CVTree:A Parallel Alignment-free Phylogeny and Taxonomy Tool Based on Composition Vectors of Genomes
Guanghong Zuo
T-Life Research Center,Department of Physics,Fudan University,Shanghai 200433,China
Abstract Composition Vector Tree (CVTree) is an alignment-free algorithm to infer phylogenetic relationships from genome sequences.It has been successfully applied to study phylogeny and taxonomy of viruses,prokaryotes,and fungi based on the whole genomes,as well as chloroplast genomes,mitochondrial genomes,and metagenomes.Here we presented the standalone software for the CVTree algorithm.In the software,an extensible parallel workflow for the CVTree algorithm was designed.Based on the workflow,new alignment-free methods were also implemented.And by examining the phylogeny and taxonomy of 13,903 prokaryotes based on 16S rRNA sequences,we showed that CVTree software is an efficient and effective tool for studying phylogeny and taxonomy based on genome sequences.The code of CVTree software can be available at https://github.com/ghzuo/cvtree.
KEYWORDS CVTree;Composition vector;Alignment-free;Dissimilarity matrix;Phylogeny;Taxonomy
Introduction
Comparative analysis of genome sequences is the fundamental approach for the phylogenetic study of biology.Traditionally,sequence comparison is based on pairwise,including global sequence alignment [1],local sequence alignment [2],and multiple sequence alignment (MSA).Software tools for sequence alignment,such as BLAST[3]and CLUSTAL[4],are the most widely used bioinformatics methods.The aligned sequences provide a very intuitive impression of the difference between sequences,especially for the sequences with high identity.However,the computation of an accurate MSA is an non-deterministic polynomial(NP)-hard problem.The MSA-based methods cannot be solved in a realistic time for the large datasets that are available today.Most MSA tools are based on heuristic algorithms.It has been found that alignment-based techniques are inaccurate in scenarios of low sequence identity [5,6].Therefore,as an alternative solution to sequence alignment,many alignment-free approaches to sequence analysis have been developed in recent decades[6-10].These methods are computationally less expensive than the alignment-based methods.Their scalability allows them to be applied to much larger datasets than conventional MSA-based methods.
Composition Vector Tree (CVTree) is a cluster of alignment-free methods based on subsequences of a defined length (named as k-string).They generated dissimilarity matrices (DMs) from a comparatively large collection of genome sequences for phylogenetic studies.The classical CVTree method was proposed by Prof.Bailin Hao and coworkers in 2004[11].In the classical CVTree algorithm,every genome sequence,including protein sequence,RNA,and DNA,was represented by a composition vector (CV),which was calculated by the difference between the frequencies of k-strings and the prediction frequencies by the Markov model.And the similarity between the two sequences was measured by the cosine of two CVs.The classical CVTree method was first developed to infer evolutionary relatedness of Bacteria and Archaea[11-14],and then successfully applied to fungi [15,16],viruses [17],chloroplasts [18],and mitochondria [19],as well as metagenomes[20,21].After the proposal of the classical CVTree method,there are three versions of CVTree web server were released successively by our group [22-24].The latest released CVTree web server,CVTree3[24],is available from http://cvtree.online (Aliyun,Shenzhen,China).
In this study,we presented the standalone software for the CVTree algorithm.Due to the flexibility of the standalone software,the CVTree software is helpful for the researchers who are interested in the intermediate results (e.g.,the collection of CVs and DMs)or unwilling to upload their data to web server,as well as bioinformatics developers.In the CVTree software,the programs were redesigned in an objectoriented model.The OpenMP technique was employed to make the main programs parallel.An inbuilt automatic workflow helps users to obtain the phylogenetic tree from the Fasta files directly,and the intermediate result can be cached to avoid redundant calculation.Based on the scheme of the CVTree algorithm,other alignment-free phylogenetic methods based on the CVs were implemented[25].Furthermore,by using CVTree software,we obtained the phylogeny of 13,903 prokaryotes based on their 16S rRNA sequences[26].Interestingly,these CVTree methods are much faster than the alignment-based methods,and they are effective to obtain a taxonomy-compactible phylogenetic tree.
Algorithms and implementations
Scheme of CVTree
CVTree includes a cluster of alignment-free methods to obtain phylogenetic relationships based on genome sequences.Figure 1showed the scheme of the CVTree algorithm.There are three steps for the algorithm,i.e.,modeling the genomes to the CVs,calculating the DM from the CVs,and inferring a phylogenetic tree based on the DM.In the CVTree software,the classical CVTree CV generative model and algorithm were named as Hao model and Hao method,respectively,to honor Prof.Bailin Hao [27].And in the Hao method,the genome sequences were cut into small k-strings.Then the CV of the genome was modeled by the frequencies of k-strings,including the lengthsk-2,k-1,andk,based on a Markov model.The dissimilarity of the two genomes was measured by the cosine of the angle between two vectors.Finally,the phylogenetic tree was inferred by the neighbor-joint algorithm.Based on the scheme,other conventional dissimilarity methods,including Jaccard,Manhattan,and Euclidean,were integrated into the CVTree software [25].Two CV models (i.e.,direct count model and Hao model) and an enhanced neighbor-joint tree method were also provided in the software.Users can compose the models and methods by the options of programs (see details in File S1).

Figure 1 Scheme of CVTree algorithm
Implementation
The CVTree software,written in standard C++,facilitates compilation compliant with CMake and execution on Linux/Unix,Macintosh,and Windows platforms.The CVTree software,including example data,documentation,and source codes,is freely available for academic use at https://github.com/ghzuo/cvtree.
The CVTree programs were designed in an objectoriented model.In the scheme of CVTree algorithm(Figure 1),there are four states for the information:genomes,CVs,DM,and phylogenetic tree.They were described by different classes in CVTree programs.In more detail,the k-strings were encoded into an unsigned long integer(64-bit length) to improve efficiency.It is obvious that for aNlength sequence consisting ofmletters,when the lengthkof k-strings is large enough,the number of k-strings in the sequence,,is much less than that of the types of k-strings,mk.That is,the CV is sparse,i.e.,most of the dimensions are zero.Thus,only the non-zero dimensions were saved as key-value pairs in CVTree programs.All CVs were handled by associated arrays in the generation and by sorted sequential arrays in the calculation,respectively.The operations on these four states were also described by classes.And to organize different methods,we designed three virtual classes as the interface to describe the three operations in the CVTree scheme.In this way,a new method can be implemented by deriving from corresponding base classes (see details in File S1).To improve efficiency,the main programs of CVTree were implemented in parallel by OpenMP techniques.And these classes were carefully designed to keep threads safe.
The input data for CVTree software are the genomes in Fasta form,in which one file contains one genome.A file containing the list of the genomes is also required.The final output of CVTree is the phylogenetic tree in Newick form.In the scheme of CVTree algorithm,there are three steps from genomes to a phylogenetic tree.Thus,there are three programs,named asg2cv,cv2dm,anddm2tree,to perform these three tasks,respectively.Apart from the step-by-step way,an integrated program,named ascvtree,is also provided in the CVTree software.Figure 2showed the flowchart of thecvtreeprogram.Instead of bundling those three programs into a command,thecvtreeprogram automatically refers to the intermediate data to reduce computing resource consumption.Therefore,besides the final phylogenetic tree,the intermediate data,including CVs and DMs,are also saved for reuse in the next calculation.And to save storage,these intermediate data are compressed into binary format,which cannot be inspected directly.Thus,we also provided the tools to handle these compressed files in the CVTree software(see details in File S1).
Results and discussion
Performance of cvtree command
The CVTree was first developed to infer evolutionary relatedness based on whole genomes to obtain real species trees instead of gene trees.The phylogenetic tree for prokaryotes based on whole genomes can be accessed on the CVTree3 web server by an interactive interface.As an example,here we showed the CVTree software by performingcvtreecommand on 13,904 RNA sequences,in which 13,903 sequences were 16S rRNA sequences from the LTPs132 of the “All-Species Living” project [26] and one sequence was from the virus,Ferret parechovirus,as the outgroup to root the phylogenetic tree.It was found that the performance ofcvtreeis very remarkable.By the acceleration of multi-core CPUs,a typical phylogenetic tree for these 13,904 sequences can be obtained in 108.8 s on our Dell PowerEdge Server(4×20-Core Intel Xeon Gold 6248@ 2.50GHz,Linux System) or in 493.4 s on our Apple MacBook Pro (8-Core Intel Core i9 @ 2.3GHz,MacOS System).Figure 3showed the elapsed time ofcvtreecommand as a function of the number of threads in our Dell PowerEdge Server.As shown in Figure 3,the speedup of parallel was very significant.The calculation was accelerated about 1.7 times when the number of threads doubled.Detailed studies showed that most of the time was spent in the last two steps,calculating the DM and inferring the phylogenetic tree,and the speedup by parallel in calculating the DM was more significant.It was about 1.9 times with double of threads.We noted that with the increase of the length of genome sequences,the complexity of calculating a DM is lower than linear complexity,while the complexity of inferring a phylogenetic tree is constant.That is,the amount of computing resource of CVTree methods is scaled with the length of the sequence below linear.And CVTree method may obtain a rich benefit by parallel.Therefore,the CVTree programs are efficient enough to obtain the all-species living tree based on whole genomes.
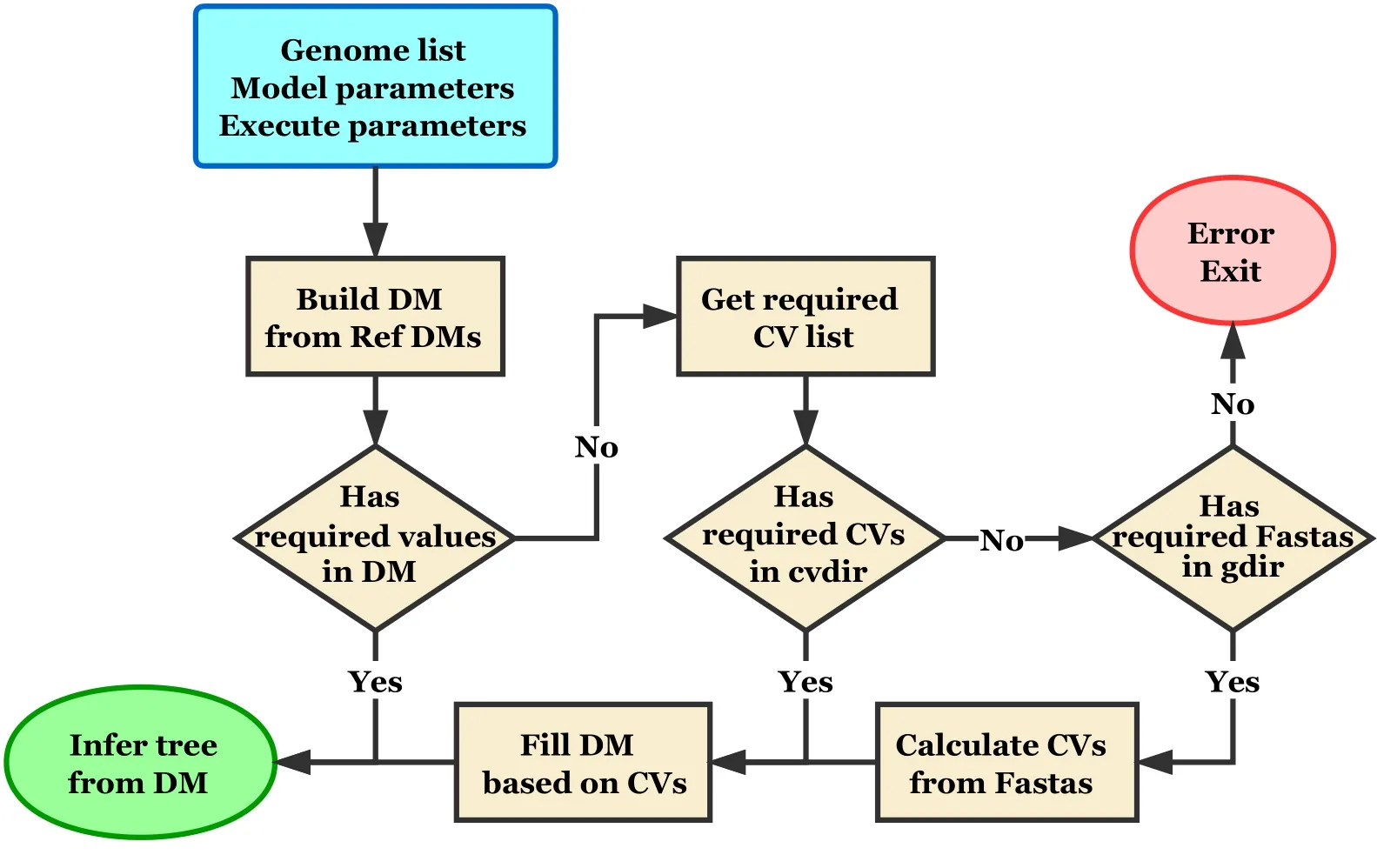
Figure 2 Flowchart of cvtree command
Compatibility with taxonomy
To examine the accuracy of CVTree,we compared the phylogenetic trees with the taxonomy system of these prokaryotes.It was a frequently asked question that what is the best length of the k-string,i.e.,how to set the parameterk.According to our studies,a reasonable length was in the range logmN<k<(logmN)+2for the Hao method,wheremis the number of the genome tyeps (i.e.,the types of amino acids or nucleotides)andNis the average length of the genome sequences.And the reasonablekfor new methods in the CVTree software should be a little bigger than that of the Hao method and has a larger value range.A detailed discussion of this problem can refer to our previous studies [28,29].In this study,we setk=6 for the Hao method andk=7 for the InterList method and the InterSet method.Figure 4showed the relative entropy difference between the taxonomy and the phylogenetic tree at every taxon level.The relative entropy difference between the taxonomy and the phylogenetic tree from the LTPs132,which was obtained by the alignment method,was also plotted in the figure as the benchmark.It was found that the results of CVTree methods and LTPs132 had similar performance at the high taxon levels of phylum,class,and order.At the low taxon levels of family,genus,and species,however,the results of CVTree methods were more consistent with the taxonomy than that of LTPs132.That is,the taxa were more monophyly in the CVTree methods.Moreover,our previous study showed that the CVTree methods may have much better performance with whole genomes for prokaryotes[30].This indicated that the CVTree was an effective tool for deducing the taxonomy system.

Figure 3 Speedup of cvtree command by multi-thread
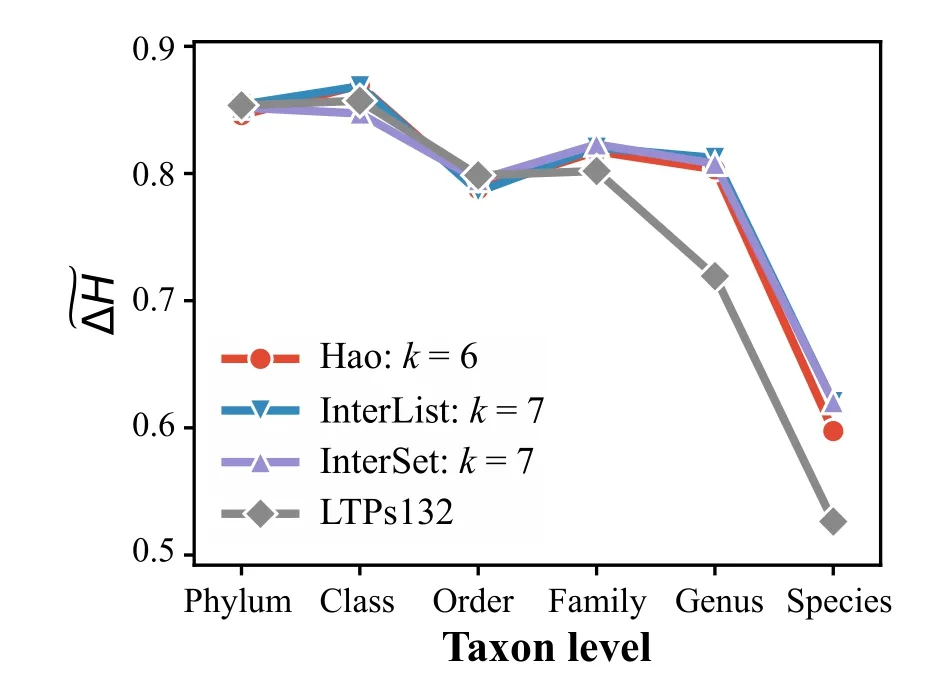
Figure 4 Relative entropy difference between phylogeny and taxonomy
Conclusion
CVTree is a cluster of alignment-free methods to infer phylogenetic relationships based on genome sequences.It has been applied to viruses,prokaryotes,and fungi with remarkable success,as well as chloroplasts,mitochondria,and metagenomes.Here we released the standalone CVTree software.The main programs of the software are parallel by OpenMP techniques.It is efficient to obtain a taxonomycompactible phylogenetic tree based on the 16S rRNA sequences.Since the complexity of the CVTree algorithm islower than linear complexity with the length of genome sequences,CVTree is efficient to handle huge whole genomes to obtain the phylogenetic relationship,especially for the prokaryotes.We believe that CVTree software is an efficient and effective tool for establishing a phylogenybased prokaryotic taxonomy.
Code availability
CVTree software code can be downloaded at https://github.com/ghzuo/cvtree and https://ngdc.cncb.ac.cn/biocode/tools/BT007094.
CRediT author statement
Guanghong Zuo:Conceptualization,Methodology,Software,Data curation,Visualization,Investigation,Supervision,Validation,Writing-original draft,Writing-review&editing.The author has read and approved the final manuscript.
Competing interests
The author has declared that no competing interests exist.
Acknowledgments
We thank Dr.Qiang Li for his helpful discussion.
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gpb.2021.03.006.
ORCID
0000-0002-7822-5969 (Guanghong Zuo)
 Genomics,Proteomics & Bioinformatics2021年4期
Genomics,Proteomics & Bioinformatics2021年4期
- Genomics,Proteomics & Bioinformatics的其它文章
- SPA:A Quantitation Strategy for MS Data in Patient-derived Xenograft Models
- RePhine:An Integrative Method for Identification of Drug Response-related Transcriptional Regulators
- NOGEA:A Network-oriented Gene Entropy Approach for Dissecting Disease Comorbidity and Drug Repositioning
- DeepCAPE:A Deep Convolutional Neural Network for the Accurate Prediction of Enhancers
- The Genome Sequence Archive Family:Toward Explosive Data Growth and Diverse Data Types
- Genome Warehouse:A Public Repository Housing Genomescale Data
