Cloning of MYB2 Gene from Dryopteris fragrans and Its Response to ABA and Drought Stress
Li Jie, Chen Ling-ling, and Chang Ying
College of Life Sciences, Northeast Agricultural University, Harbin 150030, China
Abstract: The myeloblastosis oncogenes (MYB) are important transcription factors that facilitate induction of variously developmental and stress responsive genes. They are hence, emerging as key players in improving stress tolerance of plants in response to several abiotic stresses. It was predicted that DfMYB2 contained an open reading frame (ORF) of 1 621 bp coding 377 amino acid residues with molecular weight of 41.5 ku. It had 50 potential phosphorylation sites, 44 potential N-glycosylation sites and one transmembrane domain. In neighbor-joining (NJ) phylogenetic tree, DfMYB2 was close to the branch of angiosperms. The subcellular localization of DfMYB2 was in nucleus. The expressions of DfMYB2 increased gradually in the filament, prothalli and sporophyte. The results showed that DfMYB2 played a more and more important role in its growth and development. And the expression levels in rhizomes were significantly higher than those in roots and leaves. After abscisic acid (ABA) and PEG600 treatment, DfMYB2 showed a downward trend, followed by an upward trend. The expression of DfMYB2 was inhibited in a short time under drought stress, the strong induction of DfMYB2 expression by ABA indicated that it was possibly involved in abiotic stress responses in an ABA-dependent manner.
Key words: transcription factor, DfMYB2, abscisic acid (ABA), drought stree
Introduction
Plant growth is greatly influenced by adverse environmental conditions, such as drought, salt or extreme temperature. Among these adverse factors, drought stress is commonly encountered and it is aggravated by climate. Drought stress conditions trigger changes gene expression profiles, which can be generally grouped into two classes. One class contains 'regulatory proteins' that play roles in signal transduction and regulate the expression of genes involved in the stress response. The other class includes genes that serve to protect plant cells from abiotic stress. The myeloblastosis oncogenes (MYB) transcription factors are abundant in the plant kingdom and have special sequences (Ambawatet al., 2013). They have a conserved DNA-binding domain with two or three imperfect repeats (R1, R2 and R3), each one of 51-53 amino acids (Duet al., 2012; Guoet al., 2017). R2R3-MYB is the largest number of MYB transcription factors. The researches have shown that 10.4% of MYB transcription factors is R2R3-MYB inArabidopsis thaliana(Galiset al., 2006). The roles of plant R2R3-MYB TFs have been implicated in phenylpropanoid metabolism, cell shape determination, hormonal responses, cellular proliferation and abiotic stress responses (Duboset al., 2010). R2R3-MYB has DNA binding domains and activation or inhibition domains at its C-terminal and N-terminal, which enable it to perform a variety of functions, such as high salt, low temperature and defense response (Pryeret al., 2001). Ferns are the most highly evolved sporophytes with primitive vascular tissues and have abundant medicinal functions and special evolutionary status (Mülleret al., 2001). At present, there are some understandings about evolution, development, gene regulation and so on (Gaoet al., 2019).
Dryopteris fragransis a deciduous perennial herb with aroma and dense glandular hair and mainly distributes in China, Japan, Mongolia and so on. China is located in Baikalu Mountain of Tahe County, Dabaishan Highland of Huzhong, Jingbo Lake of Mudanjiang River and the northern part of Xiaoxing'an Mountains, of which Wudalianchi area has a larger distribution area (Huanget al., 2014).D. fragranshas antioxidant, bacteriostatic, antipsoriasis and antitumor effects, grows in a special environment and lives on talc slopes or on stone baskets, on gravel slopes in forests and in magma fissures around volcanoes in alpine areas with altitudes of 1 000-1 500 m (Itoet al., 2014). Its growth cycle is longer andD. fragranscan tolerate high temperature and low temperature, strong ultraviolet radiation in summer (Tamuraet al., 2011). There are many MYB transcription factors inD. fragrans, but no one has been studied. In this study, a fragment of MYB transcription factor was obtained from the transcriptome library ofD. fragrans. A R2R3-MYB transcription factor named MYB2 was cloned by rapid amplification of cDNA end (RACE). Among abiotic stresses, MYB2 homologs fromArabidopsiswere found to be induced in response to dehydration and salt stress, suggesting their important roles in stress responsive gene regulation. Therefore, considering the importance of MYB2 TF in plants, this study aimed to analyze the sequence and expression ofDfMYB2.
Materials and Methods
Plant materials
The spores ofDryopteris fragranswere collected from Wudalianchi City, Heilongjiang Province. Spores were cultivated on the surface of sterile soil at 25±1℃ with a 12 h light/12 h dark period. When they grew into sporophyte, RNA was extracted from different parts of the sporophyte.
The total RNA was isolated using TiangenTMPlant Total RNA Kit. RNA concentration has greater than 500 ng · uL-1. Agarose gel run three distinct bands: 28S, 18S and 5S. Spectrophotometer analysis, OD260/OD280was in the range of 1.8-2.1. The first strand cDNA used the Reverse Transcription Kit (Haigene, China), according to the manufacturer's instructions. RACE Ready-cDNA used SMARTTM RACE cDNA Amplification Kit User Manual.
Cloning of DfMYB2
A unigene fragment annotated MYB2 was found in the transcriptome database (not published), which was consistent with other MYB gene sequences selected from NCBI GenBank database by BLAST program. All the primers used for the study are listed in Table 1. The cloned pMD-T18 vector was used for recovery and purification of PCR products. After transformation inE. coliDH5αcompetent cells, plasmids were isolated from positive clones and sequenced. Designed 3' and 5' primers, based on transcriptome sequences, cDNA end (RACE) PCR was done using SMARTTM RACE cDNA Amplification Kit (Invitrogen, USA). The full-length cDNA was isolated by PCR.

Table 1 Primer for MYB2 gene amplification in D. fragrans
Sequence analysis and phylogeny prediction
The full-length cDNA sequence was analyzed using online bioinformatic tools. ORF (open reading frame) was found by ORF Finder (http://www.biosoft.net/sms/). Conserved domains were detected by Conserved Domain Database (http://www.ncbi.nlm.nih.gov/structure/cdd/wrpsb.cgi). Physical and chemical parameters of proteins were determined using ProtParam tool (http://us.expasy.org/tools/protparam.html). The signal peptide was predicted using SignalP3.0 Server (http://www.cbs.dtu.dk/services/SignalP). Hydrophobicity analysis was performed on ProtScale (http://www. expasy.org/cgi-bin/protscale.pl). The transmembrane domain was predicted by TMHMM bioinformatic program (http://www.cbs.dtu.dk/services/TMHMM/). PSORT tool (http://wolfpsort.org) was used to predict subcellular localization. The predict subcellular localization was used (http://www.csbio.sjtu.edu.cn/cgi-bin/PlantmPLoc.cgi). 3-D structure was established (https://swissmodel.expasy.org). MEGA 5.05 software was used to construct a phylogenetic tree using neighborjoining (NJ) algorithm, includingArabidopsis thaliana(AT2G47190),Gossypium hirsutum(XM_016866407.1),Populus trichocarpa(NC_037285.1),Oryza sativa(XM_015772960.1),Volvocales carteri(EFJ45034.1,Triticum aestivum(AAT37168.1),Betula platyphylla(AKN79284.1),Eucalyptus pyrocarpa(BAM05602.1),Anthurium andraeanum(AML84515.1) andLithospermum erythrorhizon(AIS39993.1) (Livak and Schmittgen, 2001).
Expression pattern analysis
RT-PCR used 18S ribosomal RNA gene as internal reference. ABI7500 System and 3B Quantimix Easy SYBR Green Kit (2×with 1×ROX, 3B BlackBio Biotech, China) were used for detection (von Arnim, 2007; Lambertet al., 2002). The primer was DfMYB2-F/DfMYB2-R. The expression patterns ofDfMYB2 were analyzed in filament, prothalli and sporophyte. The expression patterns ofDfMYB2 were analyzed in leaves, roots and rhizomes. The sporophytes ofD. fragransgrowing for 70 days were treated with abscisic acid (ABA) under the same growth conditions. The leaves were sprayed with ABA at a concentration of 50 mL of 10 umol · L-1each time, watering with 50% PEG600. Distilled water sprayed with similar growth conditions was selected as the control. The treatment time was 0, 2, 4 and 6 h, respectively. Three samples of RNA were extracted three times at a time. The conditions for qRT-PCR were as the followings: one cycle of 5 min at 95℃, followed by 40 cycles of 15 s at 95℃, 30 s at 60℃ and 35 s at 72℃. All the samples were run in triplicate and repeated twice. The relative expressions of gene were analyzed using the comparative CT method (2-ΔΔCt)(Lin, 1974).
Subcellular localization
pBI121-DfMYB2-GFP expression vector was constructed. PCR products were digested with DfMYB2-ORF-F1/DfMYB2-ORF-R1 connected to pPBI121-GFP expression vector and transformed into LB4404 cells, the positive transformants were identified by PCR. The positive monoclones were cultured in Luria-Bertani medium containing 50 μg · mL-1Kanamycin and 50 μg · mL-1Rifampicin, growing at 28℃ with shaking at 170 r · min-1. When OD600reached 0.6-0.8, they were centrifuged at 5 000 g for 30 min. Used 1/2MS [Twain 20 (1/25), 10 mol · L-1MgCl2, 10 mmol · L-1acetyleugenone, 30 g sucrose] resuspension, infected with onion epidermis and observed under laser confocal microscope (Helenius and Aebi, 2001).
Results
Cloning and analysis of DfMYB2 gene
The potential partial sequence (780 bp) was selected. The full-length gene was cloned by RACE. The lengths of 5' and 3' RACE fragments were 190 bp and 718 bp. A total of 1 621 bp sequence (GenBank: AQZ26708.1) of the 377 amino acid residues was obtained. The 5'-UTR and the 3'-UTR were determined (Fig. 1).DfMYB2 was predicted as a theoretical molecular mass of 41.5 ku. Compared with other R2-R3 MYB2, Myb superfamily proteins included transcription factors and mRNA splicing factors, Myb-like DNA-binding domain. This family contained DNA binding domains from Myb proteins, SANT SWI3, ADA2, N-CoR and TFIIIB DNA-binding domains (Fig. 2). The three-dimensional (3D) structure ofDfMYB2 was established by Swiss-Model Workspace, which was similar to the structures of MYB2 from other species (Reedet al., 1994).DfMYB2 had one transmembrane domain, which was located from position 1 to positions 200 (Fig. 3) (Ambawatet al., 2013).DfMYB2 was no signal peptide (Fig. 4)and was a non-secretory protein. There were 50 potential phosphorylation sites inDfMYB2 predicted, including threonine 16, serine 31 and tyrosine 3(Fig. 5) (Liet al., 2012). The total average hydrophobic GRAVY value was -0.579, which indicated that the predicted protein was hydrophilic (Fig. 6) (Jiaoet al., 2013).

Fig. 1 Deduced amino acid sequence and nucleotide sequence of DfMYB2
Phylogenetic analysis
A NJ phylogenetic tree was constructed by using the published amino acid sequences of MYB2 in different plants. The result showed thatMYB2s in different plants were evolved from a single ancestral gene. It was close toEpimedium sagittatum,Populus trichocarpaandBetula Platyphylla(Fig. 2).

Fig. 2 Neighbor-joining phylogenetic tree constructed from amino acid sequences of DfMYB2 and MYB2s from other species using MEGA 5.05
Tissue specific expression analysis of DfMYB2
The expression levels at different developmental stages had different tissue specificities and the highest transcriptional level was detected in rhizomes, followed by leaves and roots (Fig. 3). The expression levels in filament, prothallium and sporophyte increased gradually (Fig. 4).
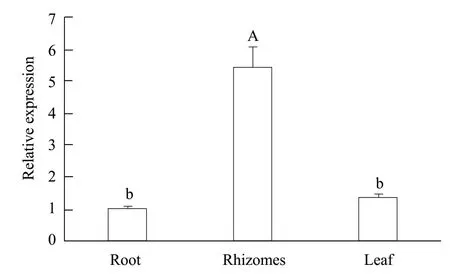
Fig. 3 Differential expressions of gene DfMYB2 in different tissues
Expression analysis of ABA treatment
It had been reported that MYB transcription factor was involved in ABA response.AtMYB2 gene inArabidopsis thalianacould be induced by ABA response and dehydration. In order to verify whetherDfMYB2 was induced by ABA, distilled water treated was used as the control. The expressions of MYB2 in these two treatments were calculated by fluorescence quantitative PCR. The results showed that the expressions of MYB2 decreased at first and then increased after treatment with ABA compared with the control group sprayed with distilled water. It decreased at 2 and 4 h after spraying ABA and increased significantly after 6 h (Fig. 5).

Fig. 5 Differential expressions in leaves of gene DfMYB2 under ABA stress
Expression analysis of PEG600 treatment
It had been reported that MYB transcription factors were involved in drought response. Compared with the control group, the expression ofDfMYB2 was significantly decreased after PEG600 simulated drought treatment. The expression ofDfMYB2 was increased after 6 h of the treatment (Fig. 6).
Subcellular localization
The results of subcellular localization of onion epidermis showed that DfMYB2 protein located at nucleus, which was consistent with the predicted results and one of the characteristics of the transcription factors (Fig. 7).
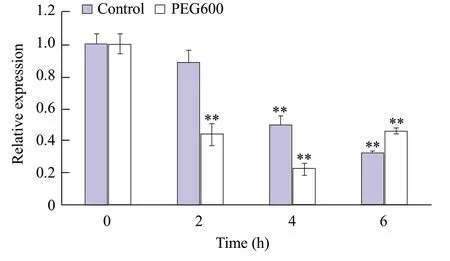
Fig. 6 Differential expressions in leaves of gene DfMYB2 under PEG600 stress
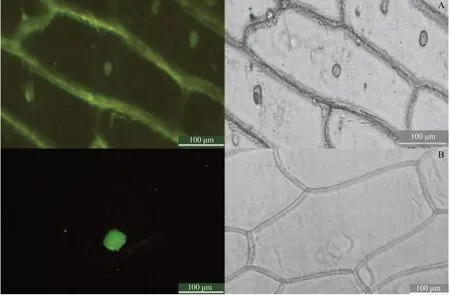
Fig. 7 Subcellular localization
Discussion
D. fragranswas a kind of traditional medicinal plant. It was mainly used to treat beriberi and acne. In recent years, it was found that some compounds ofD. fragranshad medicinal effects, such as bacteriostasis, anti-tumor and anti-inflammation (Junet al., 2015). The growth environment ofD. fragranswas very special. It lived in volcanic rocks. However, how did it resist these stresses through gene regulation had not been reported. It was considered that MYB transcription factors were related to many stresses in plants (Qiet al., 2015), but the functions of these transcription factors inD. fragranswere unknown, therefore, it was important to understand their expressions in plants and in adversities.
In this study, the full-length cDNA ofDfMYB2 was obtained by RACE. The phylogenetic tree showed thatDfMYB2 came from the same source as other MYB2s. After the branch ofVolvocales carteri,Epimedium sagittatumwas similar, followed byPulus trichocarpaandBetula platyphylla, which meant thatDfMYB2 could be similar to the amino acid sequence of angiosperms. After one branch,DfMYB2 was shorter than EsMYB2, indicating that MYB2 evolution in ferns was later.
The results showed that the expression of MYB2s had tissue specificity. The qRT-PCR results revealed that transcriptional level ofDfMYB2 was the highest in rhizomes and the lowest in roots, which was similar withMedicago truncatula(Huanget al., 2013),Scutellaria baicalensis(Guanet al., 2014),Epimedium sagittatum(Koornneefet al., 1982) andArabidopsis trichome(Webbet al., 2001). The reason for this characteristic might be the result of natural selection. The expressions ofDfMYB2 in filament, prothallus and sporophyte were increased, indicating that during the alternation of generations,DfMYB2 might play a more and more important role. Subcellular localization results showed thatDfMYB2 was expressed in the nucleus as other transcription factors.
ABA was one of the recognized plant hormones, also known as stress hormones (Uraoet al., 1996). When plants were under drought stress, ABA synthesis increased. Applying ABA to plants could increase stomatal resistance, reduce transpiration rate of leaves and decrease relative water contents in plants. Most of MYB transcription factors could be induced by ABA (Kumaret al., 2004). This study found thatAtMYB2 could be induced by drought and exogenous ABA, applying ABA and drought stress to plants could increase stomatal resistance, reduce transpiration rate of leaves and decrease relative water contents in plants. Higher growth-related genes were inhibited by ABA, while drought-related genes were activated by ABA, thus the plant drought resistance was increased. In this study, after ABA treatment, the expression ofDfMYB2 was significantly decreased, and then increased. After treatment for 2 h, the expression ofDfMYB2 was 14 times lower than that of 0 h and increased by 3.5 times after treatment for 6 h. After being treated with 50% PEG600, the expression ofDfMYB2 showed a downward trend, the expression ofDfMYB2 was increased after 6 h (Liaoet al., 2008). These results suggested that the expression ofDfMYB2 was inhibited in a short time, under drought stress and the strong induction ofDfMYB2 expression by ABA indicated that it was possibly involved in abiotic stress responses in an ABA-dependent manner.
Conclusions
In this study,DfMYB2 contained an open reading frame (ORF) of 1 621 bp coding 377 amino acid residues with molecular weight of 41.5 ku. It had 50 potential phosphorylation sites, 44 potential N-glycosylation sites and one transmembrane domain. In NJ phylogenetic tree,DfMYB2 was close to branch of angiosperms, the subcellular localization in nucleus. The expression ofDfMYB2 increased gradually in the filament, prothalli and sporophyte. The results showed thatDfMYB2 played a more and more important role in its growth and development. And the expression level in rhizomes was significantly higher than that in roots and leaves. After ABA and PEG600 treatment,DfMYB2 showed a downward trend, followed by an upward trend. The expression ofDfMYB2 was inhibited in a short time under drought stress and the strong induction ofDfMYB2 expression by ABA indicated that it was possibly involved in abiotic stress responses in an ABA-dependent manner.
 Journal of Northeast Agricultural University(English Edition)2021年4期
Journal of Northeast Agricultural University(English Edition)2021年4期
- Journal of Northeast Agricultural University(English Edition)的其它文章
- Effect of Foliar GA3 and ABA Applications on Reactive Oxygen Metabolism in Black Currants (Ribes nigrum L.) During Bud Paradormancy and Secondary Bud Burst
- Mining Heat Stress Associated Genes in Tomato Fruit (Solanum lycopersicum L.) Through RNA-seq
- Advances on Current Techniques and Methodologies in Milk Lipidomics
- NIRS Prediction of SOM, TN and TP in a Meadow in the Sanjiang Plain, China
- Screening of Feather Degrading Bacteria and Optimization of Fermentation Conditions from Poultry
- Isolation, Molecular and Phylogenetic Analysis of Porcine Encephalomyocarditis Virus Strain HLJ in China
