Identification of the Potential Function of circRNA in Hypertrophic Cardiomyopathy Based on Mutual RNA-RNA and RNA-RBP Relationships Shown by Microarray Data
Abstract—The pathogenesis of hypertrophic cardiomyopathy (HCM) is very complicated,particularly regarding the role of circular RNA (circRNA).This research pays special attention to the relationships of the circRNA-mediated network,including RNA-RNA relationships and RNA-RNA binding protein (RNA-RBP) relationships.We use the parameter framework technology proposed in this paper to screen differentially expressed circRNA,messenger RNA(mRNA),and microRNA (miRNA) from the expression profile of samples related to HCM.And 31 pairs of circRNA and mRNA relationship pairs were extracted,combined with the miRNA targeting database;145 miRNA-mRNA relationship pairs were extracted;268 circRNA-mRNA-miRNA triads were established through the common mRNA in the 2 types of relationship pairs.Thus,268 circRNA-miRNA regulatory relationships were deduced and 30 circRNARBP relationship pairs were analyzed at the protein level.On this basis,a circRNA-mediated regulatory network corresponding to the two levels of RNA-RNA and RNA-RBP was established.And then the roles of circRNA in HCM were analyzed through circRNA-mRNA,circRNA-miRNA,and circRNA-RBP,and the possible role in disease development mas inferred.
1.Introduction
Hypertrophic cardiomyopathy (HCM) is a common genetic cardiovascular disease caused by excessive hypertrophy of the myocardium and characterized by thickening of the left ventricular wall[1].
The mutation of the junctophilin-2 (JPH2) gene is the cause of HCM and the main genetic cause of left ventricular hypertrophy and myofilament disorders,and the PH2 protein is a member of the junctophilin family and is mainly expressed in the heart[2].At the transcriptome level,HCM is closely related to the expression levels of mRNA,microRNA (miRNA),and circRNA.A recent study showed that HCM is usually associated with missense mutations in the MYH6 and MYH7 genes.Silencing specific MYH6 alleles in mice can reduce the incidence of HCM[3].Jin and Chen found that abnormal miR-145-5p expression affects circRNA in oxygen glucose deprivation-induced (OGD-induced) cell damage by upregulating miRNA-145-5p and the mitogenactivated extracellular signal-regulated kinase (MEK)/extracellular signal-regulated kinases (ERK) pathway to activate the mammalian target of rapamycin (mTOR) and silence circRNA_0010729,thereby protecting against HCM damage[4].The CYTOR gene may activate the protein kinase B (PKB) and NF-kB signaling pathways through miR-155 to inhibit cardiac hypertrophy,most possibly through serving as ceRNA for miR-155 to counteract the miR-155-mediated repression of the inhibitor of nuclear factor kappa-B kinase subunit epsilon (IKBKE)[5].The circRNAs DNAJC6,TMEM56,and MBOAT2 can be used together to distinguish between healthy patients and who with HCM.In addition,circTMEM56 and circDNAJC6 can be used as indicators of disease severity in hypertrophic obstructive cardiomyopathy patients[6].Thus,the circRNAs regulate gene expression at the transcriptional and post-transcriptional levels and participate in various biological processes,leading to the occurrence of HCM[7],[8].
At the protein level,the p.I603M mutation is mapped to the C4 domain of the cardiac myosin-binding protein (cMyBPC).It was found that the stability of C4 I603M was severely impaired in HCM,so p.I603M was used as a basis for reclassification of variants[9].SerumN-terminal pro-B-type natriuretic peptide (NT-proBNP)and cardiac troponin I (cTnI) concentrations have been used to indicate the presence of a variety of heart diseases,including HCM,in various species[10].
circRNA is a non-coding RNA molecule that does not have a 5'end cap or a 3'end poly (A) tail and forms a ring structure with covalent bonds.Because circRNA is usually produced by special variable shearing,more than 80% of circRNAs contain protein exons,and has a large number of identical sequences with homologous mRNA,which acts as a sponge for adsorbing miRNA.circRNA participates in the pathological process of various diseases through spongy miRNA,but the role of circRNA in HCM is still unclear.
The currently used circRNA annotation tool,circRNADb (http://reprod.njmu.edu.cn/cgi-bin/circrnadb/resources.php)[11]is a comprehensive database of circRNA molecules in humans.It is difficult to prove the role of circRNA in disease development based on circRNA-mRNA-miRNA.Therefore,the RNA-RNA and RNA-binding protein (RNA-RBP) relationships noted in this article were used to speculate on the possible role of circRNA in HCM.The results were confirmed by Gene Ontology (GO)[12]and Kyoto Encyclopedia of Genes and Genomes (KEGG)[13]enrichment analysis and by using an experimentally verified database,which is more helpful for functional annotation of circRNA,especially in HCM research.Therefore,the method used in the present paper also has potential value for studying the role of circRNA in other complex diseases.
2.Materials
The Gene Expression Omnibus (GEO,http://www.ncbi.nlm.nih.gov/geo/) is an international public repository for high-throughput microarray datasets,we used the keywords“circRNA”and“HCM”to search for relevant information.Datasets related to HCM were obtained,including the human circRNA expression profile(ID:GSE148602),which included case (n=15 HCM samples) and control (n=7 normal samples) data,and the miRNA expression profile (ID:GSE36946),which contained case (n=107 HCM samples) and control(n=20 normal samples) data.Datasets related to HCM can be downloaded from GitHub (https://github.com/wgb2098/HCM).
3.Overview of Methods
As shown in Fig.1,the method used in this study consists of three steps:The 1st step,the construction of circRNA-mediated co-expression and regulatory relationship pairs;the 2nd step,the construction of the circRNA-mediated co-expression and regulatory network based on the 1st step and the deduction of circRNArelated relationship pairs;the 3rd step,functional analysis.In Fig.1,PCC is the Pearson correlation coefficient and SCC is the Spearman coefficient.In this section,we will describe the three steps in detail.

Fig.1.Overview of the method used in the present study.
3.1.Construction of circRNA-Mediated Pairs
The construction of circRNA-mediated relationship pairs was performed via three main steps.Firstly,differentially expressed (DE) mRNAs,miRNAs,and circRNAs were identified.Secondly,the relationship between circRNA and mRNA was established using PCC and SCC.Thirdly,circRNA-mRNA relationship pairs were extracted using circBase (http://www.circbase.org/)[14].
3.1.1.Identification of DE RNAs
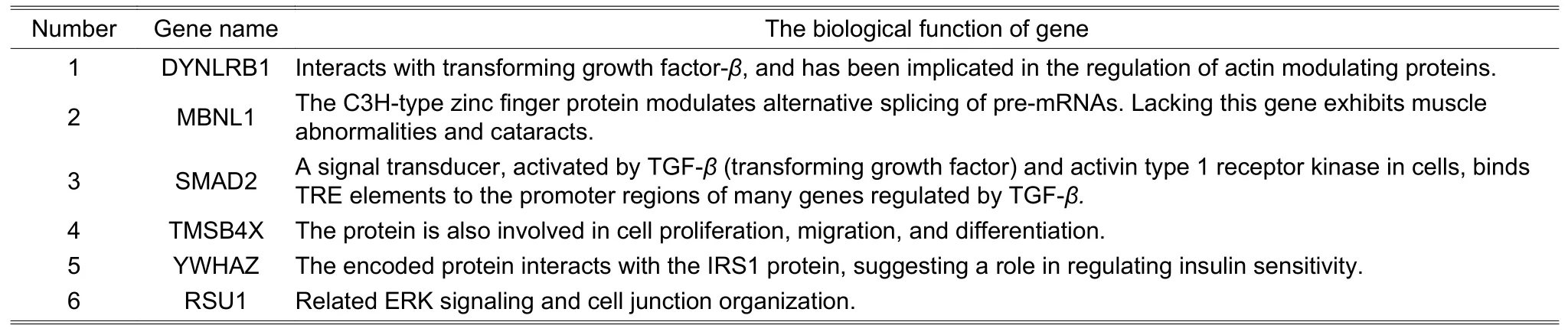
First,DE RNAs (circRNAs,mRNAs,and miRNAs) were extracted from the two datasets mentioned above using the linear models for microarray data (limma) package in R language,and the Benjamini-Hochberg methods were used to implement the analysis of variance (ANOVA) for all DE circRNAs and mRNAs(adjustedp-values≤0.05 and | log2(fold change) |≥1) and DE miRNAs (p-value≤0.05,−0.22 3.1.2.Extraction of circRNA-mRNA Co-Expression Relationship Pairs 1) Establishment of circRNA-mRNA relationships with PCC PCC reveals the degree of the linear correlation between two vectors.And PCC between the expression vectors of circRNA and mRNA is denoted by where X represents the circRNA profile value vector and Y represents the mRNA profile value vector;E(X)andE(Y) represent the means of X and Y,respectively;ρXY(PCC)represents PCC between X and Y. 2) Calculation of circRNA-mRNA correlations with SCC SCC between rank variables was calculated by whereXiandYirepresent theith element of X and Y,respectively;andrepresent the means of X and Y,respectively;ρXY(SCC)represents SCC between X and Y. 3) Extraction of circRNA-mRNA co-expression relationship pairs We used a circRNA database named circBase (http://www.circbase.org/)[14]to extract circRNA-mRNA relationship pairs. The thresholds for both PCC and SCC were set as 0.6.If PCC or SCC is larger than 0.6,it is considered to indicate a strong correlation.The correlation analysis was performed using OmicShare,a free online platform for data analysis (http://www.omicshare.com/tools). 3.1.3.Extraction of miRNA-mRNA Regulatory Relationship Pairs DE miRNAs were put into the miRWalk 3.0[15]database and then the miRNAmRNA pairs were extracted.The presence of common mRNA between miRNA-mRNA and circRNAmRNA relationships indicates that the interaction between miRNA and mRNA is related to circRNA.Those miRNA-mRNA relationship pairs related to circRNA were retained in this study. 3.1.4.Extraction of circRNA-RBP Co-Expression Relationship Pairs Next,circRNA-RBP relationship pairs were extracted to explore the interaction between RNA and protein using the CircInteractome database (https://circinteractome.nia.nih.gov/index.html)[14]to analyze the relationship between circRNA and RBP based on the circRNAs of strongly correlated circRNA-mRNA pairs. circRNA-mediated networks include circRNA-RNA and circRNA-RBP networks.The former category is divided into two main subcategories:Co-expression circRNA-mRNA networks and circRNA-miRNA regulatory networks,which are defined as interaction networks between RNA and RNA.circRNA-RBP networks were used to study the relationship between circRNA and the binding protein. Step 1:Using Cytoscape,a circRNA-mRNA co-expression network was constructed based on the extracted circRNA-mRNA co-expression relationship pairs.The starting point was circRNA and the end point was mRNA. Step 2:Since circRNA-mRNA and miRNA-mRNA share common mRNA,a circRNA-mRNA-miRNA ternary was established,called the circRNA-associated triad (circAT). Step 3:Based on the circRNA-mRNA-miRNA triad network,the circRNA-miRNA relationship pairs could be deduced.Finally,the circRNA-miRNA regulatory network was established. Step 4:The co-expression network was constructed based on the circRNA-RBP relationship pairs,revealing the interaction between circRNA and the RNA-binding protein. 1) Analysis of the biological functions of mRNAs in the circAT network The mRNAs in the circAT network were enriched and analyzed using PANTHER (http://www.pantherdb.org/)[16].Gene functions were assigned to one of three categories:Biological process,cellular component,and molecular function. 2) KEGG pathway enrichment of mRNAs in the circAT network To investigate gene functions and significant biological pathways,we used PANTHER[16]to understand the high-level functions of molecular-level information.The families and subfamilies of HCM-associated mRNAs were annotated with GO terms and sequences were assigned to PANTHER pathways. 3) Validation of predicted miRNA-mRNA pairs The miRTarBase (http://mirtarbase.mbc.nctu.edu.tw)[17],which contains the experimentally validated miRNA-target interactions,was used to confirm the miRNA-mRNA interaction relationships in the circAT network. Using the well-cited limma package in R language[18],562 DE circRNAs,179 DE mRNAs,and 203 DE miRNAs were identified after extracting valuable relationship pairs from the target gene database,and 62 miRNAs were retained after removing duplicates,as shown in Table 1. Table 1:RNA-RBP pairs obtained using circInteractome In total,43437 and 41383 circRNA-mRNA pairs were extracted using PCC and SCC (|cor| >0.6) based on the expression profiles of circRNA and mRNA,respectively.Additionally,92375 circRNA-mRNA coexpression pairs were extracted using the database circBase[19],[20](http://www.circbase.org/).Generally speaking,a correlation coefficient greater than 0.6 is considered to indicate a strong correlation.A total of 31 circRNA-mRNA pairs satisfied the circBase[20]target correspondence relationship,as shown in Table 2.We extracted 5876 miRNA-mRNA pairs based on mRNAs of the strongly correlated circRNA-mRNA pairs using miRWalk 3.0[15].Further,145 miRNA-mRNA relationship pairs were retained based on 203 DE miRNAs in HCM,as shown in Table 3.The 268 circAT pairs were deduced based on common mRNAs between circRNA-mRNA pairs and miRNA-mRNA pairs,as shown in Table 4. Table 2:Validated miRNA-mRNA pairs based on the miRTarBase dataset Table 3:Six mRNAs related to HCM and their biological functions in terms of circRNA-mRNA relationships Table 4:Five miRNAs and their biological functions in terms of circRNA-miRNA relationships 1) Constructed circRNA-mediated RNA-RNA networks Using the established circRNA-mRNA,miRNA-mRNA,and circRNA-miRNA relationship pairs,as well as the circRNA-mRNA-miRNA ternary,we constructed a circRNA-mRNA co-expression network (as shown in Fig.2 (a)),a circAT network (as shown in Fig.2 (b)),and a circRNA-miRNA regulatory network (as shown in Fig.2 (c)). 2) Constructed circRNA-mediated RNA-RBP network In this study,a total of 30 circRNA-RBP relationship pairs were extracted (in Table 1).Using the Cytoscape,a circRNA-RBP co-expression network was drawn,as shown in Fig.2 (d). 1) GO enrichment analysis of circAT-related mRNAs Gene functions (in the PANTHER Go-Slim biological process,Fig.2 (e)) were mainly enriched in the following categories:The cellular process,metabolic process,multicellular organismal process,locomotion,biological regulation,and cellular component organization of biogenesis. 2) KEGG enrichment analysis of circAT-related mRNAs The enriched pathways (in PANTHER pathway in Fig.2 (e)) were primarily the oxidative stress response,the FGF signaling pathway,the EGF receptor signaling pathway,T cell activation,and the TGF-βsignaling pathway. 3) Validation of relationship pairs by miRTarBase To validate relationship pairs,we used the experimentally validated miRNA-mRNA interactions from miRTarBase[17].As a result,6 miRNA-mRNA interactions were verified in 145 miRNA-mRNA interactions (in Table 2). Fig.2.circRNA-mediated regulatory network and functional analysis of target genes in the regulatory network:(a) analysis of circRNA-mRNA interaction relationships.Blue nodes represent mRNAs and light green nodes represent circRNAs,(b) visualization of interactions in the circRNA-mRNA-miRNA triad network.Light green nodes represent circRNA,blue nodes represent mRNAs,and red nodes represent miRNAs,(c) inferred circRNA-miRNA interaction relationships,(d) gene symbols and interactions between DE circRNAs and RBPs.Purple spots represent RBPs and blue spots represent circRNAs,and (e) results of GO functional and KEGG pathway enrichment analysis of mRNA using the PANTHER software. Research on the circAT network is progressing rapidly,and more and more evidence indicates that circRNA is involved in the regulation of human cardiovascular diseases[26].However,the potential role of the circAT in diseases is difficult to study due to a lack of large-scale public databases.Therefore,the present paper established a circRNA-mediated network of 31 circRNAs,62 miRNAs,and 14 mRNAs,including interactions between 31 circRNA-mRNA pairs,268 circRNA-miRNA pairs,and 30 circRNA-RBP pairs.It is speculated that circRNA plays a role in HCM at the transcription and protein levels[4],[26],[27]. We screen 14 mRNAs from strongly related circRNA-mRNA relationship pairs and circRNA-mediated regulatory networks;a literature search indicated that six of these mRNAs are related to HCM,including DYNLRB1,MBNL1,RUS1,SMAD2,TMSB4X (as shown in Table 3),and YWHAZ.The literature also shows that DYNLRB1 is related to the TGF-βsignaling pathways and that RUS1 is related to ERK signaling[28]. A number of studies have shown that the TGF-β/Smad signaling pathway is involved in inhibiting cardiac fibrosis.For example,Cutoloet al.found that downregulation of the Smad2/Smad3 and Erk1/2 intracellular signaling pathways inhibited fibrotic activity induced by TGF-β1[29].Therefore,we infer that circRNA may influence or change cardiac fibrosis through TGF-β/Smad signaling. The circRNA molecules are rich in miRNA binding sites.Cdr1as contains~70 conservative miR-7 (miRNA response elements,MREs) and one miR-671 MRE[30],and these binding sites act as a miRNA sponge in cells.Additionally,a circRNA called circSlc8a1 was reported to function as an endogenous sponge for miR-133a in cardiomyocytes and attenuate pressure overload-induced hypertrophy[1].Also,astragaloside IV inhibits myocardial fibrosis[24].Research found that miR-135a regulates the expression of the TGF-β/Smad pathway through the regulation of the target gene TRPM7[24].In addition,miRNAs are used as markers of disease performance.Shiet al.identified four miRNAs (hsa-miR-155-5p,hsa-miR17-5p,hsa-miR-20a-5p,and hsa-miR-181a-5p) as biomarkers for HCM diagnosis and treatment[23].Among the circRNA-miRNA relationship pairs with regulatory relationships identified in the present study,five miRNAs including (hsa-miR-195-5p,hsa-miR-373-5p,miR-17-5p,miR-15a-5p,and miR-135a) may be related to HCM (in Table 4).It is speculated that circRNA participates in the TGF-β/Smad2 signaling pathway as well as other signaling pathways through the miRNAs of strong circRNA-miRNA pairs,but more specific regulatory information is still unclear. RBPs play an important role in the formation of circRNAs.circRNA-derived proteins are RBPs that can change splicing patterns or mRNA stability.Cytoplasmic circRNAs seem to be involved in post-transcriptional regulation and sequestering the RNA-binding protein,and can even be translated into small peptides.Legniniet al.identified circ-ZNF609,which controls the proliferation of myoblasts and is translated into the protein[31].Studies have found that ADAR[20],[26]is associated with arrhythmia syndrome.The AIFM1 gene both encodes NADH oxidoreductase and acts as a regulator of apoptosis[32]-[34],and therefore plays a key role in apoptosis regulation,signal transduction,and regulation of apoptosis and mitochondrial proteins.Eight RBPs from the circRNA-RBP pairs related to HCM and the circRNA-mediated regulatory network were screened in the present study:FUS,AGO2,SFRS1,EIF4A3,FMRP,IGF2BP2,HuR,and IGF2BP3.The main function annotations of genes are obtained through GeneCards (https://www.genecards.org/)[35],as shown in Table 5.It is speculated that circRNAs also have certain relationships with the cell,signal transfer,and mitochondrial protein conversion,just like binding proteins.All of these conclusions are mere speculations,however,and need to be further confirmed through wet experiments. Table 5:Important RBPs and their biological functions in terms of circRNA-RBP relationships We selected popular or well-cited methods to study circRNA co-expression and regulation mechanisms.They are flexible methods,and users can choose any existing methods or new methods to replace one or more steps in our framework. Moreover,our framework is a parametric framework.Most parameter settings were based on commonly used principles.Specifically in the first step,identification of circRNA-mRNA pairs—the PCC and SCC cutoffs can be altered.The larger the correlation coefficient value,the closer the interaction relationship is.Given that the circRNA-mRNA interaction relationship is also subject to the limitations of interactions in other databases,we set the correlation coefficient to 0.6;if the parameter was set to 0.8 or above in the present study,the number of interconnected circRNA-mRNA in circBase would be very small,or even zero. Finally,the HCM-associated dataset is still limited,although we studied the possible roles of circRNAs in HCM based on RNA-RNA and RNA-RBP interactions,and found some evidence related to HCM.Because the regulatory network mediated by circRNA is very complicated,it is difficult to find more convincing evidence.In the future,we will analyze more HCM-related datasets to further confirm the effectiveness of this framework. This paper proposes a parametric framework.First,we identified DE RNAs (circRNAs,mRNAs,and miRNAs) by analyzing the HCM-related expression profile.Then,we combined the target genes database and the DE miRNAs and extracted miRNA-mRNA relationship pairs.As circRNA-mRNA and miRNA-mRNA share common mRNAs,a circRNA-mRNA-miRNA ternary relationship could be constructed,and circRNAmiRNA interaction relationships were deduced.Finally,circRNA-RBP interaction relationships were analyzed at the protein level,and circRNA-mediated networks were constructed based on the above relationship pairs,including a circRNA-mRNA co-expression network,a circRNA-miRNA regulatory network,and a circRNARBP co-expression network.This study considers both the transcription level and the binding protein level to investigate the possible roles of circRNA in HCM. Supplementary Materials The related data and the list of abbreviations are available as the supplementary materials at JEST website:http://www.uestc.edu.cn or at JEST ScienceDirect page:https://www.sciencedirect.com/journal/journal-of-electronicscience-and-technology. Table S1 (data):Differentially expressed circRNAs,mRNAs,and miRNAs. Table S2 (data):Identified circRNA-mRNA pairs. Table S3 (data):Putative miRNA-mRNA interactions. Table S4 (data):Identified circRNA-mRNA-miRNA regulatory pairs. Table S5 (data):List of abbreviation. Acknowledgment We would like to thank Dr.Wei Zeng for his valuable comments and advice,which helped improve the work substantially. Disclosures The authors declare no conflicts of interest.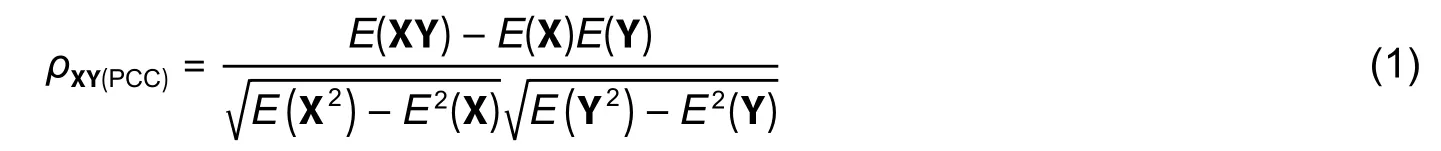

3.2.Construction of circRNA-Mediated Networks
3.3.Functional Analysis
4.Results
4.1.ldentified DE RNAs

4.2.ldentified Relationship Pairs


4.3.Constructed circRNA-Mediated Regulatory Networks
4.4.Results of Functional Analysis

5.Discussion
5.1.lnferring the Potential Functions of circRNAs from circRNA-mRNA Relationships
5.2.lnferring the Potential Function of circRNA from circRNA-miRNA Relationships
5.3.lnferring the Potential Functions of circRNAs from circRNA-RBP Relationships

5.4.Proposed Parametric Framework Technology
6.Conclusions
 Journal of Electronic Science and Technology2021年1期
Journal of Electronic Science and Technology2021年1期
- Journal of Electronic Science and Technology的其它文章
- Nonlinearity-Compensation-Free Optical Frequency Domain Reflectometry Based on Electrically-Controlled Optical Frequency Sweep
- Signal Acquisition and Processing Method for Capacitive Electromagnetic Flowmeter
- lmage Classification with Superpixels and Feature Fusion Method
- Bioinformatics Analysis on lncRNA and mRNA Expression Profiles for Novel Biological Features of Valvular Heart Disease with Atrial Fibrillation
- lmpact of Coronavirus Pandemic Crisis on Technologies and Cloud Computing Applications
- Molecules against COVlD-19: An in Silico Approach for Drug Development
