Open reading frame 3 protein of hepatitis E virus: Multi-function protein with endless potentia
Yong-Lin Yang, Yu-Chen Nan
Abstract Hepatitis E virus (HEV), a fecal-orally transmitted foodborne viral pathogen,causes acute hepatitis in humans and is responsible for hepatitis E outbreaks worldwide. Since the identification of HEV as a zoonotic agent, this virus has been isolated from a variety of hosts with an ever-expanding host range. HEV-open reading frame (ORF) 3 , the smallest ORF in HEV genomes, initially had been perceived as an unremarkable HEV accessory protein. However, as novel HEVORF3 function has been discovered that is related to the existence of a putative third virion structural form, referred to as “quasi-enveloped” HEV particles, HEV is challenging the conventional virion structure-based classification scheme,which assigns all viruses to two groups, “enveloped” or “non-enveloped”. In this review, we systematically describe recent progress that has identified multiple pathogenic roles of HEV-ORF3 , including roles in HEV virion release, biogenesis of quasi-enveloped virus, regulation of the host innate immune response, and interference with host signaling pathways. In addition, implications of HEVORF3 -associated quasi-enveloped virions are discussed to guide future development of improved vaccines against zoonotic HEV infection.
Key Words: Hepatitis E virus; Zoonosis; Quasi-enveloped virion; Hepatitis E virus-open reading frame 3 ; Innate immunity
INTRODUCTION
Hepatitis E virus (HEV), a quasi-enveloped, single-stranded positive-sense RNA virus,is classified as a member of the family Hepeviridae[1 ]. Hepeviridae is a highly diverse family that contains several HEV and HEV-like virus species with zoonotic, anthropotropic, and animal-restricted tropisms[2 ]. Currently, nearly 3 million symptomatic cases of HEV infection are reported annually, resulting in approximately 70000 deaths and 3000 stillbirths in each year[3 ]. Generally, mortality of HEV ranges from 0 .5 % to 3 % overall, but HEV mortality rates have approached 30 % in pregnant women[4 ,5 ].
The viral genome of HEV is 7 .2 kb in length and is an mRNA-like molecule (capped and poly-adenylated at 5 ' and 3 ' ends, respectively)[6 ]. To date, three well-defined open reading frames (ORFs) have been detected in all HEV genotypes studied(Figure 1 )[7 ,8 ]. HEV-ORF1 protein is translated directly from HEV genome with HEVORF2 and-ORF3 proteins translated from subgenomic RNAs[9 ]. Moreover, ORF4 ,whose expression is promoted by an atypical internal ribosome entry site (IRES)-like element, completely overlaps with ORF1 and was identified recently only in HEV-1 isolates[10 ]. In addition to ORFs, HEV genome contains at least four cis-reactive elements (CREs) that are required for viral replicationin vivo[11 -13 ]. Two of these CREs, which are located within intergenic-junctional regions between HEV-ORFs,form “stem-loop” structures that act as promoter-like elements for initiation of subgenomic RNA synthesis[11 ,12 ]. Conversely, another two CREs within ORF1 and ORF2 , function as a scaffold that generates specific signals that trigger recruitment of viral and host factors for a replication complex[13 ].
The latest classification system of HEVs includes two genera within the family Hepeviridae,Orthohepevirus(covering all HEV isolates with mammalian and avian origin) andPiscihepevirus(only HEV-like virus with cutthroat trout origin). All four previously characterized HEV genotypes (1 -4 ) that cause human infection are categorized within the speciesOrthohepevirusA[1 ], withOrthohepevirusB, C, and D species encompassing HEV isolates found in other non-human hosts[1 ]. WithinOrthohepevirusA, HEV-1 and HEV-2 isolates are anthropotropic viruses without any animal reservoirs, while HEV-3 and HEV-4 isolates are zoonotic[6 ,14 ]. The HEV isolates originally identified from Japanese wild boar, containing unique RNA sequences, are categorized into HEV-5 and 6 , whereas camel HEV isolates belong to HEV-7 and HEV-8 genotypes[1 ,15 ]. Notably, HEV isolated from a human liver transplant patient has been reported to most closely match camel HEV, suggesting that camel HEVs may be zoonotic as well, although this concept requires further confirmation[16 ]. In addition to mammalian HEVs, two unique groups of HEV-like viruses that have been isolated from avian species and cutthroat trout (Oncorhynchus clarkia) have been assigned to speciesOrthohepevirusB and to genusPiscihepevirus(as the only member), respectively.

Figure 1 Schematic illustration of hepatitis E virus genome and three well-defined open reading frames. The numbers above or below the RNA boxes indicate nucleotide numbers based on hepatitis E virus-1 prototype Sar55 strain (GenBank accession number AF444002 ). NCR: Non-coding region; ORF:Open reading frame.
Initially, HEV was assumed to be solely restricted to humans, in whom it induced self-limiting hepatitis symptoms. However, the emergence of HEV and HEV-like isolates in swine and other animal species supports a much wider HEV host tropism,with some HEV genotypes identified as zoonotic pathogens[17 ]. Currently, hepatitis E cases have been frequently reported in both developing and developed countries and have occurred in step with increasingly more frequent observations of expanding host ranges[18 -22 ]. Generally, inter-species transmission and infection of zoonotic type of virus from animal to humans is considered to be primary routing of HEV transmission within Western worlds, while fecal-oral transmission appears to be the predominant route of HEV transmission within developing countries[23 ,24 ]. Notably, immune serological surveillance data support a high prevalence rate of previous HEV infection in the general population but demonstrate a declining trend in recent years that may be due to undetected endemic HEV circulation[25 ,26 ]. Meanwhile, frequent detection of recent cases of chronic HEV, HEV-related acute hepatic failure, and extrahepatic HEV manifestations supports this speculation as well[25 ,27 -30 ]. Moreover, these observations imply that zoonotic HEV infection is a complicated pathogenic process underlying various forms of HEV-related disease. However, our understanding of HEV remains restricted due to the lack of a robustin vitroHEV cell system.Nevertheless, the HEV genome is known to contain three well-defined ORFs that have been found in all HEV genotypes[7 ,8 ]. HEV-ORF3 , the smallest ORF found in HEV genomes, encodes a unique protein with multiple indispensable functional roles associated with viral replication and pathogenesis. In this review, we discuss the recent progress toward understanding HEV-ORF3 pathological roles in detail and provide new insights.
PROTEINS ENCODED BY HEV
HEV-ORF1 polyprotein as viral replicase
The HEV-ORF1 protein is the largest protein encoded by the HEV genome and can be directly translated from the mRNA-like genome of HEV[8 ]. Bioinformatics-based protein homology analysis indicated that at least eight function domains are present within HEV-ORF1 according to similarities to counterparts from other RNA viruses(Figure 2 )[31 ]. These protein domains include methyltransferase domain, the Y domain, papain-like cysteine protease (PCP) domain, a hypervariable region containing previously assigned hypervariable domain and proline-rich domain, the X domain (also named macro-domain), RNA helicase domain, and a RNA polymerase domain (RdRp)[2 ,32 ]. The methyltransferase domain and Y domain together are thought to constitute the functional unit of RNA capping enzyme[2 ,32 ]. It remains unknown whether HEV-ORF1 protein acts alone to perform all putative viral replicase functions or is cleaved by host protease or viral protease (viaPCP domains) to generate independent units resembling viral replicases similar to other positive-sense RNA viruses[2 ,33 -35 ]. To date, available data suggests that proteolytic cleavage of the HEVORF1 product involves PCP domain encoded within HEV-ORF1 [2 ,33 -35 ]. Besides viral replication, another notable characteristic of HEV-ORF1 protein is its flexibility for insertions or deletions within the hypervariable region. The proline-rich domain within the hypervariable region was proposed to act as a linking hinge for the upstream PCP domain and downstream macro-domain, leading to formation of an unstable tertiary structure[31 ,36 -38 ]. Conversely, the hypervariable region is considered an intrinsically disordered region containing extensive gene segment insertions or deletions and may participate in viral replication[39 -42 ]. As a unique example, one HEV-3 isolate, Kernow-C1 p6 strain, which was originally isolated from an HIV patient chronically infected by HEV as well, contains a 174 -nt insertion of a human ribosomal protein S17 sequence originating from the host[43 ], which confers ORF1 protein with novel nuclear/nucleolar trafficking capability and may promote viral replicationin vitro[44 ,45 ].
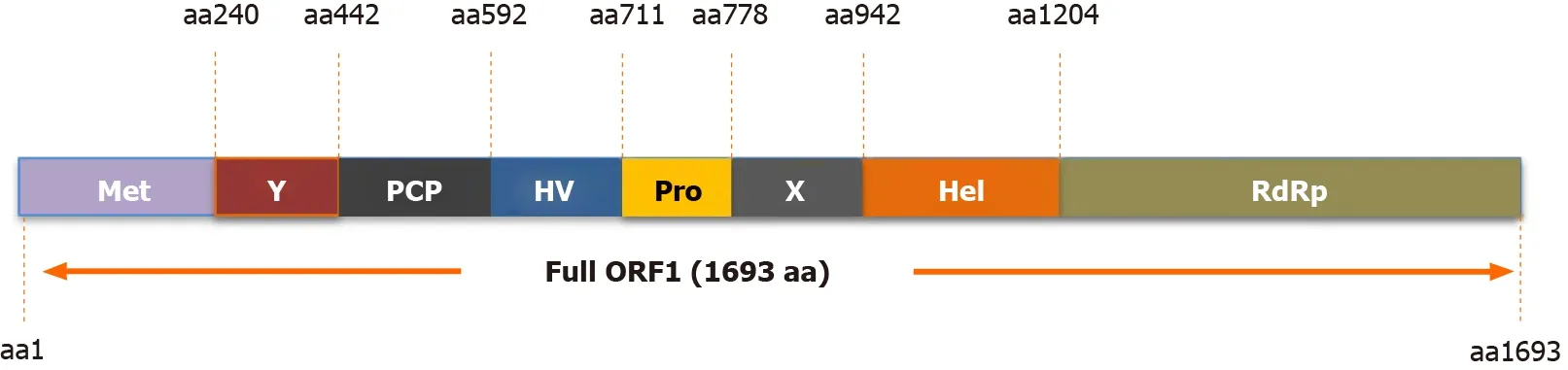
Figure 2 Function domains of hepatitis E virus-open reading frame 1 polyprotein. Putative functional domains of hepatitis E virus (HEV)-open reading frame 1 polyprotein based on HEV-1 prototype Sar55 strain are listed as follows: Methyltransferase domain; Y domain; papain-like cysteine protease;hypervariable region; proline-rich domain; X-domain; Hhelicase; RNA-dependent RNA polymerase. Met: Methyltransferase domain; Y: Y domain; PCP: Papain-like cysteine protease; HV: Hypervariable region; Pro: Proline-rich domain; X: X-domain; Hel: Helicase; RdRp: RNA polymerase; ORF: Open reading frame.
HEV-ORF2 encodes viral capsids
HEV-ORF2 encodes the putative capsid protein of HEV virions with a full-size of 660 aa residues, and has a predicted molecular mass of 72 kDa[46 ]. Notable, the full length ORF2 protein carries N-linked glycans at three putative glycosylation Asn residues at positions 137 , 310 , and 562 , as well as a 15 aa signal peptide at the N-terminus directing full length ORF2 protein to the endoplasmic reticulum (ER)[47 ]. One notable characteristic of HEV-ORF2 is the existence of various forms of ORF2 -derived proteins with multiple functions. Researchers observed very early that multiple processed ORF2 -derived products were detected when recombinant ORF2 was expressed in different systems. The mature HEV capsid protein is generated from the full-length ORF2 precursor via proteolytic processing to remove the first N-terminal 111 aa and the last C-terminal 52 aa. However, at least two other forms of HEV-ORF2 have been detected in HEV infected cells and patients[48 ,49 ]. The first was a secreted form of ORF2 protein (ORF2 s), which utilized an upstream start codon and contained a signal peptide that earmarked ORF2 for subsequent glycosylation and secretion[50 ]; the second was a capsid-associated truncated form of ORF2 (ORF2 c), which was translated beginning at an internal methionine-encoding AUG start codon (aa16 of ORF2 )[50 ].Meanwhile, a truncated form of ORF2 (ORF2 c) detected in HEV-infected cells may be secreted into the extracellular milieu, as is ORF2 s[49 ].
The mature HEV capsid that lacks N-terminal 111 aa and the last C-terminal 52 aa of full ORF2 can form virus-like particles when expressed in insect cells[51 ,52 ]. Genetic analysis of ORF2 sequences of HEV genotypes 1 -4 suggests that these proteins share greater than 85 % similarity overall, with divergence mainly observed within the first N-terminal 111 aa, which are not incorporated into final virions[53 ]. A recombinant subunit vaccine using truncated HEV-1 ORF2 protein (HEV239 ) as major immunogen is licensed in China (Hecolin®)[2 ].
A multi-functional protein encoded by HEV-ORF3
ORF3 , the smallest ORF among all HEV ORFs, partially overlaps with the N-terminus of ORF2 for about 300 nt and is translated from a different reading frame[9 ]. It was initially proposed that ORF3 protein contains 123 aa that are encoded by a subgenomic RNA distinct from the RNA encoding ORF2 [7 ]. However, it was later confirmed that ORF3 protein is translated from a bicistronic subgenomic RNA to generate a 114 -aa protein with a predicted molecular weight of 13 kDa (vp13 ), which is actually 9 aa shorter than the initially predicted length[9 ,54 ]. Basic sequence analysis of HEV-ORF3 protein indicated that there are two hydrophobic domains and two proline-rich domains present within the N-terminal half and C-terminal portion of HEV-ORF3 proteins, respectively[55 ,56 ], of which the first proline-rich domain contains a mitogen-activated protein kinase (MAPK) phosphorylation site (Ser71 )[57 ].Furthermore, two presenilin-associated protein (PSAP) motifs within the ORF3 protein were identified in HEV-1 prototype strain Sar55 , with the first PSAP motif comprised of aa 86 -89 and the second comprised of aa 95 -98 [58 ]. Although ORF3 protein is not required for viral RNA replicationin vitro[59 ], it is irreplaceable forin vivoHEV infection and required for viral particle releasing[54 ,60 ,61 ]. In fact, most studies demonstrated that HEV-ORF3 is indispensable for viral particle egress and biogenesis of lipid membrane-wrapped HEV particles, which is now recognized as quasienveloped particles. Moreover, the second PSAP motif within the HEV-ORF3 protein has been shown to be required for the formation of membrane-associated HEV particles, a process that relies on an association of ORF3 with lipids[58 ,62 ]. Currently,HEV-ORF3 is thought to form an ion channel that shares key structural features with class I viroporins that are required for virion particle release from cells during infection[63 ]. This observation aligns with the reported putative role of pORF3 [61 ] and with other evidence indicating that HEV-ORF3 protein interacts with tumor susceptibility gene 101 (TSG101 ), the key component of host endosomal sorting complex required for transport (ESCRT) pathway, which is mainly employed by enveloped virus for budding and acquiring of viral envelope. Formation of ESCRT complex has been shown to lead to biogenesis of quasi-enveloped HEV particles[64 -67 ].
HEV-1 specific ORF4
In recent years, a hidden ORF4 from HEV-1 was characterized[10 ]. Translation of HEV-ORF4 protein is promoted by a novel element located in HEV-ORF1 appearing to be an atypical IRES sequence and works in combination with a suboptimal Kozak sequence[10 ]. The exact function of HEV-1 specific pORF4 is still unclear. It was demonstrated that pORF4 stimulates ER stress upon HEV replication[10 ]. It also interacts with multiple ORF1 domainsin vitrowhich are presumably to form a complex further enhancing RdRp activity [10 ]. Furthermore, HEV-pORF4 specific antibodies are detectable in HEV-infected patients[10 ]. Nonetheless, additional investigations are needed to understand functions of the ORF4 product that are unique to genotype 1 HEVs.
REGULATION OF HOST INNATE IMMUNITY AND SIGNALING BY HEVORF3 PROTEIN
Initially, HEV-ORF3 protein did not receive much attention, due to its presumed role as an accessory protein involved in regulation of host signaling to promote HEV replication and invasion. This hypothesis was partially evidenced by the fact that ORF3 protein was dispensable for in vitro replication of HEV-RNA[59 ]. However,subsequent research studies demonstrated that HEV-ORF3 -associated putative interference mechanisms acted on multiple host cell signaling pathways, such as those involved in host innate immunity[2 ,68 ,69 ], indicating that HEV-ORF3 activities ultimately promote viral replication and pathogenesis.
As a multifunctional protein, HEV-ORF3 has been demonstrated to play both positive and negative roles in interferon (IFN) induction. In our previous research, we found that HEV-ORF3 protein could enhance retinoic acid-inducible gene I (RIG-I)activation to subsequently enhance IFN induction[68 ]. More specifically, HEV-1 ORF3 extended protein half-life of RIG-I and interacted with the RIG-I N-terminal portion to enhance ubiquitination-mediated RIG-I activation triggered by addition of the dsRNA analog poly (I:C)[68 ] (Figure 3 A). Interestingly, it is notable that genotypic differences in HEV-ORF3 -associated enhancement of RIG-I-mediated IFN induction were observed. For example, ORF3 proteins from the HEV-1 Sar55 strain and HEV-3 kernowC1 p6 strain could enhance RIG-I activation, while HEV-2 and HEV-4 ORF3 proteins could not[68 ], suggesting that HEV-ORF3 participated in genotype-specific HEV virulence and pathogenic effects. Moreover, these results also aligned with results of a more recent report demonstrating HEV-ORF3 -associated increases of IFNα/β and interferon-stimulated gene 15 levels in hepatoma cell line HepG2 /C3 A[70 ].Conversely, other reports have demonstrated that overexpression of HEV-ORF3 downregulated Toll-like receptor (TLR) 3 and TLR7 and their downstream signaling pathways[71 ,72 ] (Figure 3 B). Meanwhile, another study has demonstrated that ORF3 protein from an HEV-1 strain interacted with signal transducer and activator of transcription (STAT) 1 to block type I IFN-activated pathway[73 ] (Figure 3 C).Furthermore, another study found that ORF3 proteins blocked nuclear translocation of STAT3 to down-regulate STAT3 -dependent gene expression, including expression of acute-phase response proteins[74 ].
Besides the innate immune response, yeast two-hybrid-based screening detected that binding of ORF3 to Pyst1 , a MAPK phosphatase, led to activation of MAPK pathways[75 ]. Thus, ORF3 appears to regulate host gene expression, as MAPK is related to host gene expression and signaling. Meanwhile, HEV-ORF3 may promote expression of glycolytic pathway enzymes by enhancing phosphorylation and transactivation function of p300 /CREB-binding protein as well[76 ]. Additionally,microarray analysis of Huh7 cells has suggested that liver-specific genes may also be modulated by HEV-ORF3 , since it modulated phosphorylation of hepatocyte nuclear factor 4 [77 ]. Moreover, recent research has demonstrated that HEV-ORF3 plays a functional role in virus-cell interactions by influencing expression of integral membrane protein and basement membrane proteins to alter host cell processes associated with apoptosis and lipid metabolism[78 ]. Taken together, these data suggest that HEV-ORF3 modulates multiple signaling pathways, including those involved in host innate immunity, to ultimately promote HEV pathogenesis.
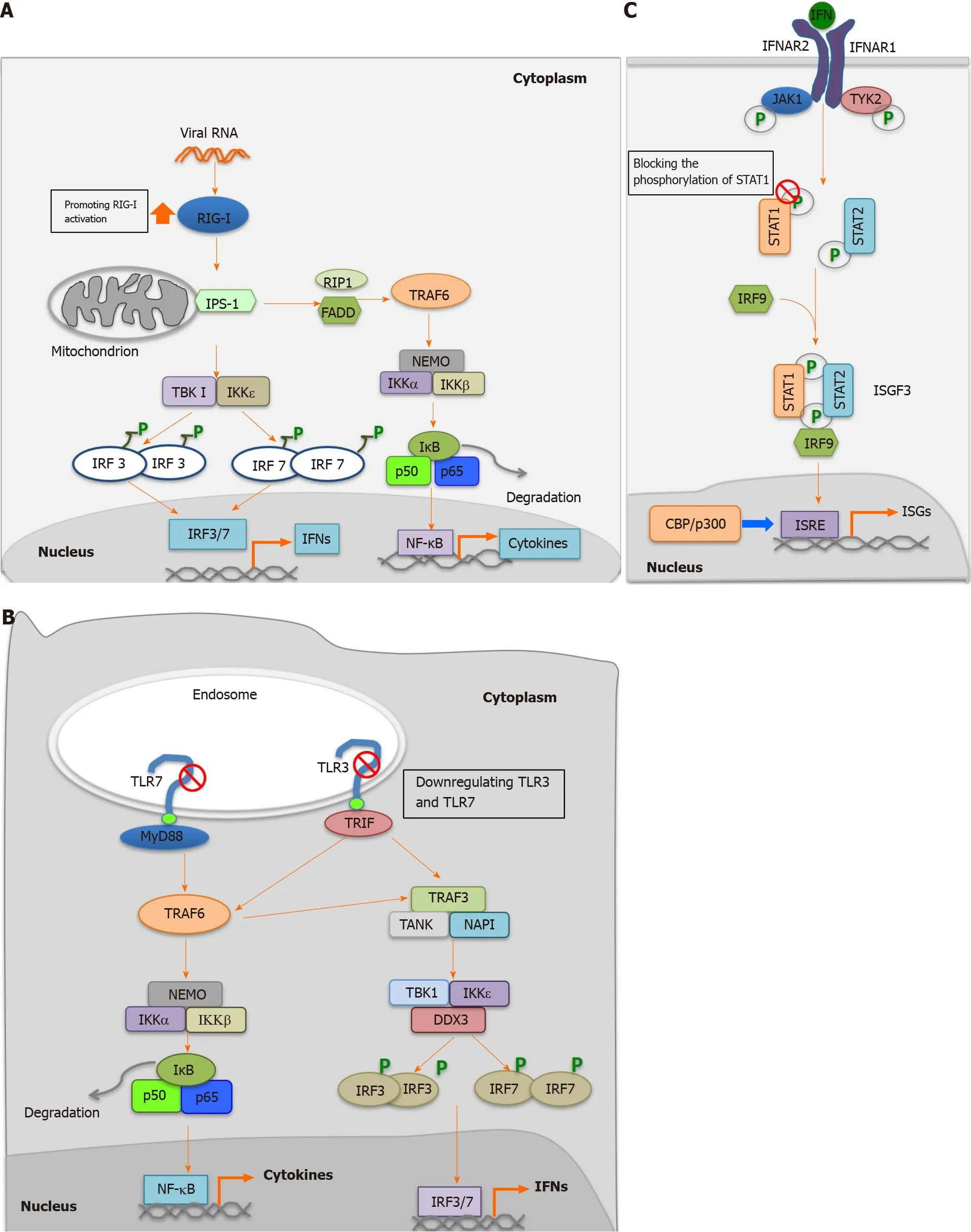
Figure 3 Regulation of host innate immune response by hepatitis E virus-open reading frame 3 proteins. A: Promotion of retinoic acid-inducible gene I mediated activation by hepatitis E virus (HEV)-open reading frame (ORF) 3 ; B: Inhibition of Toll-like receptor (TLR) 3 and TLR7 by HEV-ORF3 ; C: Blocking of the phosphorylation of signal transducer and activator of transcription (STAT) 1 to inhibit Janus kinase/STAT signaling. RIG-I: Retinoic acid-inducible gene I; RIP-I:Ribosome-inactivating proteins type I; FADD: Fas-associated protein with death domain; IKK: IkappaB kinase; TBK: Tank-Binding-Kinase; IRF: Interferon regulatory factor; NF-kB: Nuclear factor kB; IFN: Interferon; TLR: Toll-like receptor; TRIF: Toll-interleukin 1 receptor domain-containing adapter inducing interferon-beta; NAPI:Net anthropogenic phosphorus input; IFNAR: Inflammation-the type I interferon receptor; JAK: Janus kinase; TYK: Targeting tyrosine kinase; STAT: Signal transducer and activator of transcription; ISGF: Interferon-stimulated gene factor; ISRE: Interferon-stimulated response element; ISGs: Interferon-stimulated genes; CBP: CREBbinding protein.
INVOVEMENT OF HEV-ORF3 PROTEIN IN BIOGENESIS OF QUASIENVELOPED HEV PARTICLES
The presence of a lipid layer-based envelope has long been used as the basic criterion for virus classification[66 ]. The presence of a lipid layer can be assessed through treatment of virus preparations with bile salts, a process that abrogates infectivity of enveloped virions, but not non-enveloped virions, by removing their surface lipid layers[66 ]. Generally, it is believed that during the virion budding process a viral envelope is formed from membranes of infected cells that contain molecules of at least one membrane-embedded virus-encoded glycoprotein (presented as peplomers). Viral envelopes interact with corresponding virus receptors located on target cells to promote membrane fusion of cellular membrane and viral envelop after initial interactions of virions and corresponding receptors. Meanwhile, surface glycoproteins located in viral envelope serve as antibody-neutralization targets in most cases[66 ],while the lipid layer of the virus envelope prevents internal virus antigens, such as nucleocapsid proteins, from serving as viral neutralization targets[79 ]. Thus, as compared to a non-enveloped virus, a quasi-enveloped virion would be perceived by the host immune system as antigenically distinct from a naked virion. For example,hepatitis A virus (HAV) was the first non-enveloped virus which is confirmed to hijack host cell membrane similar to enveloped virus as an enveloped form[80 ].Biogenesis of enveloped HAV particles has been shown to depend on the ESCRT system[80 ,81 ], which is involved in budding of enveloped viruses. Membranewrapped or enveloped HAV particles mainly exist in circulation system during acute infection phase of HEV and envelopment confers protection of virus from recognition by neutralizing antibodies, which prevents impairment of virion infectivity[80 ].Similar to HAV, HEV was originally classified solely as a non-enveloped virus before membrane-wrapped HEV particles were discovered, with HEV-ORF3 involvement in biogenesis of quasi-enveloped HEV virions confirmed only very recently.
It was observed very early that HEV-ORF3 protein is not required for viral RNA replicationin vitro[59 ]; however, this protein is irreplaceable for HEV replicationin vivoand is required for viral particle release from HEV infected cellsin vitro[54 ,60 ,61 ].Meanwhile, antibody-capture assays of HEV virions with or without detergent treatment demonstrated that HEV virion from either serum samples of patients or supernatants of HEV-infected cells were associated with lipid layer and ORF3 protein[82 ]. Subsequent screening to detect proteins interacting with HEV-ORF3 protein pinpointed the TSG101 , a component of the ESCRT complex, as the potential interacting partner of HEV-ORF3 [62 ]. Importantly, the ESCRT complex recognizes and earmarks ubiquitinated proteins for subsequent incorporation into multivesicular bodies (MVBs), an essential step for lysosomal degradation[83 ]. The ubiquitin E2 variant domain in TSG101 recognizes and interacts with the P(T/S)AP motif present in target proteins to recruit targets to the endosomal membrane[84 ].
Many enveloped viruses are equipped with a P(T/S)AP motif within their structural proteins that interact with TSG101 to redirect assembled viral components to cell membrane for virion release from infected cells[85 ]. Originally, two PSAP motifs comprised of aa 86 -89 and aa 95 -98 were identified within HEV-ORF3 proteins, the second of which was found to be conserved among HEV genotypes[58 ]. Replacement of the PSAP motif in HEV-ORF3 proteins with heterologous domain motifs (PPPY,YPDL, and PSAA) or mutated PSAP motifs has been shown to affect HEV virion release from infected cells[58 ,86 ], including avian HEV release[87 ]. Additionally, HEVORF3 expressed in cells was found to associate with the cellular cytoskeleton fraction,with deletion of the N-terminal hydrophobic domain of vp13 abolishing this association[57 ]. Moreover, a more recent study demonstrated that green fluorescent protein-tagged ORF3 protein interacted with cellular microtubules and modulated microtubule dynamics[55 ]. This microtubule-like filament of HEV-ORF3 protein indicated that it was potentially involved in a process that promotes virus egress; this process is reminiscent of the process by which the pUL37 protein of herpes virus interacts with dystonin, an important cytoskeleton cross-linker involved in microtubule-based transport of capsids during virion egress[88 ]. Although HEV was originally defined as a non-enveloped virus, such HEV-ORF3 functions that were formerly attributed only to enveloped viruses may now be related to HEV in its recently discovered “quasi-enveloped” form[64 -67 ].
Consistent with the aforementioned roles of HEV-ORF3 in virus egress[61 ], a recent study found that HEV-ORF3 shares key features with class I viroporins, including its function as an ion channel participating in viral particle egress or release[63 ], a function that had been previously demonstrated for the well-characterized viroporin of influenza A virus matrix-2 (M2 ) protein[63 ]. Meanwhile, a putative transmembrane region identified in pORF3 may be involved in ER localization of this protein[63 ].Since viroporins of other viruses, such as M2 of influenza A virus, are components of virions, these observations imply that HEV-ORF3 is a structural HEV virion protein that exists in a membrane-associated state during the formation of envelope structures of quasi-enveloped HEV virions[89 ]. Interestingly, palmitoylation of cysteine residues within the N-terminal region of HEV-ORF3 has been shown to participate in its association with the membrane and is also required for infectious particle secretion[90 ].
The unique role that HEV-ORF3 plays during biogenesis of quasi-enveloped virus particles makes it a novel target candidate for antiviral drug development. In fact, one study has shown that the addition of a cyclic peptide inhibitor (CPI) to HEV-infected cells interrupted the interaction between the HEV-ORF3 PSAP motif and TSG101 [85 ]and reduced virion release by over 90 % when a 50 % inhibitory concentration of CPI of 2 μM was used. Thus, HEV-OR3 has potential as a novel candidate for further development as an anti-HEV drug.
POTENTIAL ROLES OF HEV-ORF3 PROTEIN IN HEV HOST TROPSIM
Since the isolation of zoonotic HEV strains from swine HEV and discovery of other HEV-like viral isolates, HEVs have been continually identified from various mammalian hosts. Based on their ability to cause inter-species infection, HEV isolates can be divided three distinct groups: HEV-1 and 2 are only restricted in human; HEV-3 , 4 , and 7 /8 are zoonotic types; whileOrthohepevirusC is animal-restricted type. Based on reports in the literature, it implies that either factors or viral determinants contribute to HEV host tropisms and cross-species transmission events.
Among all ORFs, HEV-ORF1 encodes the largest HEV protein and appears to be indispensable for determining HEV host range. Anin vitrostudy demonstrated that swapping of genetic fragment among HEV-1 and HEV-4 infectious clones indicated that chimeric virus formed from an HEV-1 infectious clone bearing surface HEV-4 ORF1 could replicate in porcine kidney cells, while the original HEV-1 virus could not[91 ]. Meanwhile, chimeric virus containing the junction region between ORF1 /2 and the 3 ' non-coding region (NCR) of HEV-3 or the 3 ' end of the HEV-1 backbone failed to infect piglets, suggesting that the 5 ′ NCR and ORF1 are involved in HEV cross-species infection[92 ]. By contrast, a recent report demonstrated,viagenetic fragment swapping of ORF1 regions between HEV-1 and HEV-3 infectious clones, that recombinant chimeric viruses could be generatedin vitro. In any case, these chimeric viruses could not infect pigletsin vivo[93 ], suggesting that ORF1 is not the only determinant that can confer cross-species infectivity of HEVin vivo.
As the viral capsid protein, HEV-ORF2 was initially thought to be an unlikely determinant of host tropism, since it is conserved among all major genotypes infecting humans[94 ]. However, results ofin vivoreverse genetics-based studies that swapped segments between different HEV genotypes indicated that HEV-3 or HEV-4 based chimeric viruses inserting ORF2 from HEV-1 was incapable to cause effective infection in swine[95 ]. Thus, it appears that HEV-ORF2 is also involved in HEV-interspecies infectivity, in agreement with results of another report demonstrating that replacement of the HEV-3 capsid region spanning aa 456 to 605 (the putative virus receptor-binding region) with corresponding region from HEV-1 prevented chimeric virus from entering and infecting swine cells[96 ]. Therefore, these data imply that in addition to ORF1 , viral capsid proteins also determine host preference. Nevertheless,until cellular receptors for HEV are identified, the link between viral capsid residues and cellular receptor determinants underlying HEV host tropism still requires further investigation.
In addition to ORF1 /2 , other reports have demonstrated that ORF3 proteins may be involved in determining host range of HEV. Up to date, the literature suggests that HEV-ORF3 protein acting as an ion channel essentially resembles viroporins involved in viral particle release during HEV infection[63 ]. This viroporin-like function depends on the highly conserved PSAP motif spanning aa 95 -98 withinOrthohepevirusA, which has been proposed to interact with host TSG101 . However, truncation analysis has indicated that the N-terminal 25 aa of HEV-1 ORF3 are required for its association with microtubules as well as virus release[55 ]. Meanwhile, alignment of ORF3 aa sequences of all eight HEV genotypes indicates that the region containing the M-terminal 25 aa of different HEV-ORF3 proteins are more conserved than the rest of HEV-ORF3 .Therefore, the conservation of this region may reflect the conserved role of HEVORF3 -dependent virion release across all genotypes. Nonetheless, a recent study demonstrated that rat HEV-ORF3 protein possessed the PxYPMP motif in place of the original PSAP motif found in human HEV-ORF3 proteins[97 ]. Intriguingly, unlike human HEV-ORF3 proteins, rat HEV-ORF3 proteins did not bind to TSG101 , but instead utilized MVB-based sorting to achieve virion release; this mechanism differed from the aforementioned TSG101 -dependent mechanism for effecting release of human HEV from infected cells[97 ]. Thus, these results taken together imply that HEV-ORF3 may have an important species-specific function.
Meanwhile, except for the abovementioned conserved motifs, less homology is observed elsewhere in the HEV-ORF3 protein, especially within its C-terminal half (aa 62 to aa 114 ) (Figure 4 ), a region that appears to be important for adaptation to various hosts. It is also notable that genomic locations of ORF3 -encoding genes vary among species ofOrthohepevirus(either of partially or fully overlapping with ORF2 , Figure 5 )[98 ,99 ], which implies a genotype-specific evolution pattern influencing functions of HEV-ORF3 that affect HEV host tropism. This speculation is in line with a genotypespecific enhancement of IFN induction by HEV-ORF3 proteins observed in our previous report[68 ]. Therefore, the mechanism by which a genotype-specific function of ORF3 product influences HEV host tropism requires further confirmation, although the accumulating literature indicates that a host-specific function exists that may influence host tropism by HEV-ORF3 proteins.
HEV-ORF PROTEIN AS VACCINE TARGET FOR HEV
Since the discovery of quasi-enveloped virions, researchers have tried to determine if these particles differ from classically enveloped virions, since the outer lipid bilayers of quasi-enveloped particles, such as those of HAV, are devoid of any viral proteins[66 ,80 ], while both HAV virion forms are equally infectious[80 ]. This apparent paradox raises the question of how membrane-wrapped particles can infect cells in the absence of viral peplomers that are generally thought to be required for enveloped virus infectivity[66 ]. Nevertheless, HEV appears to differ from HAV, since researchers observed very early before (prior to the identification of quasi-enveloped HEV particles) that HEV-ORF3 protein specific monoclonal antibody (mAb) could capture viral particles from serum samples of HEV patients or cell culture supernatants of HEV infected cells[89 ]. It is now clear that HEV-ORF3 protein associates with the lipid layer in quasi-enveloped virions produced bothin vitroandin vivo, while HEV virions from feces fail to be captured by this mAb due to the lack of the HEV-ORF3 -containing envelope[89 ]. This observation was recently confirmed by electron microscopy showing that immunogold-labeled mAb recognizing HEV-ORF3 proteins bound to quasi-enveloped HEV particles as well[100 ]. Moreover, althoughin vitroinfectivity appears to be equivalent between quasi-enveloped HAV particles and naked counterparts, quasi-enveloped HEV particles infect fresh cells in a less efficient mannerin vitro, as reflected by their need for a longer inoculation time to achieve maximal infectivity[67 ]. Meanwhile, it appears that cell entry by quasi-enveloped HEV virions depends on endosomal trafficking, which can be abrogated by blocking endosomal acidification[67 ]. Furthermore, additional investigations have demonstrated that HEVORF3 protein acts on ion channel protein and participates in the release of infectious virions from infected cells[63 ]; this role is similar to that of other well-characterized viroporins such as M2 protein of influenza A virus[63 ]. It is also notable that two hydrophobic domains located in N-terminal half of HEV-ORF3 demonstrated unique functions, whereby the first one is required associating with microtubules[55 ], while the second one contains a putative transmembrane region involved in ER localization[63 ]. Since viroporins of other viruses, such as M2 protein of influenza A virus, are components of virions, these observations imply that HEV-ORF3 is also a structural component of HEV virions, although it is not known if antibody-based neutralization mechanisms differ between the two types of HEV particles.Nevertheless, since HEV-ORF3 is present within quasi-enveloped HEV virions[89 ], it would be an interesting question to be determined if HEV-ORF3 -specific antibodies are capable to neutralize quasi-enveloped HEV viral particles since capsid specific antibodies fail to do so.
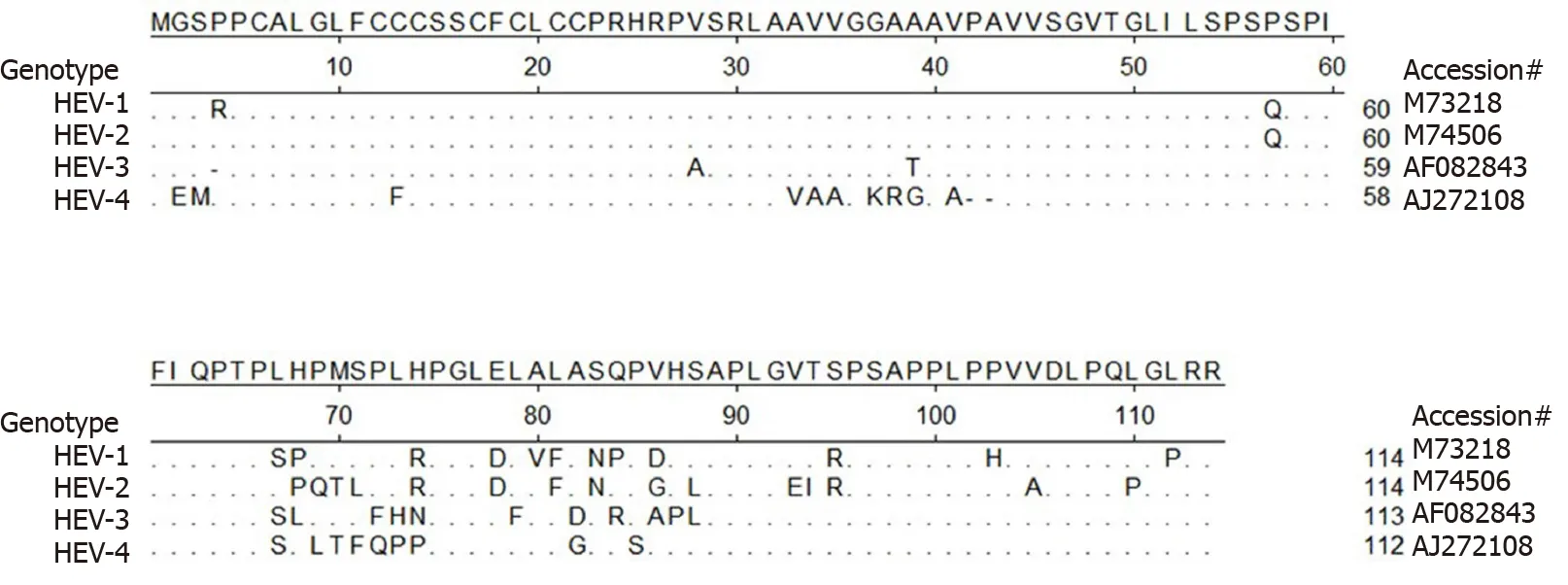
Figure 4 Alignment of amino acid sequence of hepatitis E virus-open reading frame 3 from 4 genotypes in Orthohepevirus A virus.Alignment of amino acid sequence of open reading frame 3 from all seven genotypes classified as Orthohepevirus A virus. Hepatitis E virus (HEV)-1 (reference sequence GenBank accession #M73218 ), HEV-2 (reference sequence GenBank accession #M74506 ), HEV-3 (reference sequence GenBank accession#AF082843 ), and HEV-4 (reference sequence GenBank accession #AJ272108 ) are shown. Those residues that are the same as consensus sequence are shown as“.”. HEV: Hepatitis E virus.
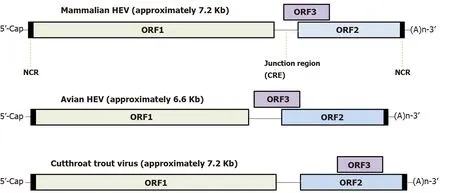
Figure 5 Hepatitis E virus genome organization. Genome location of open reading frame 3 among different hepeviruses. HEV: Hepatitis E virus; NCR: Noncoding region; CRE: Cis-reactive element; ORF: Open reading frame.
It remains unclear whether HEV-ORF3 acts as a potential neutralizing target for HEV. A previous report suggested that a recombinant vaccine candidate using HEV-4 ORF3 protein fused with interleukin-1 β might confer partial protection against virus challenge[101 ]. Moreover, similar to vaccines based on HEV-4 ORF3 , our research on avian HEV demonstrated that chickens immunized with recombinant avian HEVORF3 protein showed partial protection upon challenge and had milder disease symptoms than did controls[102 ]. However, it is notable that challenge experiments for these vaccines only employed virus stocks obtained from fecal samples which contained only naked viral particles (without envelopes containing HEV-ORF3 protein)[102 ]. Therefore, it is possible that antibodies induced by current ORF3 -based vaccines cannot prevent first-round infection during initial challenge with naked virus,since naked HEV virions cannot be neutralized by antibodies specific for ORF3 -HEV.Nevertheless, partial protection observed in both experiments may be due to ORF3 -specific antibody-based neutralization of newly synthesized quasi-enveloped HEV virion entering into circulation after initial infection caused by naked HEV virion used for challenging. In any case, these observations raise the interesting question of whether an ORF2 -based vaccine could protect hosts from challenge with quasienveloped HEV particles, a concept that warrants further investigation.
Up to date, available data indicated that ORF3 proteins (including avian HEVORF3 ) is highly immunogenic to evoke host humoral response, with most B-cell epitopes located at the C-terminal of HEV-ORF3 [103 -106 ]. Thus, the C-terminal half(about 60 aa) of HEV-ORF3 proteins appears to be a promising candidate as a recombinant subunit vaccine. However, an obstacle to employing this region for a vaccine candidate is the potential antigen variation issue as predicted by aa sequence alignment of HEV-ORF3 s from the four reference strains of HEV-1 to -4 (Figure 4 )[68 ].Therefore, systematic mapping of antigenic epitopes within HEV-ORF3 proteins of different HEV genotypes may be required before using immunogenic ORF3 epitopes as additional components of HEV subunit vaccines.
CONCLUSION
Although nearly three decades have elapsed since the identification and characterization of the complete genome sequence from the first HEV isolate, the full spectrum of this virus remains unclear and HEV infection now is a public health concern in developed countries as well. Currently, cross-species infection and host tropisms of different HEV genotypes remain elusive, due to the lack of easy-to-handle animal model and a robustin vitrosystem for studying HEV. Although many details about this virus and its pathology have been revealed in recent years, it is notable that HEVORF3 protein, the smallest ORF encoded by HEV, appears to have diverse functions and key roles in HEV virion release, biogenesis of quasi-enveloped virus, regulation of the host innate immune response, and neutralization of quasi-enveloped virus. These advances will guide further studies to reveal the basic biology of HEV, functions of HEV proteins, and HEV pathogenic factors toward the development of effective therapeutics and an improved vaccine.
 World Journal of Gastroenterology2021年20期
World Journal of Gastroenterology2021年20期
- World Journal of Gastroenterology的其它文章
- Deep learning for diagnosis of precancerous lesions in upper gastrointestinal endoscopy: A review
- State of machine and deep learning in histopathological applications in digestive diseases
- COVID-19 in normal, diseased and transplanted liver
- Upregulation of long noncoding RNA W42 promotes tumor development by binding with DBN1 in hepatocellular carcinoma
- Development and validation of a prognostic model for patients with hepatorenal syndrome: A retrospective cohort study
- Inflammatory bowel disease in Tuzla Canton, Bosnia-Herzegovina: A prospective 10 -year follow-up
