cells following treatment with vehicle or LLM.The results shown are mean ± SD (n = 3).Significantly different(P < 0.05) compared with *ethanol-treated control, and #between EV and cMyc groups by one-way ANOVA followed by Bonferroni’s test (A).Trypan blue dye exclusion assay using EV and cMyc cells following treatment with vehicle or LLM.The results shown are mean ± SD (n = 3).Significantly different (P < 0.05) compared with *ethanol-treated control, and #between EV and cMyc groups by one-way ANOVA followed by Bonferroni’s test (B).Cell migration assay using EV and cMyc cells following treatment with vehicle or LLM.The results shown are mean ± SD (n = 3).Significantly different (P < 0.05) compared with *ethanol-treated control, and#between EV and cMyc groups by one-way ANOVA followed by Bonferroni’s test (C).Each experiment was performed twice, and the results were generally consistent.
Effect of LLM treatment on protein levels of cMyc regulated proteins
Next, we determined the effect of LLM treatment on the levels of cMyc target proteins.Expression of several glycolysis-related proteins was decreased by LLM treatment in 22Rv1 and LNCaP cells [Figure 3A].There was a decrease in circulating level of lactic acid, the end product of glycolysis, in LLM-treated mice than in control mice but the difference was not significant.The 22Rv1 xenograft level of lactate was also not suppressed by LLM treatment [Figure 3B].The expression of cMyc and some of its target proteins was also not suppressed to a significant extent by LLM administration to 22Rv1 xenograft bearing mice[Figure 4A and B].
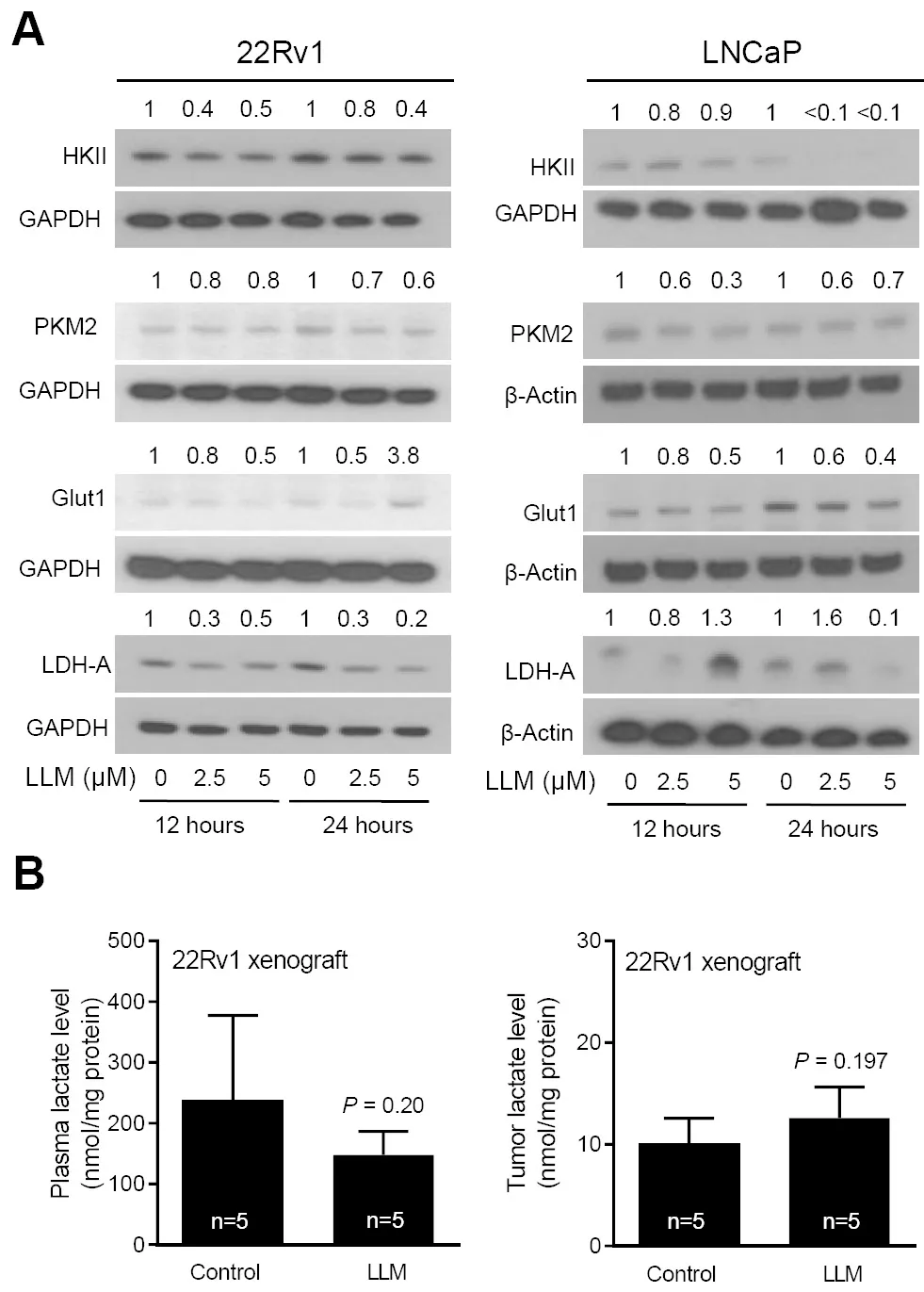
Figure 3.The effect of leelamine (LLM) treatment on glycolysis-related proteins.Western blots showing the effect of LLM treatment on expression levels of glycolysis-related proteins.The numbers above the bands represent quantitation of the protein level change relative to corresponding ethanol-treated control.The experiment was performed twice, and the results were generally consistent (A).Levels of lactate in the plasma and 22Rv1 xenograft tumors of control and LLM-treated mice (n = 5).The results shown are mean ± SD and statistical significance was determined by Student’s t-test (B).
Effect of LLM treatment on cancer incidence in Hi-Myc mice
Initially, we used a cell line (Myc-CaP) derived from the prostate tumor of a Hi-Myc transgenic mouse to determine the effect of LLM treatment on cMyc protein level.Similar to human prostate cancer cells[Figure 1A and B], western blot analysis revealed a decrease in the protein level of cMyc following LLM treatment in the Myc-CaP cell line [Figure 5A].Confocal microscopy confirmed LLM-mediated downregulation of cMyc protein level [Figure 5B].Similar to our previous studies in mice[27], LLM administration was also well-tolerated by Hi-Myc mice [Figure 5C].Figure 5D shows the H&E stained prostate tissue sections from control and LLM-treated Hi-Myc mice.The incidence of HG-PIN, ACin situ,and Mic-inv was lower in LLM-treated mice compared to vehicle-treated control mice but the difference was not statistically significant [Figure 5E].
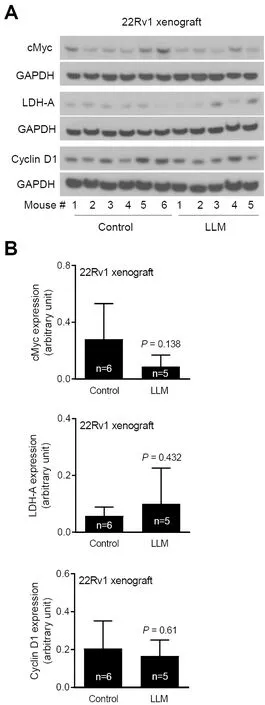
Figure 4.The effect of leelamine (LLM) treatment on cMyc and its targets using lysates from 22Rv1 xenografts.Western blots for cMyc, LDH-A, Cyclin D1, and GAPDH using tumor lysates from control and LLM-treated mice (A).Quantitation of cMyc, LDH-A, and Cyclin D1 expression.The results shown are mean ± SD (n = 5-6).Statistical significance was determined by Student’s t-test (B).

Figure 5.The effect of leelamine (LLM) treatment on cancer incidence in Hi-Myc mice.The effect of LLM treatment on the protein level of cMyc in Myc-CaP cell line as determined by western blotting.The blot was probed with an anti-GAPDH antibody to correct for differences in protein loading.The numbers above bands represent quantitation of the cMyc protein level change relative to corresponding ethanol-treated control.Similar results were obtained in repeated experiments (A).Confocal microscopic images of cMyc protein (green fluorescence) in vehicle-treated control and LLM-treated Myc-CaP cells.DRAQ5 (blue fluorescence) was used for nuclear staining.Similar results were obtained in repeated experiments (B).Body weights of control and LLM-treated Hi-Myc mice (n =10).Statistical significance was determined by Student’s t-test.LLM treatment did not cause a significant weight loss in Hi-Myc mice(C).Representative H&E-stained sections of the prostate from control and LLM-treated Hi-Myc mice (40x objective magnification,scale bar = 50 µm) (D).The effect of LLM administration on incidence of different histopathologic stages of prostate cancer in Hi-Myc mice (n = 10).Statistical significance was determined by Fisher’s test (E).
DISCUSSION
The present study shows the inhibitory effect of LLM on cMyc expression and activity in prostate cancer cells.The cMyc inhibition is functionally significant because the anticancer effects of LLM, including clonogenicity, cell viability, and cell migration, are partially attenuated by cMyc overexpression.It is also clear that cMyc suppression alone is not responsible for the growth inhibitory effects of LLM on prostate cancer cells.Several other oncogenes have been shown as targets of LLM in other cancer cell types likemelanoma, including phosphatidylinositol 3-kinase, mitogen-activated protein kinase, and STAT3[24].All these oncogenic signalling pathways are known to contribute to the pathogenesis of human prostate cancer[34-36].For example, analysis of the Oncomine database reveals upregulation ofc-Jun N-terminal kinasesin prostate cancer[35].Likewise, STAT3 plays an important role in the progression of prostate cancer to incurable metastatic castration-resistant disease by integrating multiple signalling pathways involved in the reactivation of AR pathway, stem-like cells, and the epithelial to mesenchymal transition[36].It is conceivable that LLM inhibits these signalling pathways to, in turn, inhibit the growth of prostate cancer cells.However, additional work is necessary to systematically explore this possibility.Chemoprevention represents a worthwhile strategy for reducing the mortality and morbidity associated with prostate cancer[20-22].The feasibility of prostate cancer chemoprevention has been explored clinically with 5α-reductase inhibitors (PCPT and REDUCE trials) as well as a novel combination of selenium and vitamin E (e.g., SELECT trial)[17-19].Whereas administration of 5α-reductase inhibitors resulted in ~23%-25%decrease in overall relative risk, these agents were not approved by the FDA for chemoprevention of prostate cancer due to high-grade tumors in the treatment arm[17,18].In an 18-year follow-up study, highgrade prostate cancer was still more common in the finasteride treatment group[37].Similarly, the SELECT trial did not show any preventative benefit of vitamin E and selenium supplementation[18].Therefore, a safe
and efficacious intervention for chemoprevention of prostate cancer is still a clinically unmet need.In this study, we explored the possibility of chemoprevention by LLM using a transgenic mouse model in which prostate cancer is driven by prostate epithelial-specific overexpression of cMyc.There was a trend for a decrease in the incidence of HG-PIN, ACin situ, and Mic-inv but the difference was not significant at the 10 mg/kg body weight dose of LLM.These results are surprising because, in melanoma models, LLM doses of 5 and 7.5 mg/kg body weight were effective forin vivoinhibition of tumor growth[24].
In conclusion, the present study revealsin vitro, the inhibitory effect of LLM on cMyc expression and activity in prostate cancer cells.However, LLM doses higher than 10 mg/kg body weight might be required to achieve chemoprevention of prostate cancerin vivo.Thus, similar to phytochemicals like curcumin,resveratrol, and neem extracts, LLM may be useful for the prevention of prostate cancer[38-40].In one such study, supercritical extract of neem leaves (SENL) was shown to inhibit integrin β1, calreticulin, and focal adhesion kinase activation in LNCaP-luc2 and PC3 prostate cancer cells[40].The growth of LNCaP-luc2 xenograft was also reduced significantly by SENL administration that was associated with the formation of hyalinized fibrous tumor tissue, reduction in the prostate-specific antigen, and increase in AKR1C2 levels[40].
DECLARATIONS
Authors’ contributions
Study design: Singh KB, Hahm ER, Singh SV
Performed experiments: Singh KB, Hahm ER
Study supervision: Singh SV
Data interpretation: Singh KB, Hahm ER, Singh SV
Final manuscript preparation: Singh KB, Hahm ER, Singh SV
Availability of data and materials
Not applicable.
Financial support and sponsorship
This study was supported by the National Cancer Institute grant R01 CA225716 (to S.V.S.).The following UPMC Hillman Cancer Center core facilities partly supported by the National Cancer Institute grant P30 CA047904 were utilized in this study: Animal Facility, and Cell and Tissue Imaging Facility.
Conflicts of interest
All authors declared that there are no conflicts of interest.
Ethical approval and consent to participate
The care of Hi-Myc mice was consistent with the University of Pittsburgh Animal Care and Use Committee guidelines.
Consent for publication
Not applicable.
Copyright
© The Author(s) 2021.
 Journal of Cancer Metastasis and Treatment2021年3期
Journal of Cancer Metastasis and Treatment2021年3期
- Journal of Cancer Metastasis and Treatment的其它文章
- Diverse roles of bitter melon (Momordica charantia)in prevention of oral cancer
- Seaweeds’ nutraceutical and biomedical potential in cancer therapy: a concise review
- Role of 18F-FDG PET/CT in detection of hematogenous metastases of advanced differentiated thyroid carcinoma: a systematic review and meta-analysis
- Targeting EZH2 for the treatment of soft tissue sarcomas
