Understanding the genetic basis of rice heterosis:Advances and prospects
Sinn Zhng ,Xuehui Hung ,*,Bin Hn *
a Shanghai Key Laboratory of Plant Molecular Sciences,College of Life Sciences,Shanghai Normal University,Shanghai 200234,China
b National Center for Gene Research,CAS Center for Excellence in Molecular Plant Sciences,Institute of Plant Physiology and Ecology,Shanghai Institutes for Biological Sciences,Chinese Academy of Sciences,Shanghai 200233,China
Keywords:Hybrid rice Heterosis Genetic basis Breeding
ABSTRACT Heterosis,which describes the superior vigor and yield of F1 hybrids with respect to their parents,is observed in many rice hybrid crosses.The exploitation of heterosis is a great leap in the history of rice breeding.With advances in genomics and genetics,high-resolution mapping and functional identification of heterosis-associated loci have been performed in rice.Here we summarize advances in understanding the genetic basis of grain yield heterosis in hybrid rice and provide a vision for the genetic study and breeding application of rice heterosis in the future.
1.Development of hybrid rice breeding
Although the concept of hybrid rice breeding has a long history,the approach was not widely applied at first owing to the difficulty of large-scale hybridization in rice.Only when in the 1970s Longping Yuan and his assistants discovered wild abortive cytoplasmic male sterility(WA-CMS)and developed the three-line hybrid system consisting of a sterile,a restorer,and a maintainer line[1]did hybrid rice breeding begin to be technically feasible.The elite hybrid rice Shanyou 63,generated from Zhenshan 97 as a sterile line and Minghui 63 as a restorer line,has been widely grown in Southern China for the past 40 years thanks to its high yield and stress resistance[2].
The three-line hybrid system can be divided into several types,including WA-CMS,honglian-type(HL)CMS[2]and boro-type(BT)CMS[3].However,restorer lines face a severe germplasm limitation,because they must carry specific nuclear Rf alleles for a given sterile line.With the discovery of photoperiod-sensitive genic male sterility(PGMS)[4]and temperature-sensitive genic male sterility(TGMS)[5,6],rice breeders constructed a two-line hybrid system consisting of a sterile line and a restorer line.In this system,because both PGMS and TGMS alleles are nuclear-recessive and male fertility can be restored by nearly all normal cultivars[7,8],there is no germplasm limitation for the restorer lines,and nearly all rice accessions can be used as restorer lines.Thus,the two-line hybrid system has facilitated rice hybrid breeding compared with the three-line system[9].But the two-line system is generally less stable than the three-line system,because the fertility of PGMS/TGMS is vulnerable to uncontrollable weather fluctuations[10].
Third-generation hybrid rice technology offers the advantages of both no germplasm limitation and stable sterility[11].Breeders transformed an expression vector containing a fertility restoration gene,CYP703A3[12],a pollen lethality gene,orfH79[13],and a seed color gene,DsRed2[14]into a CYP703A3-deficient recessive genic male-sterile(RGMS)line to generate a maintainer line.CYP703A3 restores male fertility and DsRed2 marks transgene seeds with red color.Thus,seeds resulting from self-pollination of the maintainer line may be distinguished by color,assorting into half male-sterile non-transgene seeds as the sterile line and half red transgene seeds as the maintainer line.Besides RGMS lines,dominant genic male-sterile(DGMS)lines have also been applied in hybrid breeding[15,16].These strategies confer several advantages,among them the elimination of the release of transgenic pollen into the environment.However,DGMS hybrid seeds containing transgene elements would be under strict control in some countries.
Very recently,it was reported[17,18]that defective function of meiosis and fertilization genes(REC8,PAIR1,OSD1,and MTL)induced rice clonal propagation through seeds.This strategy makes it possible to construct a one-line hybrid system via selfpropagation of elite hybrid rice.Because defective function of suchgenes may also affect seed setting rate of the hybrid cross,further technical improvements are needed before its practical application.
2.Researching the genetic basis of heterosis via omics
The omics field was developed to address biological questions from a global view via cost-efficient and high-throughput analyses,and offers a powerful method for researching the genetic basis of heterosis.To address the key questions in heterosis(how heterosis forms,what genes are associated with heterosis,and how they work),various omics technologies have been applied,including genomics[19,20],transcriptomics[21,22],metabolomics[23,24],and phenomics[25].
Genome-wide association studies[26]of rice heterosis have established links between agronomic traits and candidate loci[19,20].Heterosis-associated genes influencing 38 agronomic traits were identified[19]using a high-density genome map in a large number of rice lines(divided into three types:indica-indica crosses from a three-line hybrid system,indica-indica crosses from a twoline hybrid system,and indica-japonica crosses).Heterosisassociated genes were involved mainly in heading date and plant architecture.Alleles from the female parents explained much of the heterosis advantage in hybrid rice.
RNA-sequencing data from Shanyou 63 and its parents using three tissues(shoot,flag leaf,and panicle)[22],revealed 3270 genes in Shanyou 63 that showed allele-specific expression(ASE)and were suggested to play a role in heterosis.These genes could be classified into two patterns:consistent and inconsistent ASE genes(ASEGs).In the hybrid line,the expression of a consistent ASEG was biased toward one parental allele in all of 12 cases(three tissues under four growth conditions),whereas the expression of an inconsistent ASEG was biased toward one parental allele in some but not all cases.
Besides genomics analysis and transcriptomics analysis,phenomics analysis concentrating on rice yield and yield-component phenotypes has been employed to identify causal traits underlying rice heterosis[25].Metabolomics analysis has been applied to investigate the relationship between metabolism and heterosis in maize [23,24].Omics analyses offer increasing potential in researching genetic basis of heterosis.
3.Genetic basis of rice heterosis
3.1.Genetic mechanism of rice heterosis
The classical hypotheses advanced to explain heterosis include single-locus overdominance,dominance complementation,and epistatic interaction.In overdominance(Fig.1A),a single heterozygous locus displays a more extreme or better adapted phenotype than either homozygous parental locus.In dominance complementation(Fig.1A),two non-allelic loci(A and B)play complementary roles in control of the same phenotype.For example,the genotype of parent 1(P1)is aaBB,while that of parent 2(P2)is AAbb(where lower-case letters represent recessive and uppercase letters dominant alleles).When P1and P2are crossed,the F1(with genotype AaBb)shows a more extreme phenotype than either P1or P2because of the complementary effect of loci A and B.Pseudooverdominance is a special case of dominance complementation,when two closely linked QTL display heterosis.Under the epistasis hypothesis(Fig.1A),genetic interaction between two non-allelic loci results in heterosis.
In rice,genetic studies have shown that overdominance,incomplete dominance,and epistasis may all contribute to heterosis[27-29].Hua et al.[30]constructed an immortalized F2population(IMF)by pairwise crossing of RILs from Shanyou 63.By investigating all the genetic components in the IMF,they determined the contributions of dominance and overdominance effects in heterosis of Shanyou 63.In a recent study[19],we investigated the genetic basis of heterosis via an integrated genomic approach using 17 F2populations to identify key heterosis-associated loci and their contributions to heterosis.The results showed that incomplete dominance and overdominance effects played respectively primary and secondary roles in rice heterosis.Using an F4population derived by crossing Lemont and Teqing,Li et al.[31]found that epistasis influenced rice yield,especially kernel number per panicle and grain weight per panicle.Yu et al.[32]also identified the importance of epistasis in heterosis of hybrid rice by dissecting the genetic components of Shanyou 63.
3.2.Heterosis-associated genes
Random crosses between inbred lines may result in different degrees of heterosis,including its absence[33,34].Accordingly,for hybrid breeding especially in maize,it is desirable to identify heterotic groups and patterns[35].For hybrid rice,indica and japonica rice have been recognized as two distinct heterotic groups.Genetic dissection of heterosis-associated genes[20]may lead breeders to select suitable lines and to understand the molecular mechanism of heterosis.Many key heterosis-associated genes in the rice genome,such as Ghd8,Hd3a and TAC1,have been characterized[20,36],and the corresponding heterosis-associated alleles have been characterized in various parental lines of elite hybrid rice(Fig.1B).Hd3a,controlling heading time,is a rice homolog of the Arabidopsis thaliana FT gene[37].Yield overdominance of SFT,its homolog in tomato,has been observed in the heterozygous state[38,39].TAC1 is a major QTL regulating tiller angle in rice[40]and influences planting density in agricultural production.Ghd8 regulates kernel number,plant height,and heading date[41]in the classical two-line hybrid system that developed Liang-you-pei 9(LYP9)from the cross of Peiai64S(PA64S)and 93-11[20,25].Besides Ghd8,several alleles with positive partial dominance from female parental lines in indica-indica crosses of the two-line hybrid system have been identified,including OsMADS51[42]for heading date and LAX1[43]for kernel number as well as GW3p6(OsMADS1)[44]for grain weight.Because the genetic sources used in the two-line hybrid system are relatively diverse,many parental lines in this system contribute special superior alleles.
Wide-compatibility alleles,which can break the fertility barrier between the indica and japonica subspecies,have been identified and applied in breeding[45],promoting the development of indica-japonica hybrid rice cultivars.Several heterosis-associated genes have been characterized in indica-japonica rice crosses as well.For example,GW6a[46],influencing plant height and grain weight in indica,and Hd1[47]influencing heading time in japonica show together a dominance complementary effect,which functions in the heterosis of indica-japonica rice crosses.OsSPL14(also named IPA1)[48,49]regulating plant architecture has been identified as a heterosis-associated gene in an indica-japonica hybrid.The rare allele ipa1 from japonica accessions conferred higher kernel number per panicle,while the wild-type allele IPA1 conferred higher panicle number and seed-setting rate.The IPA1/ipa1 genotype showed single-gene overdominance with respect to grain yield per plant.
Given that multiple alleles with different genetic effects contribute to heterosis,the formation of heterosis in elite hybrid rice is so complex that it is necessary to evaluate the influence of each allele separately.One approach is to construct near-isogenic lines(NILs)[50]containing single heterosis-associated alleles,so that the genetic effect and contribution of each candidate gene can be determined without the influence of other loci[20].
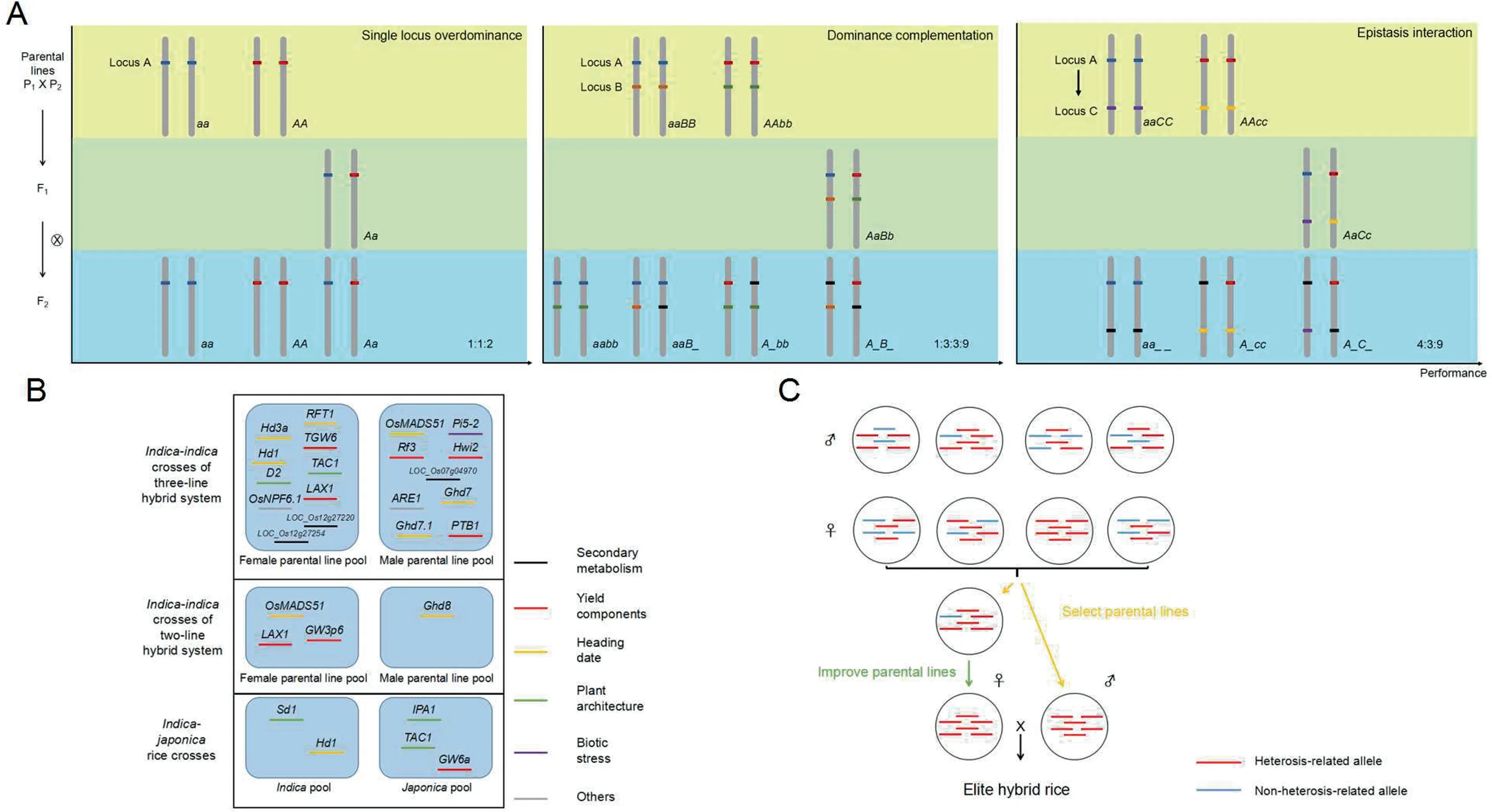
Fig.1.Understanding the genetic basis of rice heterosis.(A)Classical hypotheses for genetic mechanisms of heterosis in simplified circumstances.In single-locus overdominance,a heterozygous locus A(Aa)confers a more extreme phenotype than either AA or aa;and in an F2 population resulting from self-pollination of F1(Aa),three phenotypes appear in a 1:2:1 proportion.In dominance complementation,A and B are dominant alleles resulting in a high phenotype and a and b are recessive alleles resulting in a low phenotype.Hybrid lines(AaBb)derived from crossing AAbb and aaBB show a higher phenotype than the parental lines because the combination of A and B has an additive effect.In F2 populations derived from selfing of F1(AaBb),four phenotypes occur in a 1:3:3:9 proportion.For epistatic interaction,A and C are dominant alleles conferring a high phenotype,while a and c are recessive alleles resulting in a low phenotype.A hybrid line(AaCc)derived by crossing AAcc with aaCC shows a higher phenotype than its parental lines because of the epistatic interaction between A and C.In an F2 population derived by selfing of F1(AaCc),three phenotypes occur in a 4:3:9 proportion because effects of both C and c are suppressed by aa.Each colored bar represents a specific allele,except that black bars represent any alleles at a specific locus.(B)Heterosis-associated alleles identified in elite hybrid rice.(C)Developing new elite hybrid rice cultivars with knowledge of heterosis.Select parental lines:use molecular biology knowledge for rapid selection of suitable parents for hybrid rice breeding.Improve parental lines:introduce useful alleles or even create new alleles by genome editing.
Genome-editing technology based on clustered regularly interspaced palindromic repeats(CRISPR)and the Cas9 system has been applied extensively in plant research,including for crop improvement[51-54].It is now possible to characterize heterosisassociated candidate genes directly by generation of heterosisassociated alleles using genome editing technology,including gene knockouts,base editing[55],and even fragment knock-ins[56].Using this technology,the effect of a single heterosis-associated candidate gene may be identified more easily.With NILs,it is likely to be hard to distinguish single-gene overdominance from pseudooverdominance because mapping techniques cannot precisely determine map positions,whereas genome editing can avoid this hurdle by direct generation of a single heterosis-associated allele.
4.Perspectives for hybrid rice breeding
Several novel strategies may advance the application of heterosis in rice breeding.One strategy,mentioned above,is the development of a one-line breeding system via knockout of four meiosis and fertilization genes.Another strategy based on de novo domestication of wild allotetraploid rice[57],offers promise.Polyploids are common in nature and many economically important crops are polyploid,including wheat,tobacco,cotton,and strawberry.One accepted opinion[58,59]explaining the prevalence of polyploidy is that polyploid crops show vigor and environmental adaptability because polyploidy stabilizes heterosis.Thus,successful de novo domestication of wild allotetraploid rice could lead to stable heterosis in allotetraploid rice.
With knowledge of rice heterosis,it is also possible to develop new elite hybrid rice cultivars by making use of heterosisassociated genes(Fig.1C).In conventional hybrid rice breeding,selecting ideal elite hybrids by phenotypic screening of many hybrid crosses is time-consuming.With advances in genomics technology,molecular breeding can be adopted to improve crops[60].Many heterosis-associated alleles that have been characterized may be used to improve hybrid rice more easily and efficiently than by phenotypic screening alone,with assistance from nextgeneration sequencing[61]and/or genome editing.The genetic effects,genetic interactions,and gene-by-environment interactions of heterotic genes in hybrid rice await further study.
In very recent work[36],we constructed a comprehensive quantitative genomics map and developed a navigation system,RiceNavi,for rice quantitative-trait nucleotide pyramiding and breeding optimization,with the intention of integrating rice genomic knowledge and improvement needs.For full exploitation of gene resources in hybrid rice breeding,it is desirable in future to develop a special navigation system for hybrid rice breeding.This system may not only save time and labor in hybrid rice breeding,but allow the selection of hybrid lines superior to those selected by conventional methods.
It is also possible to use known information about heterosis to generate elite inbred lines in rice breeding.Research into fixing the advantage of heterosis from hybrids into inbreds is desirable.An increase of grain yield relative to Fuhui 676 was observed in the inbred line NIL-FH676::GW3p6[44],which was generated by introgressing the heterosis-associated allele GW3p6Guangzhan63-4Sfrom Guangzhan 63-4S into Fuhui 676(Guangzhan 63-4S and Fuhui 676 are respectively female and male parental lines of hybrid rice).The yield phenotype of NIL-FH676::GW3p6 demonstrates that some heterosis-associated genes can be used to improve inbred rice lines directly,without need of constructing hybrid lines.
CRediT authorship contribution statement
Sinan Zhangwrote the manuscript.Xuehui Huangconceptualized and supervised this work,and edited the manuscript.Bin Hansupervised the review and edited the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Dr.Jie Liu for helping to polish the language.This work was funded by the National Key Research and Development Program of China(2016YFD0100902).
- The Crop Journal的其它文章
- Breeding by design for future rice:Genes and genome technologies
- Innovation and development of the third-generation hybrid rice technology
- CRISPR/Cas systems:The link between functional genes and genetic improvement
- Genomic selection:A breakthrough technology in rice breeding
- Target chromosome-segment substitution:A way to breeding by design in rice
- Breeding by selective introgression:Theory,practices,and lessons learned from rice

