Expression and function of WNT4 involved in larvae development and limb regeneration in Portunus trituberculatus*
Zhiming REN , , Yuanyuan FU, , Lei LIU, , , Xiao LIU , , Chunlin WANG, ,
1 Key Laboratory of Applied Marine Biotechnology, Ministry of Education, Ningbo University, Ningbo 315832, China
2 Collaborative Innovation Center for Zhejiang Marine High-eき ciency and Healthy Aquaculture, Ningbo University, Ningbo 315832, China
Abstract The wingless-related integration site (WNT) proteins are a family of secreted glycoproteins that are evolutionarily conserved and are believed to be involved in evolution in vertebrates and invertebrates. WNT signaling pathways may be associated with limb regeneration and development in crustaceans. However, the detail mechanisms remain unclear. Therefore, the distribution of WNT4 in the hepatopancreas, muscle, hemocyte, ganglion, heart, eyestalk, gill tissue, and different larvae development stages of the swimming crab ( Portunus trituberculatus) were characterized using immunofluorescence, real-time PCR, and Western blotting. Significant PtWNT4 expression was detected in heart and eyestalk. In addition, PtWNT4 was expressed in all larval stages of P. trituberculatus with a dynamic expression pattern, especially in the eyestalk and other organs in the carapace area. The injection of WNT4 dsRNA into regenerative limbs significantly decreased PtWNT4 mRNA levels in the eyestalk, heart, and muscle, resulting in 1.9-fold, 2.2-fold, and 2.7-fold decreases compared with those detected in the group injected with crab saline ( P <0.05), respectively, indicating successful gene silencing. Overall, expression analysis on the WNT4 using RNAi provides an insight to its functional mechanism during limb regeneration in P. trituberculatus. The results not only demonstrated the requirement for WNT4 in limb regeneration of Crustaceans, but also suggested its ability to promote larval development at specific stages.
Keyword: wingless-type MMTV integration site family member-4 (WNT4); limb regeneration; larvae development; Portunus trituberculatus; expression pattern; RNAi
1 INTRODUCTION
Regeneration is a complex biological process by which organisms restore or repair damaged organs, and/or the shape, structure, and function of body parts that have been lost to injury or experimentally amputated. To some extent, all animals have the capability to regenerate particular tissues or organs. A smaller group of organisms in the animal kingdom, take regeneration to a different level and show very high regenerative capabilities, even regenerating the entire body from small fragments (Technau et al., 2000; Agata et al., 2003; Bely and Nyberg, 2010). Crustaceans probably represent the clearest examples of regeneration because of their segmented body plan, which provides natural breakage planes (Fleming et al., 2007). Within the subphylum Crustacea, 35 genera possess limb regeneration capabilities (Maginnis, 2006). Among them, the swimming crab ( Portunus trituberculatus), a commercially important crab species in China and other Asian countries, has been used as an important model for studies of limb regeneration because of its remarkable ability to regenerate limbs after loss or injury (He et al., 2016; Fu et al., 2018; Liu et al., 2018).
Previous studies have remarkably advanced our understanding of limb regeneration in crabs at the macroscopic and microscopic levels; however, knowledge of the molecular mechanisms and associated signaling pathways remain limited (Hopkins, 1993; Stueckle et al., 2008). Moreover, the genes and the proteins associated with crab limb regeneration have received little attention. In vertebrates, for example, salamanders, zebrafish, flatworms (Planaria), and Hydra, molecular studies have been performed in some organisms that exhibit strong regenerative capabilities. The C-X-C motif chemokine receptor 4 (CXCR4), sonic hedgehog (SHH), wingless-related integration site (WNT), basal cell carcinoma (BCC), and Hippo signaling pathways have been proven to be important for limb regeneration (Hunger et al., 2012; Singh et al., 2012). However, their roles in limb regeneration in crustaceans are poorly understood.
WNT signaling has been highly implicated in tissue regeneration across numerous phyla and tissue types. The WNT proteins are a family of evolutionarily conserved, secreted glycoproteins that are believed involved in extensive developmental processes in vertebrates and invertebrates. In mammals, the WNT family comprises 19 members that share homologies in their amino acid sequences, but often have fundamentally distinct signaling properties. The effects of WNT signaling include a wide range of cellular responses, including cell fate determination, proliferation, and differentiation (Liu et al., 2018). More recently, WNT genes and the WNT canonical pathway have been proven having a role during regeneration. The WNT family member 4 (WNT4) plays an important role in embryogenesis and regeneration in both vertebrates and invertebrates (Kobayashi et al., 2005). Studies have shown that WNT4 is expressed in overlapping and complementary patterns in the joint region of the developing synovial joint (Guo et al., 2004), particularly in Xenopus laevis (Satoh et al., 2005). Extensive studies of embryogenesis have shown that all the WNT3a, WNT4, WNT5a, and WNT7a affect cartilage development during bone healing and fracture repair regeneration (Hartmann and Tabin, 2000; Chen and Alman, 2009). Comparatively, little is known about the expression and function of WNT genes in crustaceans.
Our previous transcriptome sequencing study showed that WNT4 is differentially expressed between the regenerative limb and normal limb in P. trituberculatus. However, its expression in regenerative limb tissue remains poorly characterized. Therefore, it was necessary to perform comprehensive expression analyses, because previous studies indicate that regeneration restored all of the cell types that could be observed in adult limbs, including epidermis, neurons, and muscles (Iglesias et al., 2011; Zhang et al., 2016). Moreover, the expression of WNT4 in different larval development stages requires further study. Several processes that occur during development also take place during regeneration. Therefore, it is important to determine if the same gene (or gene pathways) could mediate both the embryonic and the regenerative events. In this study, expression of the WNT4 gene from P. trituberculatus was analyzed, and the result demonstrates that PtWNT4 may be involved in larval development, and its differential expression in tissues before and after RNA interference (RNAi) was important for understanding the molecular mechanisms of limb regeneration in crustaceans.
2 MATERIAL AND METHOD
2.1 Animal
Portunus trituberculatus (10.0±0.5 g) specimens and larvae from five zoea developmental stages (Z1, Z2, Z3, Z4, and M phases) (Supplemental Fig.S1) (Sun et al., 1984) were collected from a commercial farm in Xiangshan, China. All crabs were collected in compliance to the China’s national codes of animal use and care authorized by the Bureau of Fisheries of China. The crabs were fed for a week and adapted to the laboratory conditions. For the analysis of PtWNT4 expression in different tissues of the normal P. trituberculatus, we collected the hepatopancreas, muscle, hemocyte, ganglion, heart, eyestalk, and gill tissue from P. trituberculatus. In total, nine crabs were sampled, with three biological replicates bythree crabs per biological replicate for each tissue. For the analysis of PtWNT4 expression in different tissues of limb regenerative P. trituberculatus, crabs were fed for a week before experimental treatment, of which 90 were randomly sampled for autotomy treatments (removal of the first claw) and separately reared in 18 tanks (average 5 crabs per tank) in circulating water, and 90 normal crabs were reared in the same conditions as control. We collected the hepatopancreas, muscle, hemocyte, ganglion, heart, eyestalk, and gill tissue from all the crabs at 1, 3, 5, 7, 9, and 11 d after autotomy treatments during the limb regeneration. The tissues were stored at -80 °C and in 4% paraformaldehyde for RNA isolation and immunohistochemical analysis, correspondingly, to detect PtWNT4 expression in different tissues of the normal crab.

Table 1 Primer sequences used in this study
Larvae were also collected from three ponds at each development stage. Three parallel samples were taken from each period, and 20 larvae were randomly selected in each parallel group, which were used separately to determine the mRNA expression of PtWNT4 in different developmental stages. The larvae were stored at -80 °C and in 4% paraformaldehyde for RNA isolation and Bromodeoxyuridine (BrdU) staining, correspondingly. All experiments were performed in strict accordance with the Animal Protection Law of China.
2.2 Total RNA extraction and cDNA synthesis
Total RNA was extracted separately from different tissues using the RNAiso Plus reagent (TaKaRa, Shiga, Japan). Total RNA was treated with RQ1 RNase-Free DNase (Promega, Madison, WI, USA) to remove contaminating DNA. The RNA quality was assessed using electrophoresis on 1% agarose gel. About 1 μg of total RNA was used to synthesize cDNAs using a PrimeScript™ 1ststrand cDNA Synthesis Kit with random six-mers (TaKaRa, Japan).
2.3 Double stranded RNA (dsRNA) synthesis and injected
According to the open reading frame (ORF) sequence of the obtained PtWNT4 cDNA (GenBank: MK441658) and GFP plasmid sequence (Cloning vector pBB-GFP, GenBank accession No. ADQ43426.1), then we used the primers WNT4dsRNA-F and WNT4dsRNA-R linked with the T7 promoter sequence (5'-TAATACGACTCACTATAGGG-3') (Table 1). DsRNA of WNT4 and GFP was synthesized and injected were described previously (Fu et al., 2019).
Crab ( P. trituberculatus, 10.0±0.5 g) were collected and 50 crabs divided into two groups; dsPtWNT4, and saline (Grau and Cooke, 1992) for injection. Thus, there were 25 crabs per group, with five biological replicates (5 crabs per biological replicate). The dosage of injected dsPtWNT4 was set to 6.7 μ g/g, which was proven high eき cient to interfere limb regeneration in our previous study (unpublished data). The injection was performed at 24 h after autotomy. At 12 h after injection (36 h after autotomy), three crabs in each group were collected separately, and 7 detected tissues were isolated and frozen in liquid nitrogen for RNA extraction and western bolting.
2.4 Expression and purification of rPtWNT4 and preparation of the antibody
The recombinant PtWNT4 protein (designated as PtWNT4) were performed and purified as described previously (Zhu et al., 2018).
2.4.1 Western blotting
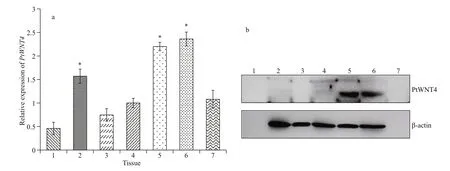
Fig.1 mRNA expression patterns (a) and protein distribution (b) of PtWNT4 in different tissues of P . trituberculatus
Western blotting was performed using a 15% separating gel and 5% stacking gel, and the protein samples were transferred from the gel to polyvinylidene fluoride (PVDF) membranes. After washing three times in TBS with 0.1% Tween-20 (TBST), the membranes were blocked in 1% casein blocking buffer with for 2 h. Then, the membranes were incubated with the primary and secondary antibodies at 37 °C for 1 h each. We used the Western Luminescent Detection kit (Vigorous Biotechnology Beijing Co., Ltd., Beijing, China) in the membranes and detected using Chemiluminescence Imaging System. After exposing films in the laboratory-grade X-ray, we selected β-actin as the control and analyzed the level of PtWNT4 using Quantity One software (BioRad, Munich, Germany) and ImageJ software (NIH, Bethesda, MD, USA).
2.4.2 Immunohistochemistry
Tissues from different developmental stages of the regenerative limb and larvae were fixed and embedded in paraき n. Sections (4 μm) were cut, dewaxed, rehydrated, and subjected to immunohistochemistry, as described previously (Zhu et al., 2018). Bromodeoxyuridine (BrdU) incorporation was used to analyze cell proliferation in the larvae, including several modifications of a previously published protocol (Harzsch et al., 2006).
2.5 Quantitative real-time PCR analysis
The quantitative real-time PCR (qPCR) assay was performed using a LightCycler®480 SYBR Green I Master kit (Roche, Branchburg, NJ, USA) to determine the distribution of PtWNT4 in the developmental stages of limb regeneration and larvae of P. trituberculatus. The PtWNT4 was amplified using the primers PtWNT4-F and PtWNT4-R (Table 1). Primers for ACTB (encoding Actin) as an internal control (Actin-F and Actin-R) (Table 1), were used. The qPCR was carried out as described previously (Fu et al., 2019). The qPCR was conducted in the procedure of: 40 cycles of 95 °C for 15 s, 55 °C for 30 s and 72 °C for 30 s. The relative expression levels of PtWNT4 were calculated using the 2-ΔΔCtmethod (Livak and Schmittgen, 2001).
All data are presented in mean±SD. For the RNA expression changes, data were subjected to use oneway analysis of variance with SPSS software (SPSS 19.0, IBM Corp., Armonk, NY, USA) and a value of P <0.05 was considered statistically significant.
3 RESULT
3.1 PtWNT4 expression and PtWNT4 protein localization in different tissues of the normal P. trituberculatus
The GenBank accession number of PtWNT4 is MK441658. The qPCR analysis showed that PtWNT4 was expressed in the hepatopancreas, muscle, hemocyte, ganglion, heart, eyestalk, and gill of P. trituberculatus. The expression levels in the heart, muscle, and eyestalk were significantly higher than those in the hepatopancreas, hemocyte, ganglion, and gill ( P <0.05; Fig.1a). Western blotting of proteins from normal crabs showed very strong PtWNT4 levels in the heart and eyestalk, and no expression was evident in the hepatopancreas, hemocyte, ganglion, muscle, and gill (Fig.1b).

Fig.2 Location of PtWNT4 in the hepatopancreas (a, b), muscles (c, d), ganglion (e, f), heart (g, h), gill (i, j), and eyestalk (k, l) of P. trituberculatus (hematoxylin and eosin (HE) staining)
The tissue distribution and expression of PtWNT4 in P. trituberculatus were analyzed using immunohistochemistry to understand its spatial expression pattern (Fig.2). The whole mount samples showed that PtWNT4 was not detected in the hepatopancreas and ganglion tissues (Fig.2a & e). In the muscle fiber and heart, a light blue-brown color appeared, which showed immunoreactivity of PtWNT4 (Fig.2d & h). The eyestalk also showed weak PtWNT4 staining, but with a more punctate pattern (Fig.2l). A clear blue-brown color appeared around the gill fibroin cells, showing positive PtWNT4 immunoreactivity (Fig.2j).
3.2 Expression profiles of PtWNT4 in different tissues of limb regenerative P. trituberculatus
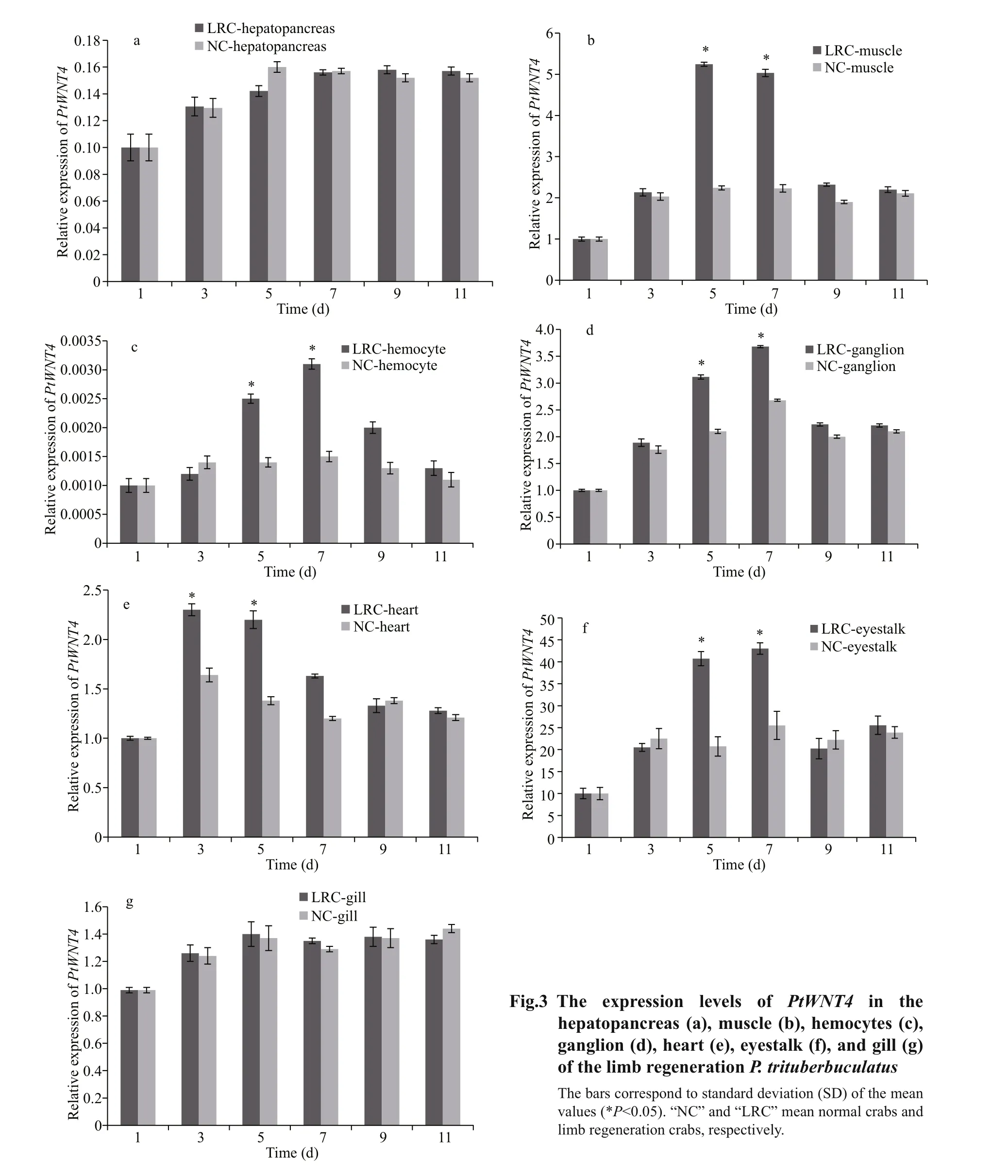
To learn the function of PtWNT4 during limb regeneration, we examined PtWNT4 expression levels in different tissues of P. trituberculatus at different time points after autotomy treatments (Fig.3). The mRNA expression levels of PtWNT4 changed and showed different expression patterns in detected tissues after autotomy treatments. There were no significant changes of PtWNT4 expression in hepatopancreas and gill at different stages of limb regeneration (Fig.3a & g). When compared with the control, the mRNA expression level of PtWNT4 was significantly upregulated ( P <0.05) on Days 5 and 7 in the muscles, hemocyte, ganglion, and eyestalk (Fig.3b, c, d, f). The mRNA expression level of PtWNT4 in the heart was significantly upregulated ( P <0.05) on Days 3 and 5, followed by a downregulation on Day 7 (Fig.3e).
3.3 Expression profiles of PtWNT4 in different tissues of limb regenerative P. trituberculatus with PtWNT4 silenced
PtWNT4 transcription was significantly downregulated at 24 h after dsPtWNT4 injection in the eyestalk, heart, muscle, resulting in a 1.9-fold, 2.2-fold, and 2.7-fold decreased levels relative to that detected in the group injected with crab saline ( P <0.05). There were no significant changes in expression of PtWNT4 in the hepatopancreas, ganglion, hemocyte, and gill between dsPtWNT4 injection group and the crab saline group (Fig.4a). Western blotting in tissue samples from the PtWNT4 interfered crab showed that the PtWNT4 protein level in the heart (Fig.4b) was significantly lower compared with that in the crab saline group (Fig.4c).
3.4 Localization of PtWNT4 mRNA and protein in larval developmental stages
PtWNT4 was expressed in all larval stages of P. trituberculatus. The highest expression was observed in the Z4stage (Fig.5a). Interestingly, the Western blotting results indicate that the PtWNT4 protein level was significantly decreased in stage M (Fig.5b), which is similar to the mRNA expression pattern.
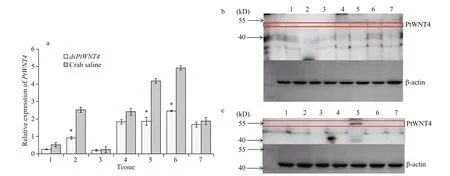
Fig.4 Expression prof iles of PtWNT4 in different tissue of P. trituberculatus groups injected with dsPtWNT4 and crab saline (a); distribution of the PtWNT4 protein in different tissues of P. trituberculatus injected with dsPtWNT4 (b); distribution of the PtWNT4 protein in different tissues of P. trituberculatus injected with crab saline (c)
PtWNT4 immunoreactivity was detected in proliferating (BrdU-positive) cells in all the larvae stages (Fig.5c). The results clearly showed that positive reactions of PtW NT4 first increased and then descended from Z1-a to Z4-a, and showed the strongest signal at phase Z3(Z2-a, Z3-a are indicated by arrows in the figure). There is no signal to be detected at phase Z4-a (Fig.5c: Z4-a). PtWNT4 moved away from the eye tissue and gradually migrated to the carapace area (Fig.5c: Z4, Z4-a, and Z4-b), which is the appendage formation site for young crabs (Supplementary Fig.S1) (Sun et al., 1984) that is initially formed at M-stage by comparison with other larval stages (Fig.5c: M). Overall, the positive reactions of PtWNT4 were strongest in the Z4-b and M-b stages.
4 DISCUSSION
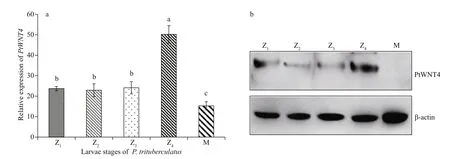
Fig.5 Expression patterns and localization of PtWNT4 in different larval development stages of P. trituberculatus
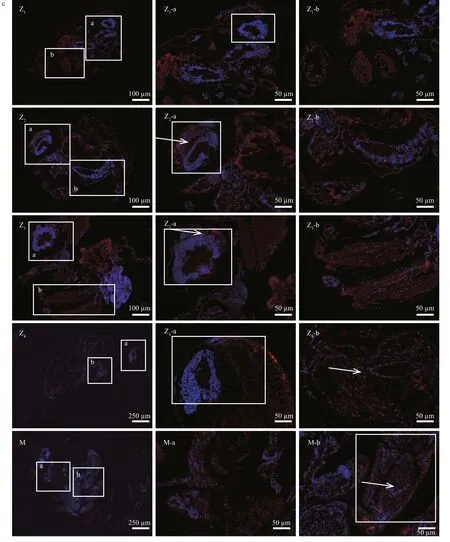
Fig.5 Continued
During the last decade, studies on limb regeneration in crustaceans have focused on morphology (Hopkins, 1993), histology (Cooper, 1998), neuroendocrinology (Hopkins, 2001), evolution (Fleming et al., 2007), morphology (Stueckle et al., 2008), and ecology (He et al., 2016). However, only recently, as technology has advanced, have studies been performed at the molecular level, although the molecular basis remains unclear. The Wnt/β-catenin signaling pathway has a pivotal function in cell proliferation, differentiation, growth, regeneration, self-renewal, and cell fate determination. This pathway also plays important roles throughout development (Majidinia et al., 2018). To date, the analysis of regeneration in different vertebrate and invertebrate contexts has stressed the highly conserved roles of the WNT family, because they share homologies in their amino acid sequence for the signal sequence for secretion, several glycosylation sites, and a characteristic distribution of 22 cysteine residues (von Maltzahn et al., 2012; Zhang et al., 2016). In vertebrates, WNT4 has long been known to regulate organ formation during embryogenesis and to play an important role in regeneration (Caprioli et al., 2015). However, the gene expression profile of the WNT4 in limb regeneration and organ development in crabs is limited. This study aimed to fill this gap and to determine the requirement for WNT4 in limb regeneration and larval development of P. trituberculatus. Gene expression data in different larval developmental stages and tissues of P. trituberculatus were obtained. Regulation of PtWNT4 expression using by RNAi was performed. The results provided novel information of WNT4 gene expression in P. trituberculatus, and suggested that PtWNT4 plays a role in limb regeneration and larval development.
Limb regeneration has been described as a biological process that shares many similarities with embryonic and larval development (Roy and Gardiner, 2002). Moreover, recent developmental biology studies in Drosophila showed that WNT4 is required in the developing and regenerating heart (Tauc et al., 2012; Chen et al., 2016) and nervous system (Schubert et al., 2000; Fradkin et al., 2010). The strong positive reaction in the carapace area in our BrdU immunofluorescence staining results also suggested that PtWNT4 was closely related to organ development in P. trituberculatus, because many organs are concentrated in the carapace area in larvae, such as the heart, nervous system, hepatopancreas, and gill. Furthermore, the high expression of PtWNT4 detected in the eyestalk (Fig.5c(Z2-a)) was similar to the result of a previous study in Xenopus laevis (Hamilton et al., 2016), which found that WNT4 was expressed in the corneal epithelium during lens regeneration and development. Our findings suggested that limb regeneration in P. trituberculatus might utilize the same molecular mechanisms involved in organ and larval development in other invertebrate animals, such as Drosophila and X. laevis.
To understand the function of WNT4 in living P. trituberculatus, we firstly detected the WNT4 mRNA expression profiles in different tissues. We found that PtWNT4 expressed highest in the eyestalk, which implied that it is necessary to maintain the normal life activity of P. trituberculatus. In Oncorhynchus mykiss, WNT4 was expressed highly in brain and pituitary, which are part of the nervous system (Nicol et al., 2012). Both the mRNA expression and immunohistochemistry results indicated that WNT4 was expressed in ganglia, but at a significantly lower level than in the muscle, heart, and eyestalk. We inferred that the difference in the expression pattern between fish and crab might be related to their different cell differentiation and life activity styles, because, in contrast to fish, crabs mostly depend on responses by the muscle, heart, and eyestalk during stress.
Most RNAi studies in crustaceans performed to date have used commercially important decapod species and have been applied to study development, differentiation, metabolism, and the innate immune system (Sagi et al., 2013). The dose of the injected dsRNA may significantly affect the results of RNAi. Our results suggests that the PtWNT4 gene silencing only partially depleted its mRNA expression in different tissues, and resulting in a 1.9-fold, 2.2-fold, and 2.7-fold decreased relative to levels detected in the crab saline group ( P <0.05) at 24 h in the eyestalk, heart, and muscle, respectively. Our study indicated the possibility of elucidating the upstream and downstream factors underlying the WNT signaling pathway in crabs during limb regeneration using RNAi.
A previous study used transcriptome analysis and found that WNT4 might be one of the key genes for limb regeneration (Liu et al., 2018). However, the source of the regenerated cells was unknown. The results of the present study showed that PtWNT4 was also distributed in different developmental stages of the larvae of P. trituberculatus. Furthermore, the WNT4 protein appeared during the process of the initial formation of ocular and crustacean tissues. Interestingly, the expression level of PtWNT4 decreased gradually during the developmental process of larvae. In the axolotl, it was observed that the role of transdifferentiation of tissues in limb regeneration was minimal, with the cells of the regenerating tissues largely arising from cells of embryonic origin (Kragl et al., 2009), and each tissue produced progenitor cells with a restricted potential. Our analysis revealed that in different larvae tissues, PtWNT4 had the similar function with unique expression signatures of early cells dividing of regenerative limb.
The regenerative capacity requires the formation of the blastema, a transient cellular structure that is a hallmark of limb regeneration (Haas and Whited, 2017). However, the transcriptional programs that drive blastemal progenitors remain unknown. Currently, we can only state that the WNT4 gene identifies embryonic progenitor cells and plays a certain role in regeneration. Such research in crabs is limited because of the very large size of the crab genomes, and no crab genome has been fully sequenced to date. In addition, cell culture technology has not been successful in this species of crab. However, leveraging mRNA expression as the starting point, together with functional experimentation, has been made up for this defect to a certain extent, and these data are rapidly enabling researchers to uncover the molecular mechanisms underpinning the biological process of limb regeneration.
5 CONCLUSION
The expression pattern analysis of the PtWNT4 suggested that it plays a significant role in limb regeneration and larval development. Overall, our results not only suggest an involvement for WNT4 in the early stages of Crustacean limb regeneration, but also it could promote limb regeneration and larval development at specific stages by regulating cell proliferation. The present study provided valuable insights into limb regeneration in crustaceans.
6 DATA AVAILABILITY STATEMENT
All data generated and/or analyzed during this study are available from the corresponding author, LIU Lei, upon reasonable request.
7 ACKNOWLEDGMENT
The authors are grateful to all the laboratory members for their technical advice and helpful discussion.
8 AUTHORS’ CONTRIBUTION
Zhiming REN, Yuanyuan FU, and Lei LIU carried out the immunofluorescence, real-time PCR, and Western blotting experiments, Xiao LIU performed the RNAi experiments and data analysis. Experiments were planned and analyzed by Lei LIU and Chunlin WANG. Manuscript preparation was done by Zhiming REN, Yuanyuan FU, and Lei LIU.
 Journal of Oceanology and Limnology2021年1期
Journal of Oceanology and Limnology2021年1期
- Journal of Oceanology and Limnology的其它文章
- Influence of sequential tropical cyclones on phytoplankton blooms in the northwestern South China Sea*
- Simulated perturbation in the sea-to-air flux of dimethylsulfide and the impact on polar climate
- Performance of ecological restoration in an impaired coral reef in the Wuzhizhou Island, Sanya, China*
- Investigating factors driving phytoplankton growth and grazing loss rates in waters around Peninsular Malaysia
- Effects of oxytetracycline dihydrate and sulfamethoxazole on Microcystis aeruginosa and Chlamydomonas microsphaera*
- Reproductive cycle of Ophiopholis mirabilis (Echinodermata: Ophiuroidea) in Zhangzi Island area, northern Yellow Sea*
