Morphology and molecular phylogeny of three species of Coolia (Dinophyceae) from Hainan Island, South China Sea*
Hua ZHANG , Songhui LÜ , , Jingyi CEN, Yang LI, Qun LI, Zhen WU
1 College of Life Science and Technology, Jinan University, Guangzhou 510632, China
2 Shenzhen Academy of Environmental Science, Shenzhen 518001, China
3 Southern Marine Science and Engineering Guangdong Laboratory, Zhuhai 519000, China 4 College of Life Science, South China Normal University, Guangzhou 510631, China
Abstract Marine benthic dinoflagellates in the genus Coolia were reported in both tropical and temperate regions. We conducted a morphological and phylogenetic characterization of Coolia species from Hainan Island, South China Sea. The morphologies of three Coolia species were similar to those of the original descriptions of Coolia canariensis, C. tropicalis, and C. malayensis. In phylogenetic analyses based on the LSU rDNA and ITS regions (ITS1-5.8S-ITS2), the Hainan strains of C. canariensis, C. tropicalis,and C. malayensis clustered within the clades of these species with other isolates from diff erent areas. No diff erences (p-distance) in LSU rDNA sequences were found between the Hainan C. malayensis strains and the strains from New Zealand, Florida of USA, Malaysia, Japan, Dominican Republic and Guangxi and Hong Kong of China. For C. canariensis and C. tropicalis, no diff erences (p-distance) in the LSU rDNA sequences were found between the Hainan strains (D1C2 and DS5F4, respectively) and the Australian strains (NQAIF252 and NQAIF90, respectively). Our study reveals the morphological and genetic diversity of Coolia species from Hainan Island, South China Sea, which provides a detailed understanding of Coolia species of this area.
Keyword: Coolia; Hainan Island; morphology; phylogeny; rDNA
1 INTRODUCTION
Benthic dinoflagellate species in the genusCooliaMeunier are distributed from temperate to tropical waters around the world (Meunier, 1919; Fukuyo,1981; Faust, 1992; 1995; Ten-Hage et al., 2000; Fraga et al., 2008; Leaw et al., 2010; Karafas et al., 2015;David et al., 2020). These species live attached to various substrates, including seaweeds, rocks, corals,mangroves, sands, and also be found in water column(GEOHAB, 2012; Hoppenrath et al., 2014; Leung et al., 2017).
Currently, there are eightCooliaspecies, including the recently reportedC.santacroceKarafas, Tomas and York,C.palmyrensisKarafas, Tomas & York andC.guanchicaH. David, Laza-Martínez, F. Rodríguez& S. Fraga (Meunier, 1919; Faust, 1995; Ten-Hage et al., 2000; Fraga et al., 2008; Leaw et al., 2010; Karafas et al., 2015; David et al., 2020).C.monotisMeunier,C.tropicalis, andC.areolataL. Ten-Hage, J. Turquet,J. P. Quod and Couté were originally described on the basis of morphological data (Meunier, 1919; Faust,1995; Ten-Hage et al., 2000), while the other species were described with both morphological and molecular data (Fraga et al., 2008; Leaw et al., 2010;Karafas et al., 2015; David et al., 2020).
Species in this genus are diff erentiated by their main morphological features, including shape, size,thecal plate arrangement, and ornamentation (Ten-Hage, 2000; Fraga et al., 2008; Leaw et al., 2010).However, the range of cell sizes amongCooliaconspecifics is wide and overlapping, with the exception of the recently reported small speciesC.palmyrensis(Karafas et al., 2015).C.tropicalis,C.areolata,C.canariensisS. Fraga, andC.guanchicashare the feature that the 1′ plate is the largest in the epitheca, located centrally on the cell (Faust, 1995;Ten-Hage et al., 2000; Fraga et al., 2008; David et al.,2014, 2020), whileC.monotiscomplex (C.monotis,C.malayensis,C.palmyensis, andC.santacroce)have a 1′ plate positioned on the left side of the epitheca and a 6″ plate as the largest (Leaw et al.,2010; David et al., 2014; Karafas et al., 2015; Lewis et al., 2018). The thecal plate ornamentation is the principal trait distinguishingC.tropicalis,C.areolata,andC.canariensis(Nascimento et al., 2019). While cell surface is smooth inC.tropicalis, it is strongly areolated inC.areolataand thecal ornamentation is restricted to the hypotheca forC.canariensiscomplex(Ten-Hage et al., 2000; Fraga et al., 2008; David et al., 2014; Karafas et al., 2015). Molecular genetic analyses have been widely used to recognize and delineate species ofCoolia(Penna et al., 2005;Dolapsakis et al., 2006; Leaw et al., 2010). TheC.monotiscomplex have similar morphologies but distinct phylogenies (Leaw et al., 2010, 2016; Karafas et al., 2015; Karafas and Tomas, 2015; Leung et al.,2017). Species in theC.monotiscomplex generally can be diff erentiated fromC.areolata,C.tropicalis,andC.canariensison the basis of both morphology and genetics (Karafas et al., 2015; Karafas and Tomas,2015; Leaw et al., 2016; Nascimento et al., 2019).
Recently, we isolated severalCooliastrains from macroalgae collected from seaweed in the waters around Hainan Island, South China Sea. Cultures were established for morphological and molecular analyses. Three benthic dinoflagellates,C.canariensis,C.tropicalisandC.malayensis, were analyzed using D1/D2 large ribosomal subunit (LSU)rDNA, internal transcribed spacer (ITS) region (ITS1-5.8S-ITS2) sequences and scanning electron microscopy (SEM) micrograph acquisitions. Our report on the morphological and phylogenetic diversity ofCooliaspecies from Hainan Island, South China Sea, provides a detailed understanding ofCooliaspecies of this area.
2 MATERIAL AND METHOD
2.1 Sampling and culturing
Samples of macroalgae (mainlySargassumsp.)were collected from seaweed beds by divers at depths of 1–3 m at Sanya (109°29′47″E, 18°12′33″N),Lingshui (109°58′37″E, 18°18′38″N), and Qionghai(110°49′15″E, 19°18′59″N) on Hainan Island, South China Sea from March, 2013 to August, 2014.Samples of macroalgae and seawater were placed into wide-mouthed polyvinylchloride bottles and immediately brought to the laboratory. These samples were vigorously shaken to detach the dinoflagellates,then filtered through 120- and 20-μm mesh filters.The residue retained on the second mesh was resuspended in filtered seawater for cell isolation.Individual cells were isolated using a single-cell capillary pipette using an inverted research microscope ECLIPSE TE2000-U (Nikon, Yokohama, Japan).Isolated cultures were established in the L1 medium(Guillard and Hargraves, 1993) at a salinity of 30 and incubated at 25 °C in a 12-h:12-h light:dark regime under 150 μmol photons/(m2·s) provided by fluorescent lamps.
2.2 Morphological observation
Morphometric features (dorsal-ventral length and width; anteroposterior length of ≥30 cells) were determined from newly preserved cultures using a QImaging Retiga 4000R digital camera (Qimaging,Surrey, BC, Canada) and IMG Pro plus 6.0 image acquisition and analysis software (Media Cybernetic Inc., Rockville, MD, USA) at ×400 magnification.For SEM, live samples of the exponential growth phase were fixed with 4% glutaraldehyde for 1 h. The samples were then filtered through nucleopore filters(diameter of 13 mm, pore size of 3 μm, Whatman,Little Chalfont, UK), washed with distilled water, and dehydrated in ethanol with a series of concentrations from 10% to 99.9%, increasing by 10% each grade,once for 15 min at each concentration and three times for 99.9%. The samples were then critical point dried with CO2(CPD 030, Bal-Tec, AG, Balzers,Liechtenstein). The filters were stub mounted, coated with gold (SCD 005, Bal-Tec) and observed with a field emission SEM Ultra 55 (Zeiss, Jena, Germany).The Kofoid tabulation system was adopted to name the thecal plates (Kofoid, 1909).
2.3 Molecular analysis
Total genomic DNA was extracted using the Omega HP Plant DNA kit (Omega Bio-Tek, Norcross,GA) following manufacturer’s instructions. The amplification reactions were conducted using a Bio-Rad PCR Thermal Cycler (Bio-Rad, Hemel Hempstead, UK). Subsequent PCR reactions used PCR master mix (TaKaRa, Dalian, China), template DNA and primers in a final volume of 50 μL. TheD1–D3 regions of the LSU were amplified with the primers D1R and D3B (Scholin et al., 1994; Nunn et al., 1996), and the ITS region was amplified using the universal primers of ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′). The PCR amplification reactions for the LSU and ITS regions were performed as previously described (Zhang et al., 2015). Sequencing was performed using the ABI 3730XL Big Dye V3.1 Mix technique (Applied Biosystems, Foster City, CA,USA). Sequences data that support the findings of this study have been deposited in GenBank with the accession number from KR229994 to KR229997 for LSU rDNA and from MF805767 to MF805770 for ITS regions.
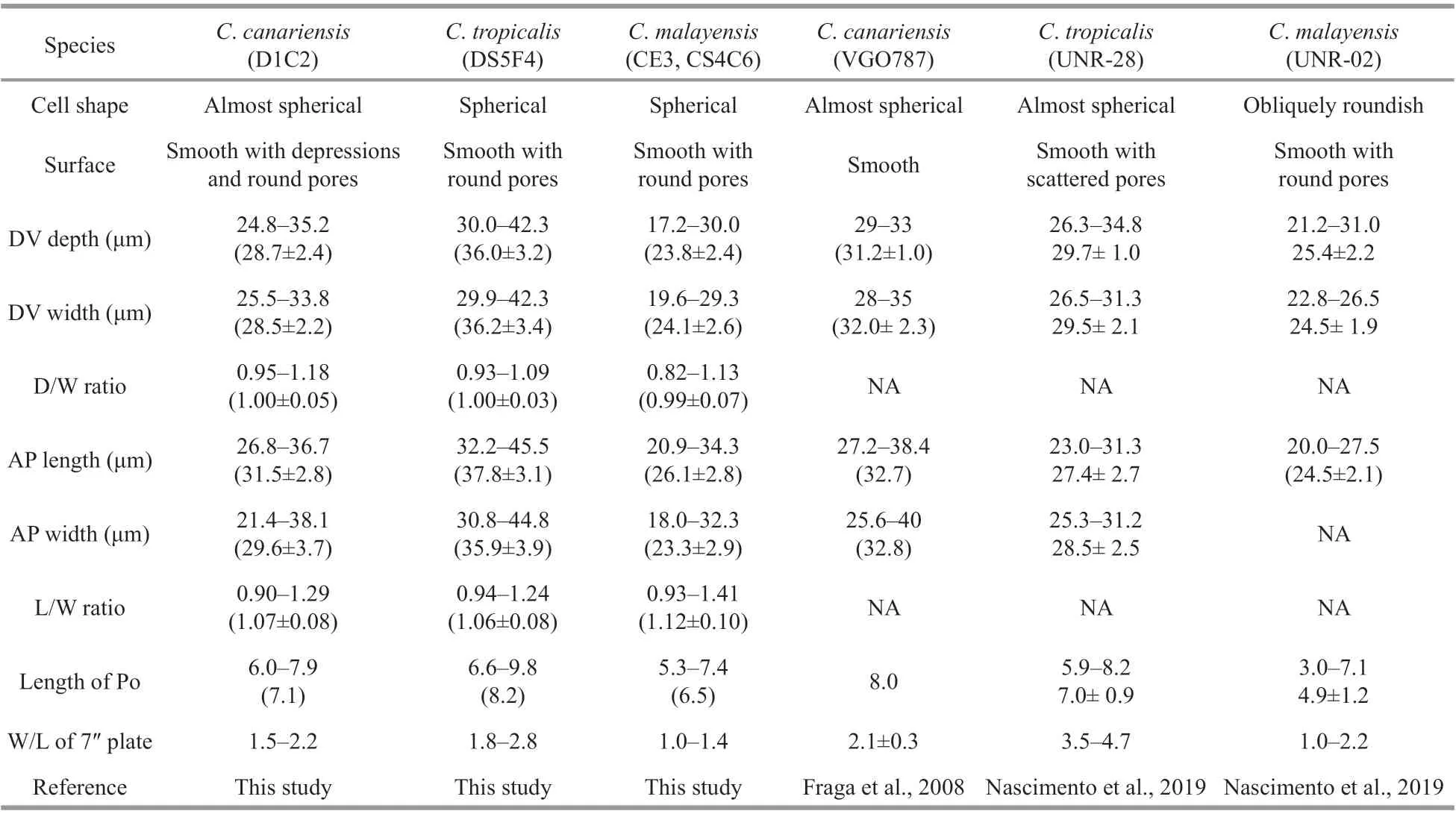
Table 1 Characterization of Coolia species
There were 52 and 46 sequences included in the LSU rDNA and ITS region phylogenetic analyses,respectively. These were aligned with other relatives retrieved from the GenBank database using ClustalW(Thompson et al., 1994). Only D1/D2 LSU rDNA sequences were analyzed.Ostreopsisspp. were used as outgroups for both the LSU rDNA and ITS region sequence analyses. A maximum likelihood (ML)phylogenetic tree was inferred for each region using Mega X (Kumar et al., 2018). Bayesian analyses were performed with MrBayes 3.1.2 using the best-fitting model (Ronquist and Huelsenbeck, 2003). The bestfitting models for the D1/D2 LSU rDNA and ITS regions were GTR+I+G and HKY+I+G, respectively,which were selected by hLRT in MrModeltest 2.3(Nylander, 2004). Identical tree topologies were inferred from the ML and Bayesian analyses for both regions. Genetic distance (p-distance) was assessed using MEGA X (Kumar et al., 2018). All positions containing gaps and missing data were eliminated.
3 RESULT
Morphological and phylogenetic analyses indicated that the four strains (D1C2, DS5F4, CE3, and CS4C6)ofCooliaspecies isolated from Hainan Island wereCooliacanariensis,C.tropicalis, andC.malayensis.
3.1 Morphology of the Hainan strain of C.canariensis
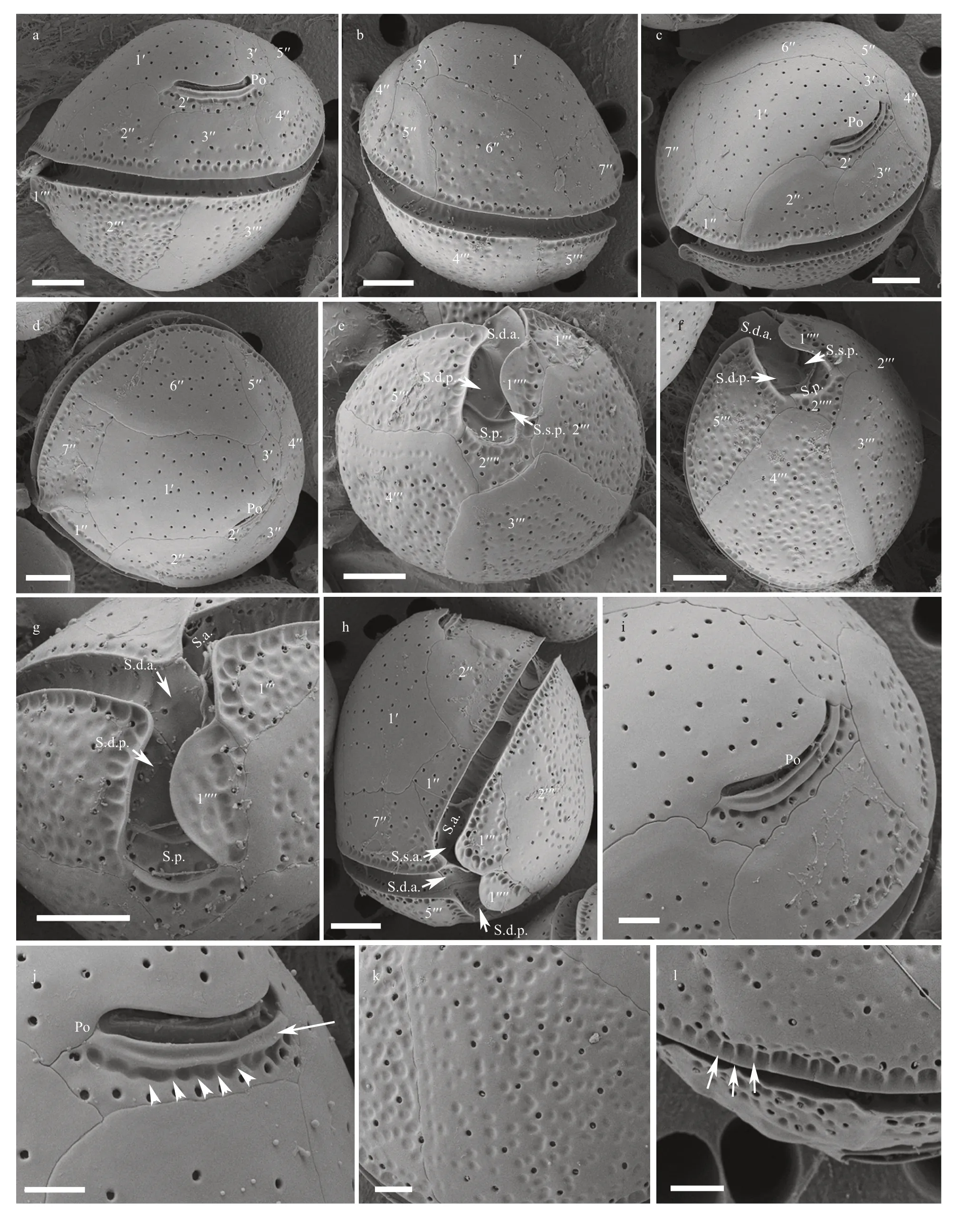
Fig.1 SEM micrographs of C. canariensis
The cells of theC.canariensisHainan strain(D1C2) were almost spherical (Fig.1a–b) with a dorsoventral (DV) depth ranging from 24.8 to 35.2 μm(28.7±2.4 μm,n>30), width from 25.5 to 33.8 μm(28.5±2.2 μm,n>30), and anteroposterior (AP) length from 26.8 to 36.7 μm (31.5±2.8 μm,n>30) (Table 1).The plate formula was Po, 3′, 7′′, 6c, 6s, 5′′′, 2′′′′(Fig.1c–f). The width/length (W/L) ratio of the 7′′plate ranged from 1.5 to 2.2 (Table 1). The cingulum was deep and slightly displaced (Fig.1g–h). The sulcus was short and there were six sulcal plates observed: the posterior sulcal plate (S.p.), the left posterior sulcal plate (S.s.p.), the right posterior sulcal plate (S.d.p.), the right anterior sulcal plate (S.d.a.),the left anterior sulcal plate (S.s.a.), and the anterior sulcal plate (S.a.) (Fig.1e–h). The Po plate was 6.0–7.9 μm (average 7.1 μm) in length (Table 1). The apical pore complex (APC) was elongated and slightly curved, and was surrounded by three apical plates: 1′,2′ and 3′ (Fig.1c–d & i–j). The 1′ plate was hexagonal and centrally located in the epitheca (Fig.1c–d). The edge of plate 2′ where embraced the APC was a protuberant, lip-like structure (Fig.1i–j). A row of hard depressions appeared along the lip-like structure(Fig.1j). The cell surface was smooth with round pores and depressions (Fig.1k). Round depressions were found especially in the hypotheca and precingular plates (Fig.1c–f & h). The margin in contact to cingular plates was ridged (Fig.1l).
3.2 Morphology of the Hainan strain of C. tropicalis
The cells of theC.tropicalisHainan strain (DS5F4)were spherical (Fig.2a–b) and the epitheca was smaller than the hypotheca (Fig.2a–b). Cell sizes ranged from 30.0 to 42.3 μm (36.0±3.2 μm,n>30) in DV depth, 29.9 to 42.3 μm (36.2±3.4 μm,n>30) in width and 32.2 to 45.5 μm (37.8±3.1 μm,n>30) in AP length (Table 1). The plate formula was Po, 3′, 7′′, 6c,6s, 5′′′, 2′′′′ (Fig.2c–f). The 1′ plate was pentagonal and centrally located in the epitheca (Fig.2c–d). The W/L ratio of the 7′′ plate ranged from 1.8 to 2.8 (Table 1). The cingulum was deep and narrow (Fig.2g–h).The sulcus was short and only five sulcal plates could be observed: S.p., S.d.p. S.d.a., S.s.a., and S.a.(Fig.2g–h). The Po plate was 6.6–9.8 μm (average 8.2 μm) in length (Table 1). The APC was slightly curved with the apical pore and round pores inside(Fig.2i–j). The edge of plate 2′ where contacted the APC was ridged like a collar (Fig.2j). The cell surface was smooth with round pores (Fig.2k–l).
3.3 Morphology of the Hainan strains of C. malayensis
The cells of the HainanC.malayensisstrains (CE3/CS4C6) were spherical (Fig.3a–b) with a DV depth of 17.2–30.0 μm (23.8±2.4 μm,n>30), width of 19.6–29.3 μm (24.1±2.6 μm,n>30), and AP length of 20.9–34.3 μm (26.1±2.8 μm,n>30) (Table 1). The plate formula was Po, 3′, 7′′, 6c, 6s, 5′′′, 2′′′′ (Fig.3c–f). The 1′ plate was oblong and positioned to the left of centre(Fig.3c–d). The W/L ratio of the 7′′ plate ranged from 1.0 to 1.4 (Table 1). The cingulum was wide(Fig.3g–h), while the sulcus was short with six sulcal plates: S.p., S.d.p., S.s.p., S.d.a., S.s.a., and S.a.(Fig.3g–h). The Po plate was 5.3–7.4 μm (average 6.5 μm) in length (Table 1). The APC was almost straight with a narrow apical pore and a line of round pores inside (Fig.3i–j). The cell surface was smooth with round thecal pores (Fig.3k). There was very fine perforation in the thecal pores (Fig.3l).
3.4 Molecular analysis
Based on the phylogenetic analysis, the LSU rDNA sequences were clustered into seven major clades:C.monotis,C.santacroce,C.malayensis,C.palmyrensis,C.guanchica,C.tropicalis, andC.canariensis(Fig.4).Two Hainan strains (CS4C6 and CE3) belonged toC.malayensis, as did other strains from the Pacific(Japan, China, Australia, New Zealand, Korea, and Malaysia) and Atlantic (Virgin Island, USA, Brazil,Puerto Rico, Dominican Republic and Belize) with good support (1.00/99). The Hainan strain DS5F4,one Belizean strain from the West Atlantic and several strains from the Pacific, including Hong Kong of China, Thailand, Australia, and Indonesia formed cladeC.tropicaliswith well support (1.00/99).Hainan strain D1C2 and strains from Hong Kong of China, Korea, Australia, and Spain formed cladeC.canariensiswith high support (1.00/99).
The phylogenetic analysis of ITS regions (Fig.5)led to the categorisation of five clades:C.monotis,C.malayensis,C.tropicalis,C.guanchicaandC.canariensis. The strain DS5F4 ofC.tropicalisfrom Hainan Island was included in the same clade as the strains from Belize, Puerto Rico, and Brazil with good support (1.00/99). TheC.malayensisclade was formed with strains from Malaysia, Florida, Puerto Rico, New Zealand, Japan, Brazil, and China,including the strains CE3 and CS4C6 from Hainan Island (0.98/87). TheC.canariensisclade comprised strains from Hainan Island, Spain, and Korea with full support.
3.5 Genetic distances
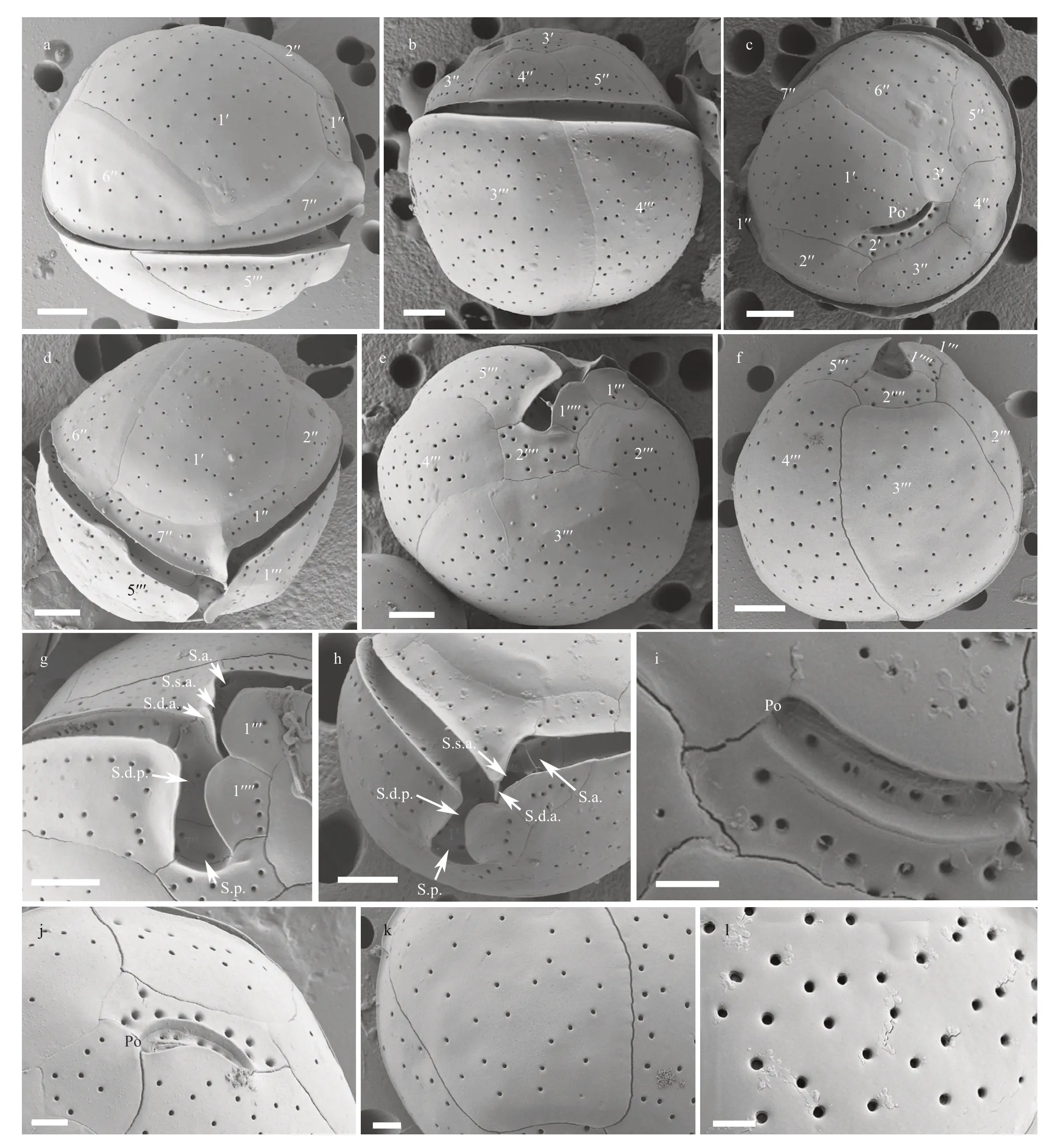
Fig.2 SEM micrographs of C. tropicalis
The highest estimated genetic distance (p-distances)values of 0.420 4–0.436 5 between the LSU rDNA sequences for selected species ofCooliawere observed betweenC.canariensisandC.tropicalis, followed by values betweenC.tropicalisandC.monotisat 0.412 6–0.418 5, and betweenC.tropicalisandC.palmyrensisat 0.392 6–0.412 0 (Supplementary Table S1). The lowest value of 0.103 0–0.132 9 was observed betweenC.monotisandC.santacroce. An estimated value of evolutionary divergence of approximately 0.4 was found betweenC.tropicalisandC.malayensis.The values observed betweenC.canariensisandC.tropicalis,C.monotis,C.santacroceandC.palmyrensisvaried from 0.35 to 0.43. Values betweenC.guanchicaandC.canariensisvaried from 0.234 8 to 0.243 0. The values among diff erent strains within species were 0–0.025 5, 0–0.013 4, 0–0.015 9,0–0.005 8, 0.041 6, 0.024 8– 0.104 7 and 0.006 5 forC.malayensis,C.canariensis,C.tropicalis,C.monotis,C.santacroce,C.palmyrensisandC.guanchica, respectively. No diff erences in the LSU rDNA sequences were found between the HainanC.malayensisstrains and strains from New Zealand,Florida of USA, Malaysia, Japan, Dominican Republic and Guangxi and Hong Kong of China. ForC.canariensisandC.tropicalis, no diff erences in the LSU rDNA sequences were found between the Hainan strains (D1C2 and DS5F4, respectively) and Australian strains (NQAIF252 and NQAIF90, respectively).
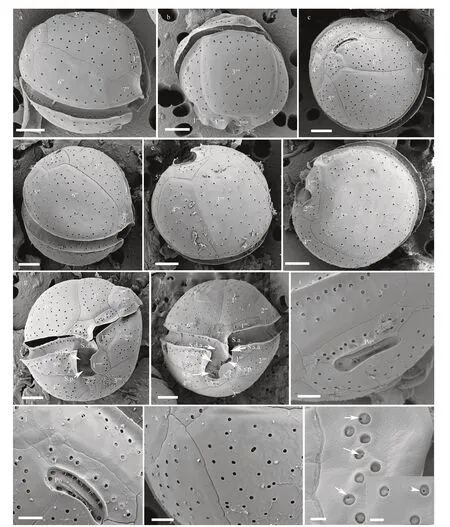
Fig.3 SEM micrographs of C. malayensis
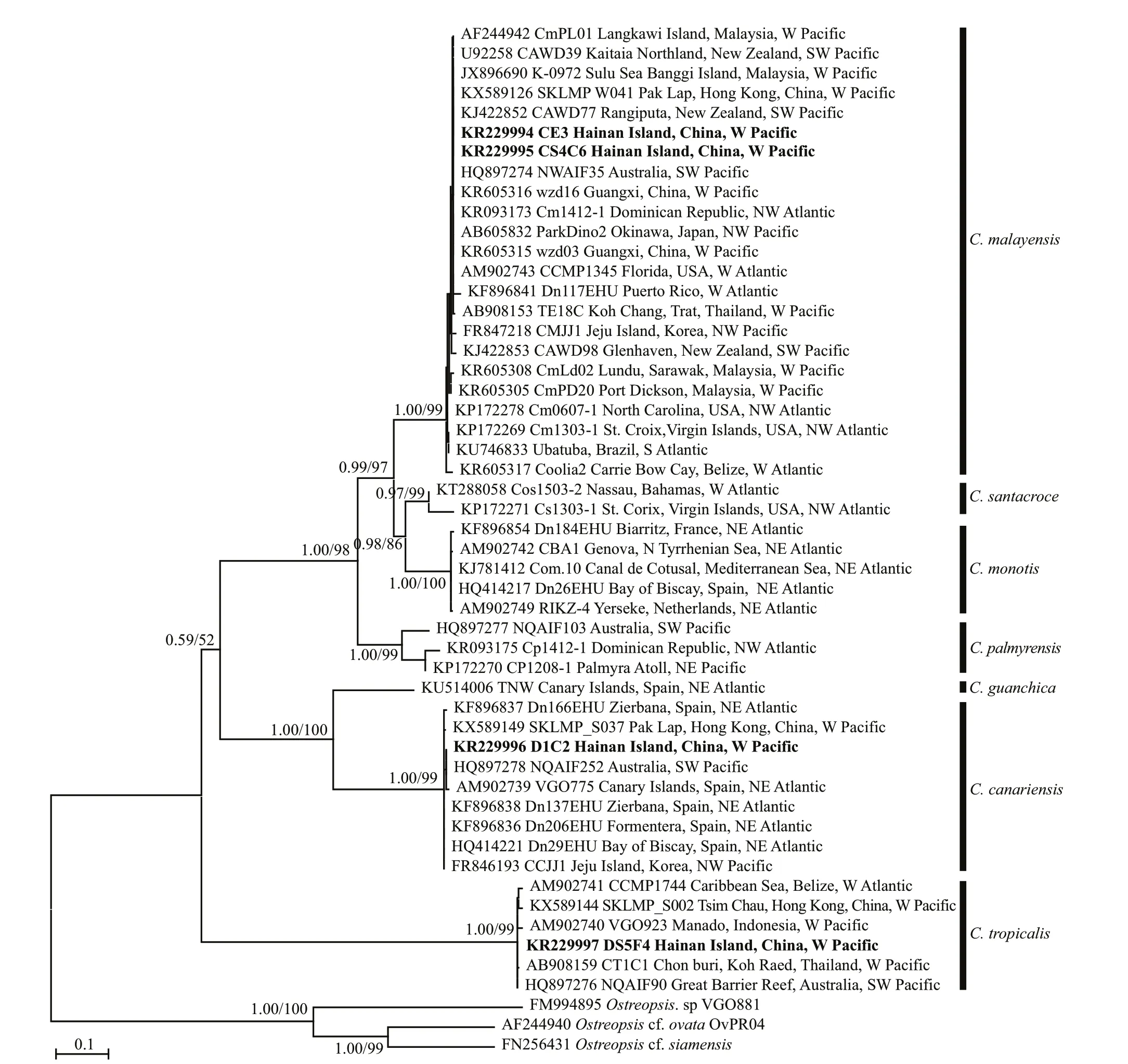
Fig.4 Phylogenetic analysis of Coolia species inferred from D1/D2 LSU rDNA sequences using Bayesian inference (BI) and Maximum-likelihood (ML)
For the ITS region, genetic distance betweenCooliaspecies ranged from 0.153 8 to 0.634 1. The lowest and highest values was observed inC.monotisvs.C.malayensisandC.canariensisvs.C.tropicalis(Supplementary Table S2). A value of 0.41–0.56 was found betweenC.malayensisandC.tropicalis/C.canariensis/C.guanchica, as well as that ofC.monotis. The values betweenC.guanchicaand the other species was 0.4–0.6. The values among strains within species were 0.161 6–0.320 3, 0–0.018 5, and 0–0.029 8 forC.canariensis,C.malayensis, andC.tropicalis, respectively. Based on ITS region sequences, the values between the Hainan strains and others were 0.253 9–0.320 3, 0.002 6–0.013 2, and 0.008 1–0.029 8 forC.canariensis,C.malayensis,andC.tropicalis.
4 DISCUSSION
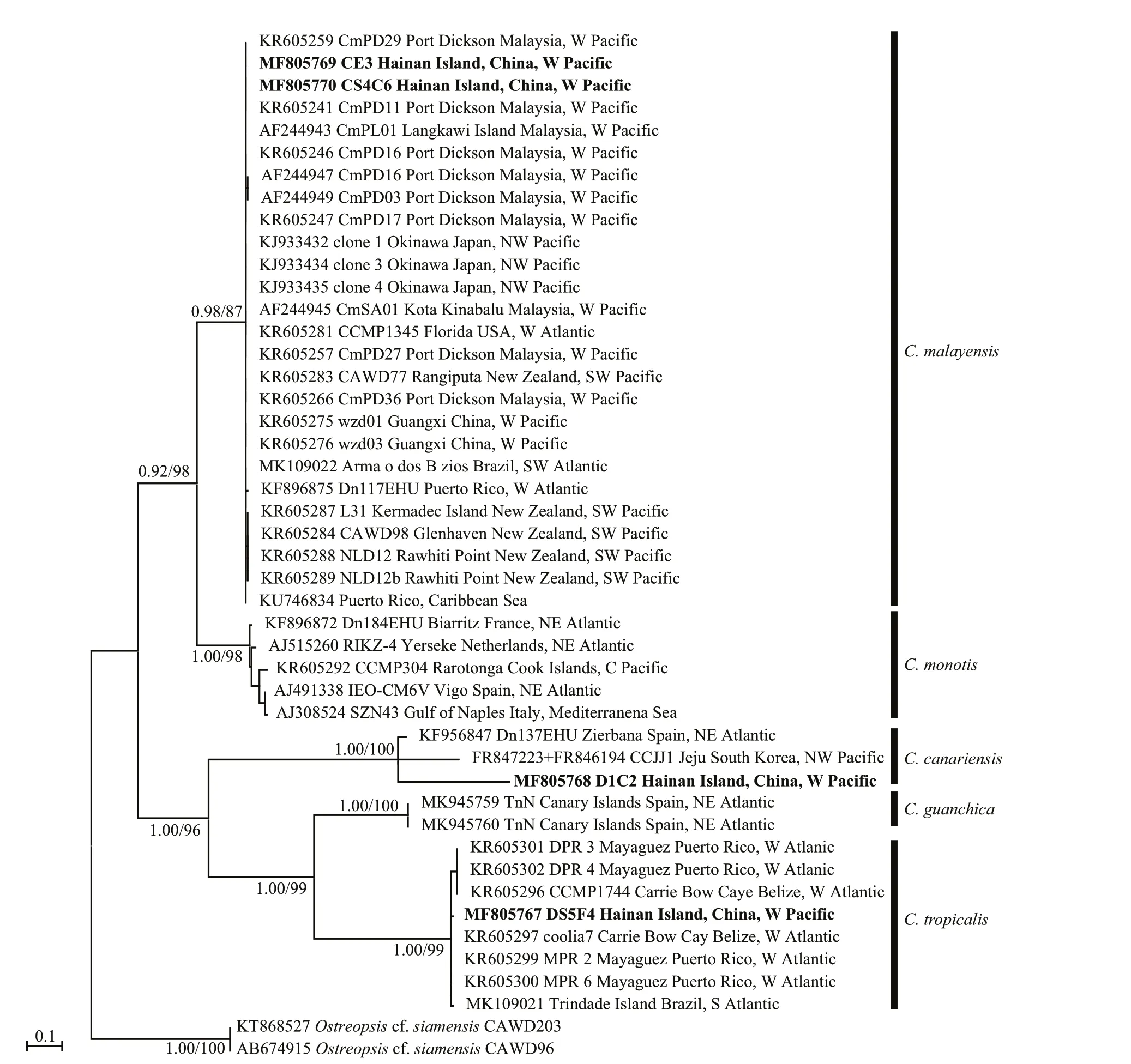
Fig.5 Phylogenetic analysis inferred from ITS region (ITS1-5.8S-ITS2) sequences using Bayesian inference (BI) and Maximum-likelihood (ML)
The overall shape of theC.canariensisHainan strain was similar to that of the original description(Fraga et al., 2008). The AP length (31.5±2.8 μm) and width (29.6±3.7 μm) of theC.canariensisHainan strain were similar to those of the Canary Islands strains at 32.7 μm and 32.8 μm (Fraga et al., 2008).There was considerable cell variation in DV dimensions among strains. TheC.canariensisKorean strain had a smaller cell size, both in DV depth and width (28.3 μm and 27.1 μm, respectively) (Jeong et al., 2012), than those of the Biscayan strains (35.2 μm and 35.4 μm) (Laza-Martinez et al., 2011), and the Hainan strain had a moderate cell size in terms of the DV length and width among the three strains.
The cell shape of theC.tropicalisHainan strain generally fit the original description from Belize(Faust, 1995). The cell size of theC.tropicalisHainan strain, at 32.2–45.5 μm in AP length and 30.8–44.8 μm in AP width, was slightly larger than that of the original description, in which Faust (1995) reported that theC.tropicaliscells varied from 23 to 40 μm in length and 25 to 39 μm in width. The cell size ofC.tropicalisstrains collected from four sites by Mohammad-Noor et al. (2013) were 35–47 μm in length and 30–45 μm in width, larger than originally described by Faust (1995). Cell size ofC.tropicalisBrazil strain UNR-28 was very similar to the Hainan strain (Table 1) (Nascimento et al., 2019).
The cell shapes of the HainanC.malayensisstrains were similar to those of the Malaysian strains described by Leaw et al. (2010). The ranges of AP length and width (respectively 20.9–34.3 μm and 18.0–32.3 μm) of the HainanC.malayensisstrains were broader than those of the Malaysian strains (28–33 μm and 27–32.3 μm; Leaw et al., 2010) and Thailand strains (22–28 μm and 23–25 μm; Tawong et al., 2015), but similar to those of the Korean strain(20–34 μm and 18–30 μm; Jeong et al., 2012).However,C.malayensisfrom Okinawa had a slightly greater width of 22–33 μm compared with a length of 20–32 μm (Wakeman et al., 2015).
Cells ofC.monotis,C.canariensis,C.tropicalis,C.areolata, andC.malayensiswere very similar in cell shape, size, and plate arrangement (Meunier,1919; Faust, 1995; Ten-Hage et al., 2000; Fraga et al.,2008; Leaw et al., 2010). However, these species could be distinguished by several morphological features (Ho and Nguyen, 2014). All of these species had a smooth thecal surface (Faust, 1995; Leaw et al.,2010; Laza-Martinez et al., 2011; Karafas et al., 2015)exceptC.canariensisandC.areolata(Ten-Hage et al., 2000; Fraga et al., 2008). There were numerous round depressions in the hypotheca and precingular plates ofC.canariensis(Fraga et al., 2008; Jeong et al., 2012) and areolates were distributed in all plate surfaces except the 1′ plate ofC.areolata(Ten-Hage et al., 2000; Fraga et al., 2008). Although theC.canariensisKorean strains were found to have round non-perforated dents in the hypotheca,cingulum, and bottom of the precingular plates (Jeong et al., 2012), theC.canariensisstrains from Hainan had round depressions in the hypotheca and precingular plates. Therefore, the Hainan strains ofC.canariensissupported the diff erence betweenC.canariensisandC.areolata. Plate 1′ was the largest of the epitheca inC.tropicalis,C.canariensis,C.areolata, andC.guanchica(Fraga et al., 2008;Jeong et al., 2012; David et al., 2014, 2020), while plate 6′′ was the largest for the otherCooliaspecies(Ho and Nguyen, 2014; Karafas et al., 2015; Leaw et al., 2016; Leung et al., 2017). In addition, plate 1′ was oblong and narrow inC.monotisandC.malayensisbut pentagonal inC.tropicalis(Mohammad-Noor et al., 2013; Momigliano et al., 2013; Ho and Nguyen,2014), and elongated inC.palmyrensisandC.santacroce(Karafas et al., 2015).C.canariensis,C.areolataandC.guanchicahad a hexagonal 1′ plate(Ten-Hage et al., 2000; Fraga et al., 2008; David et al., 2019). Plate 1′ was located in the centre of the epitheca inC.tropicalis(Faust, 1995; Mohammad-Noor et al., 2013; Momigliano et al., 2013),C.canariensis(Fraga et al., 2008), andC.areolata(Ten-Hage et al., 2000) andC.guanchica(David et al., 2019), but was situated to the left of the epitheca inC.monotis,C.malayensis,C.palmyrensis, andC.santacroce(Leaw et al., 2010, 2016; Momigliano et al., 2013; David et al., 2014; Karafas et al., 2015)
The W/L ratio of plate 7′′ has been suggested as way to distinguishCooliaspecies (Leaw et al., 2010;Jeong et al., 2012; Ho and Nguyen, 2014). The following W/L ratios were observed: 1 forC.monotis(Fraga et al., 2008), 1.2 to 1.5 forC.malayensis(Leaw et al., 2010), 2 forC.areolataandC.canariensis, and 4 forC.tropicalis(Fraga et al.,2008). However, considerable variation in the W/L ratio of plate 7′′ was observed among strains from diff erent geographic regions, ranging from 1.1 to 1.5 forC.monotis, 0.8 to 2.1 forC.malayensis, and 1.7–2.2, 1.5–3.4, and 1.2–3.5 forC.canariensisfrom Biscay (Laza-Martinez et al., 2011), Jeju Island(Jeong et al., 2012) and Brazil (Nascimento et al.2019), respectively. The W/L ratio of plate 7′′ was similar (W/L ratio ~1) amongC.monotiscomplex(Karafas et al., 2015; Wakeman et al., 2015). However,it is still useful in diff erentiating species inC.monotiscomplex from species ofC.tropicalisorC.canariensis(David et al., 2019).
Perforations within the large pores are commonly reported inCoolia, as we found in the HainanC.malayensisstrains. Jeong et al. (2012) observed perforations within the large pores in the Korean strains ofC.canariensisandC.malayensis. However,this structure was not found in the HainanC.canariensisstrains. Additionally, Laza-Martinez et al. (2011) and Leaw et al. (2010) found perforations within the large pores inC.monotisandC.malayensis,respectively. Such structure could also be observed inC.guanchica(David et al., 2019) andC.palmyrensis(Karafas et al., 2015). Therefore, these perforations may be common features and, as such, would not be reliable for species identification.
Resolving the relationships amongCooliaspecies requires the analysis of molecular data. The LSU rDNA phylogenies supportC.monotis,C.malayensis,C.tropicalis,C.canariensis,C.santacroce, andC.palmyrensisas monophyletic lineages consistent with distinct species (Fraga et al., 2008; Leaw et al.,2010; Mohammad-Noor et al., 2013; Momigliano et al., 2013; Rhodes et al., 2014; Karafas et al., 2015;Leaw et al., 2016). Additionally, Molecular data in the previously literatures displayed the same branching patterns and main clades forCooliaspecies withC.monotis,C.malayensis, andC.santacroceas closely related species andC.palmyrensisas a basal lineage to those species (Leaw et al., 2010; Rhodes et al.,2014; Karafas et al., 2015; Wakeman et al., 2015).
Mohammad-Noor et al. (2013) reported thatC.canariensiscould be clustered into two clades in the LSU rDNA phylogeny, despite the strains originating from the same area. TwoC.canariensisclades also formed based on 28S LSU rDNA sequences of 44Cooliataxa (Wakeman et al., 2015).Nascimento et al. (2019) found that both morphological and phylogenetic data supported cryptic species existence inC.canariensiscomplex. The recently reportedC.guanchicaformed a well-supported sister clade withC.canariensisbased on phylogenetic analyses and might be included in theC.canariensiscomplex (David et al., 2019). Nascimento et al. (2019)also suggested that additional molecular and morphology data of strains from diff erent regions should be acquired to strength our knowledge ofC.canariensiscomplex.
Estimates of the evolutionary divergence amongCooliadepended on sequences analyzed by previous authors (Leaw et al., 2010; Momigliano et al., 2013;Karafas et al., 2015). Moderate values of 16.1%–16.5% betweenC.malayensisand otherCooliaspecies were observed by Wakeman et al. (2015),while a value of 3.42%–32.88% was observed in this work. Karafas et al. (2015) reported the smallest intraspecific distance of D1/D2 LSU rDNA to betweenC.monotisandC.santacroce. However, we found the lowest divergence of both LSU rDNA and ITS region was betweenC.monotisandC.malayensis. This agree with Leaw et al. (2010). Compensatory base changes between ITS2 secondary structures was considered as species level divergency though lowest divergence distances betweenC.monotisandC.malayensiswithin the genus (Leaw et al., 2010).Although Litaker et al. (2007) proposed genetic distances of 0–2.1% within dinoflagellate species; we suggest that the genetic divergence might be related to the species/isolates selected for analysis.
Cooliaspecies are highly morphologically similar and exhibit high phylogenetic diversity. Further studies are needed to clarify the taxonomic ambiguity ofCoolia. Moreover,C.tropicalisproduces cooliatoxin (Holmes et al., 1995; Mohammad-Noor et al., 2013) andC.malayensisis also toxic (Mohammad-Noor et al., 2013; Rhodes et al., 2014). Consequently,toxicity and species distributions ofCooliaare important for understanding the structure of the marine food web in Chinese coastal waters.
5 CONCLUSION
Marine benthic dinoflagellates in the genusCooliaare reported in both tropical and temperate regions.ThreeCooliaspecies, includingCooliacanariensis,C.tropicalis, andC.malayensiswere observed in the coastal waters of Hainan Island. In phylogenetic analyses based on the LSU rDNA and ITS regions(ITS1-5.8S-ITS2), Hainan strains ofC.canariensis,C.tropicalis, andC.malayensisclustered within the clades of these species with other isolates from diff erent areas. Our study reveals the morphological and genetic diversity ofCooliaspecies from Hainan Island, South China Sea, which provides a detailed understanding ofCooliaspecies of this area.
6 DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author.
7 ACKNOWLEDGMENT
We thank Dr. Hualong WANG for his help with sampling.
 Journal of Oceanology and Limnology2021年3期
Journal of Oceanology and Limnology2021年3期
- Journal of Oceanology and Limnology的其它文章
- Steady increase in water clarity in Jiaozhou Bay in the Yellow Sea from 2000 to 2018: Observations from MODIS*
- Phylogenetic diversity and bioactivity of culturable deepsea-derived fungi from Okinawa Trough*
- Allelopathic eff ects of mixotrophic dinoflagellate Akashiwo sanguinea on co-occurring phytoplankton: the significance of nutritional ecology*
- Investigation of the decline of Ulva prolifera in the Subei Shoal and Qingdao based on physiological changes*
- Effi ciency of phosphorus accumulation by plankton,periphyton developed on submerged artificial substrata and metaphyton: in-situ observation in two shallow ponds*
- Petroleum exploitation enriches the sulfonamide resistance gene sul2 in off shore sediments
