Oral microbiome and risk of malignant esophageal lesions in a high-risk area of China:A nested case-control study
Fangfang Liu ,Mengfei Liu ,Ying Liu ,Chuanhai Guo ,Yunlai Zhou ,Fenglei Li ,Ruiping Xu,Zhen Liu,Qiuju Deng,Xiang Li,Chaoting Zhang,Yaqi Pan,Tao Ning,Xiao Dong,Zhe Hu,Huanyu Bao,Hong Cai,Isabel Dos Santos Silva,Zhonghu He,Yang Ke
1Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing),Laboratory of Genetics,Peking University Cancer Hospital &Institute,Beijing 100142,China;2Novogene Co.,Ltd,Beijing 100080,China;3Hua County People’s Hospital,Anyang 456400,China;4Anyang Cancer Hospital,Anyang 455000,China;5 Department of Non-communicable Disease Epidemiology,London School of Hygiene &Tropical Medicine,London WC1E 7HT,UK
Abstract Objective:We aimed to prospectively evaluate the association of oral microbiome with malignant esophageal lesions and its predictive potential as a biomarker of risk.Methods:We conducted a case-control study nested within a population-based cohort with up to 8 visits of oral swab collection for each subject over an 11-year period in a high-risk area for esophageal cancer in China.The oral microbiome was evaluated with 16S ribosomal RNA (rRNA) gene sequencing in 428 pre-diagnostic oral specimens from 84 cases with esophageal lesions of severe squamous dysplasia and above (SDA) and 168 matched healthy controls.DESeq analysis was performed to identify taxa of differential abundance.Differential oral species together with subject characteristics were evaluated for their potential in predicting SDA risk by constructing conditional logistic regression models.Results:A total of 125 taxa including 37 named species showed significantly different abundance between SDA cases and controls (all P<0.05 &false discovery rate-adjusted Q<0.10).A multivariate logistic model including 11 SDA lesion-related species and family history of esophageal cancer provided an area under the receiver operating characteristic curve (AUC) of 0.89 (95% CI,0.84-0.93).Cross-validation and sensitivity analysis,excluding cases diagnosed within 1 year of collection of the baseline specimen and their matched controls,or restriction to screenendoscopic-detected or clinically diagnosed case-control triads,or using only bacterial data measured at the baseline,yielded AUCs>0.84.Conclusions:The oral microbiome may play an etiological and predictive role in esophageal cancer,and it holds promise as a non-invasive early warning biomarker for risk stratification for esophageal cancer screening programs.
Keywords:Early warning biomarker;esophageal squamous cell carcinoma;oral microbiome;risk prediction
Introduction
Esophageal cancer is the seventh most common cancer worldwide (1).Fifty-five percent of new cases occur annually in China,and 90% of these are esophageal squamous cell carcinoma (ESCC) (2).Tobacco and alcohol consumption are well-established risk factors for esophageal cancer in western countries,but contribute little to ESCC incidence in high-risk areas such as Anyang of China (2).The etiology of ESCC needs to be further investigated (2).Most ESCC cases are diagnosed at an advanced stage which confers an unfavorable prognosis.Early detection has been shown to improve survival and reduce mortality from the disease (2), with upper gastrointestinal endoscopic screening being widely accepted as an optimal secondary prevention strategy for esophageal lesions of severe dysplasia and above (SDA),including severe squamous dysplasia,carcinomain situ(CIS),and ESCC,in high-risk populations.However,this approach has disadvantages such as its potential for complications (3).Identification of high-risk individuals in the general population through the use of minimallyinvasive biomarkers could help to maximize the benefits of endoscopic screening by targeting those most likely to benefit.
Emerging evidence has linked the human microbiome with diseases such as cancer.The development of microbiome-based risk prediction models for some types of cancer,such as colorectal cancer,has opened new research avenues,by demonstrating that the microbiome may be a valid non-invasive biomarker of risk (4,5).Oral bacteria,in particular periodontal pathogens,together with indicators of oral health (e.g.,tooth loss) have been reported to be associated with ESCC and its precursor lesions (2,6,7).The anatomic proximity of the esophagus to the oral cavity likely renders the esophagus vulnerable to the effects of oral dysbiosis (8).We thus hypothesized that the oral microbiome is associated with the risk of developing SDA lesions and thus may be useful as a non-invasive biomarker of risk for SDA lesions.Only a few studies have so far characterized the oral microbiome in SDA lesions and interpretation of their findings has been hampered by the fact they relied on a single measurement taken from a oneoff oral specimen collection,did not use optimal methods of statistical analysis for differential comparison of taxa,and did not evaluate the predictive value of oral bacteria (7,9).Given the dynamics of the human microbiome (5),prospective follow-up studies with repeat specimen sampling are warranted to better understand the role of oral microbiome in malignant esophageal lesions.
The present case-control study,nested within a population-based cohort in a high-risk area for esophageal cancer in rural China with collection of multiple (up to 8)oral swabs over an 11-year follow-up period (10,11),aims to assess the association between oral microbiome and the risk of esophageal SDA and to investigate the potential value of this biomarker in predicting risk.
Materials and methods
Study design and participants
The subjects for this nested case-control study were selected from the prospective population-based endoscopic Anyang Esophageal Cancer Cohort Study (AECCS;9,035)and its oral sub-cohort (4,073) in rural Anyang,China,as previously described (10,11).Eligible participants (i.e.permanent residents in cluster-sampled villages,aged 25-65 years,with no prior history of cancer,cardiovascular illness,or infection with Hepatitis B,Hepatitis C,or Human Immunodeficiency Viruses) were visited in their villages a maximum of 8 times for collection of oral swabs including 3 visits at 2.5-year intervals from 2006 to 2013(endoscopic inspection of the esophagus was also performed at each visit),and 5 bi-annual visits from 2013 to 2015 (Figure 1).
Cases and controls were selected from AECCS who had provided a baseline oral swab at enrollment into the cohort(Figure 1).Cases included both screen-endoscopic-detected SDA cases and clinically diagnosed SDA cases diagnosed after collection of the baseline oral swab,but prior to July 2017,when follow-up of the cohort for the present analysis ended.The clinically diagnosed SDA cases were identified through annual active door-to-door interviews and through passive linkage with claims data from the New Rural Cooperative Medical Scheme.For each case,two controls were randomly selected among cohort members who did not have SDA at the time of diagnosis of the case(incidence density sampling) matching on gender,village of residence,age at cohort entry (5-year intervals),and number and timing (±1-year) of oral swab collection and endoscopic examination.
The follow-up time for the included cases and controls was estimated from time of enrolment into the cohort to the time of a SDA diagnosis for cases,and corresponding time for their two matched controls.The median follow-up time was calculated by using the reversed Kaplan-Meier method (12).
The study was performed in accordance with the Declaration of Helsinki.Research protocols were approved by the Institutional Review Board of the Peking University Cancer Hospital &Institute.All participants provided written informed consent.
Oral specimen and questionnaire data collection
Using saline-moistened cotton swabs,exfoliated oral cells were collected from the upper and lower lips,left and right sides of the hard palate,the buccal mucosa,the top and the bottom of the tongue,and the surface of the gingiva.Cells were rinsed with 0.9% saline solution and frozen at -80 ℃pending testing after centrifugation (10,11).A total of 143 pre-diagnosis oral specimens were provided by the 84 cases(48 cases provided only one specimen;26 provided two;6 provided three;and 4 provided five or more specimens).Of these 143 specimens,49 were collected within 15 d before the diagnosis of the SDA lesion,which were all provided by screen-endoscopic-detected SDA cases (Figure 1).
A one-on-one computer-aided interview on demographic characteristics and potential risk factors for esophageal cancer (~50 items) was administered by a trained interviewer at the baseline visit conducted at enrolment into the cohort.
Laboratory handling and bioinformatics
DNA was extracted using the E.Z.N.A.Mag-Bind Tissue DNA Kit (Omega Bio-Tek,Inc.,Norcross,USA).The 16S ribosomal (rRNA) gene V3-V4 regions were amplified using universal primers (341F 5’-CCTAYGGGRBGCA SCAG-3’ and 806R 5’-GGACTACNNGGGTATCTA AT-3’) and sequenced on the Ion S5 XL sequencing platform.
Multiplexed and barcoded sequences were deconvoluted.High-quality sequences were obtained according to the Cutadapt (V1.9.1) quality-controlled process.Chimera sequences were detected using the UCHIME algorithm and then removed.Filtered sequence reads were clustered into operational taxonomic units (OTUs).OTUs with a mean relative abundance ≥0.001% were assigned to taxa using the expanded Human Oral Microbiome Database(eHOMD) with ≥97% sequence similarity.From 428 oral specimens (143 from cases;285 from controls),we obtained 32,917,908 (,76,911±10,444) high-quality sequence reads,with similar numbers of reads per specimen for both case and control groups (Supplementary Table S1).A total of 15 phyla,44 classes,79 orders,147 families,324 genera,and 720 species were identified and included in our analysis.
Quality control
Specimens from any given case-control triad were included in the same batch and tested blindly.Ten replicate aliquots of oral cell DNA from eight volunteers were mixed and included in the 5 sequencing batches (2 replicates per batch) as quality control samples.The intra-plate and interplate coefficients of variation (CV) for the Shannon diversity index and observed-species of the quality control samples were all <7.0% (Supplementary Table S2).Rarefaction curves and the species-accumulation boxplot indicate sufficient sequence depth and adequate sample size,respectively (Supplementary Figure S1,S2).
Statistical analysis
Dataset description
To obtain stable measurements of bacterial populations and to account for heterogeneity in the number and timing of specimens from different subjects,a full averaged dataset was produced for bacterial abundance comparison and prediction model establishment.This dataset contained a total of 84 SDA cases providing 143 oral specimens and 168 matched controls providing 285 oral specimens(Figure 1).The bacterial population values for each specimen provided by an individual were averaged to produce single values at each taxonomic level (e.g.,class,species) for that individual.
Overall diversity comparison
Trends of α diversity (Shannon index) with years of specimen collection prior to diagnosis of malignant esophageal lesions were evaluated using linear mixed-effects (LME)regression (LME function in R) by treating the subject as a random effect.Differences in α diversity between cases and controls were also analyzed by LME regression.Differences in overall bacterial community composition (β diversity) according to case and control status were assessed with permutational multivariate analysis of variance(PERMANOVA;adonis function in R) by treating matched case-control triads as strata.
Association analysis
To compare relative abundance of taxa in SDA cases and controls at each level (phylum to species),DESeq (DESeq2 package,R) with variance and mean linked by local multivariable regression,which is an optimal method for microbiome data analysis,was performed based on the full averaged dataset (13,14). Taxa were considered significantly differentially abundant between groups if P<0.05 &the false discovery rate (FDR)-adjusted Q<0.10.
Prediction analysis
To establish a final prediction model for risk of SDA lesions and determine which species should be retained in the final prediction model,analysis was carried out based on the full averaged dataset as follows (Supplementary Figure S3).For each of the named and cultured differential species selected by DESeq analysis,multiple speciesspecific classifiers (low carriagevs.high carriage),derived from a series of cut-off points ranging from quantiles 5% to 95% (5% per step) of the relative abundance in the control group,were evaluated in separate univariate conditional logistic regression models (dependent variable: SDA status).Taking both error probability and effect size into consideration,the optimal classifiers for each species with the lowest sum of odds ratio rank and reverse P value rank,together with subject characteristics were included in the multivariate conditional logistic model.Their retention in the final prediction model was determined using the Akaike information criterion (step AIC function,MASS package,R).The area under the receiver operating characteristic curve (AUC) and the DeLong test were adopted to evaluate the performance of the prediction model.Leave-one triadout cross-validation was used to estimate the generalization error on the basis of predicted probabilities for each casecontrol triad from models built on all the remaining triads.
Temporal stability assessment
To assess the temporal stability of the relative abundance of oral species within and between individuals,we used the metrics of mean,standard deviation,and CV as employed by Utteret al(15).A total of 128 specimens provided from 10 cases and 18 controls (each with three or more serial specimens) were included in this analysis.For each species,means and CVs for each individual were calculated based on the relative abundance of three or more specimens from this individual.The mean CV (intra-individual CV) was the mean of all the CVs calculated from all included individuals;the overall CV was calculated based on the relative abundances from all specimens provided by all included individuals.
Sensitivity analysis
To reduce the likelihood of reverse causation,the following sensitivity analyses were performed:1) including only cases diagnosed more than 1 year after collection of the baseline specimen and their matched controls,but using the average microbiome data from all their collected oral specimens(strictly averaged dataset;55 cases with 87 specimens and 110 controls with 176 specimens);2) including all enrolled study subjects,but using only microbiome data from their oral specimens collected at baseline (full baseline dataset;84 cases and 168 controls,each with a single baseline specimen);and 3) including only cases diagnosed more than 1 year after collection of the baseline specimen and their matched controls,and using only microbiome data from their oral specimens collected at baseline (strict baseline dataset;55 cases and 110 controls,each with a single baseline specimen).Also,stratified analysis was carried out by separating screen-endoscopic-detected and clinically diagnosed case-control triads.Model performance was also recalculated using 75th quantile cut-off points instead of optimal thresholds for classification of lowvs.high carriage of the predictive bacteria.
All multivariate models included level of education,type of employment,cigarette smoking,alcohol consumption,and family history of esophageal cancer unless otherwise specified.All analysis was carried out using R statistical software (Version 3.4.3;R Foundation for Statistical Computing,Vienna,Austria).P values less than 0.05 (twosided) were considered to be statistically significant.
Results
Participant characteristics
Median follow-up time for study participants was of 8.7(interquartile range:5.2-9.7) years.Cases and controls were of a similar age and gender (matching variables) and had a similar educational level,type of employment,and cigarette smoking and alcohol intake habits.Cases were,however,more likely to have a family history of esophageal cancer than controls (15.5%vs.7.1%,P=0.037) (Table 1).
Overall microbiome diversity in relation to malignant esophageal lesions
No significant trend over years of specimen collection prior to diagnosis of malignant esophageal lesions in the Shannon diversity index was found for SDA cases (P=0.124)or controls (P=0.425) (Supplementary Figure S4).Between groups,cases showed a slightly higher Shannon diversity index than controls (P=0.044).Cases differed significantly from controls in overall oral microbiome composition (β diversity) neither when measured by unweighted (P=0.248)nor when measured by weighted UniFrac distances(P=0.590) (Supplementary Figure S4).
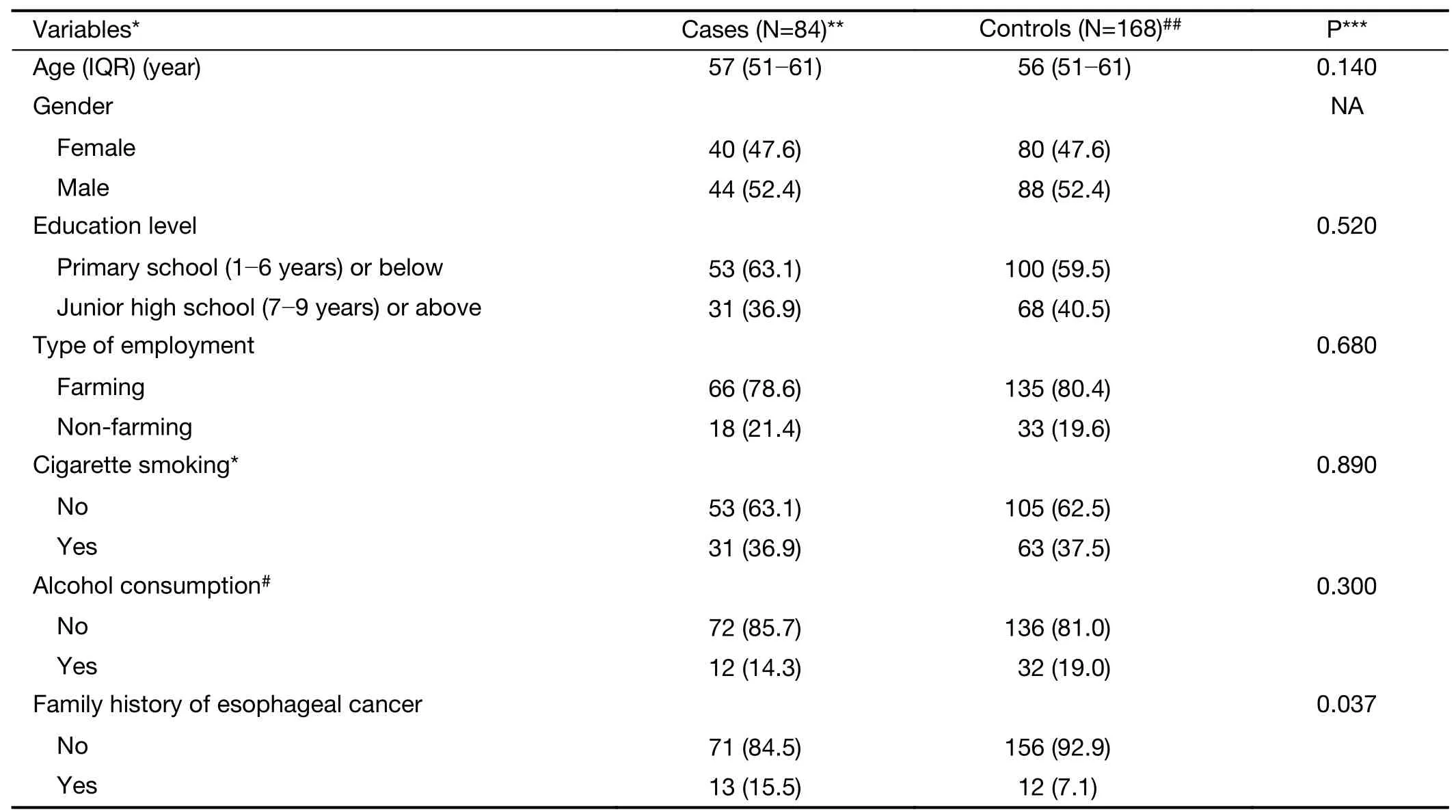
Table 1 Selected demographic and baseline behavioral characteristics of cases of malignant esophageal lesions and matched controls from Anyang,China,2006-2017
Taxa associated with malignant esophageal lesions
Based on DESeq analysis,a higher abundance of 15 taxa including 6 species was found to be associated with decreased risk of SDA lesions,and a higher abundance of 110 taxa including 66 species was associated with increased risk of SDA lesions (all P<0.05 &Q<0.10).Of the 72 species with differential abundance between cases and controls,37 were named and cultured according to eHOMD (Figure 2;Supplementary Table S3).The species Fusobacterium nucleatum which is known to be associated with periodontal diseases (16,17),and all of its higher taxonomic levels were among the above taxa with positive associations.
Species-level prediction model for malignant esophageal lesions
A total of 11 species of 37 named and cultured differential species selected by DESeq analysis,together with family history of esophageal cancer were retained in the final model predicting risk of SDA lesions.These species and their corresponding optimal cut-off points for relative abundance are shown inTable 2.Higher carriage of the predictive species was associated with increased risk of SDA lesions,with adjusted ORs ranging from 1.98 (Prevotella baroniae) to 10.93 (Lachnoanaerobaculum umeaense).For Fusobacterium nucleatum,the adjusted OR was 3.85 (95%CI,1.12-13.24).
The AUC was 0.89 (95% CI,0.84-0.93) for the final model,which was constructed based on the full averaged dataset (Figure 3).Leave-one triad-out cross-validation provided similar AUC statistics (AUC,0.89;95% CI,0.88-0.89).After exclusion of cases which were diagnosed within 1 year of collection of the baseline specimen and matched controls for these cases,the AUCstrictlyaverageddatasetwas 0.85 (95% CI,0.80-0.91).When stratifying by case type,the AUC for screen-endoscopic-detected cases and matched controls was 0.90 (95% CI,0.86-0.95) and the AUC for clinically diagnosed SDA cases and matched controls was 0.88 (95% CI,0.81-0.94) (SupplementaryFigure S5).Additionally,when analysis was limited to baseline specimens,the AUCs were also similar [AUCfullbaselinedataset:0.84 (95% CI,0.79-0.89);AUCstrictbaselinedataset:0.85 (95% CI,0.79-0.91)].When the 75th quantile was used as the cut-off point,the AUCs remained above 0.78(Supplementary Figure S6).
Temporal stability of predictive species
For the 11 predictive species,shifts in the relative abundance over time (~8 years) within a single individualwere generally fluctuations around an individual mean,which did not exhibit any increasing or decreasing trend(Supplementary Figure S7).These species had lower intraindividual CVs within each subject (average of intraindividual CVs=107.9%) than overall CVs across all specimens provided by all included subjects with multiple sampling (average of overall CVs=251.2%),resulting in an average ratio of intra-individual CV and overall CV of 0.4(Figure 4;Supplementary Table S4),and no appreciable discrepancy was found in cases and controls.
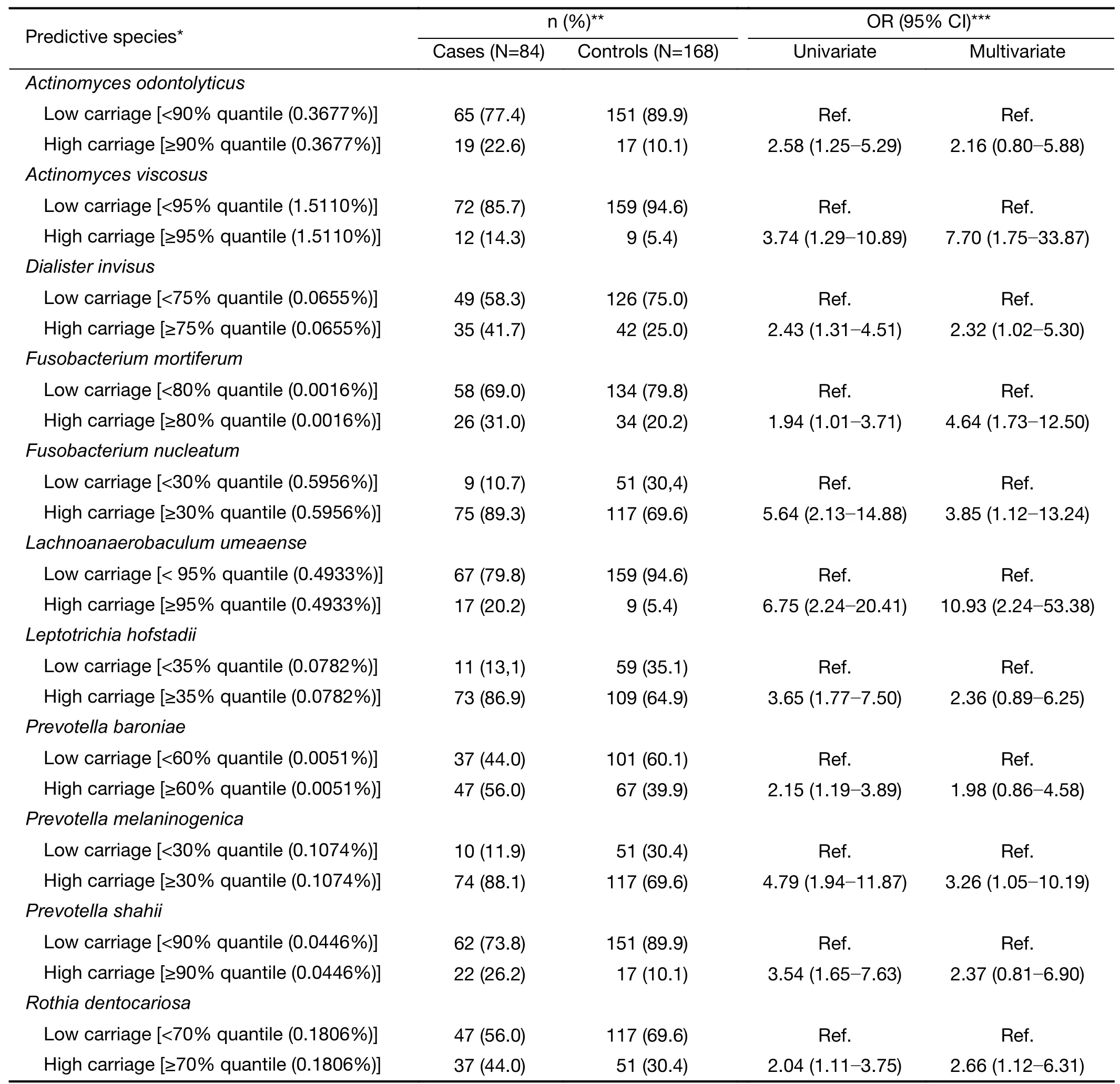
Table 2 Structure and OR of oral microbiome-based prediction model for risk of malignant esophageal lesions in Anyang,China,2006-2017
Discussion
One of the key problems in current microbiome-oncology research is the lack of prospective longitudinal studies,and the execution of such studies within the microbiome field is challenging but is urgently needed to provide direct evidence of causation (18). In this first dynamic longitudinal investigation of the causative and predictive role of oral microbiome in malignant esophageal lesions,we show that specific oral species are differentially abundant with respect to disease status,and a panel of 11 bacteria can accurately distinguish SDA cases from healthy controls.It seems likely that the oral microbiome has an etiological role in esophageal cancer,and it holds promise as a non-invasive early warning biomarker for risk stratification for esophageal cancer screening programs.The oral microbiome presents an opportunity to better understand esophageal cancer and how it might be prevented.
Cross-sectional studies and case-control studies have reported distinct differences in upper digestive tract microbiome between gastroesophageal reflux disease (19-21),Barrett’s esophagus (19-22),esophageal adenocarcinoma (EAC) (19,23),esophageal squamous dysplasia(24),or ESCC (6,7,9) cases and controls.Additionally,poor oral health,including poor periodontal health,tooth loss,and irregular teeth brushing,has repeatedly reported to be linked with the risk of malignant esophageal lesions(2,6,7,25,26),supporting the hypothesis that oral healthrelated microbial environment (e.g.oral dysbiosis) may play a role in the carcinogenesis of esophageal epithelium.However,only one study to date has prospectively examined whether upper digestive tract microbiome influences risk for subsequent esophageal cancer.In a nested case-control study conducted in USA,Peterset al.evaluated oral bacteria using 16S rRNA gene sequencing in prediagnostic mouthwash specimens from n=81/160 EAC and n=25/50 ESCC cases/matched controls(7).They found that several specific species were associated with cancer risk (For EAC,Tannerella forsythiaandStreptococcus pneumoniaewith P<0.05;For ESCC,Prophyromonas gingivaliswith a P value of 0.09).In our study,at the species level,we found that dozens of oral bacteria were associated with malignant esophageal lesions.Using a larger sample size and a more appropriate statistical method for abundance comparison (Deseqvs.Conditional logistic regression) may partially explain the larger number of cancer related-species we found.Our results are in keeping with the current concept that mixed communities of pathogens collectively drive disease progression,rather than individual species working in isolation (13,27).The molecular mechanisms by which the microbiome may be involved in the aetiopathogenesis of cancer have been extensively discussed.All the proposed mechanisms,including genomic integration,genotoxicity,inflammation,immunity and metabolism,seem to ultimately converge on final common pathways of enhanced capacity of replication and dedifferentiation,and prolonged host cell survival (18).Further study about the oncogenic mechanisms by which the oral microbiome,alone or alongside with environmental and host factors,may initiate and/or drive the carcinogenesis of esophageal cancer is warranted.
All 11 SDA lesion-associated oral species included in this prediction model were anaerobic bacteria.Four of these(Actinomyces odontolyticus,Actinomyces viscosus,Lachnoanaerobaculum umeaense,andRothia dentocariosa)were Gram-positive,and all the others were Gramnegative.For the most part,these bacteria live in harmony with the host,generally in a commensal state.However,under certain circumstances,this commensal relationship may break down,and these bacteria may be involved in human disease.While most of these 11 bacteria have been reported to have associations with dental cavities and periodontal diseases,some have a linkage with autoimmune diseases (e.g.,Lachnoanaerobaculum umeaenseinduces animal models of celiac disease),and some are correlated with cancer (e.g.,Prevotella melaninogenica shows increased abundance in oral cancer patients) (28,29).
Fusobacterium nucleatumis a well-known periodontal pathogen identified as one bacterium among these 11 predictive bacteria (17).This bacterium has frequently been found to be enriched in colorectal cancer tissues,and it has been suggested that it influences colorectal carcinogenesis through activation of cellular proliferation pathways,and by suppression of the antitumor immune response (30).Given the proximity of the esophagus to the oral cavity,Fusobacterium nucleatummay also play a role in esophageal cancer.Using real-time polymerase chain reaction (PCR),Yamamuraet al.reported that 23% of ESCC tumor tissues containFusobacterium nucleatumDNA,which is greater than that in normal adjacent esophageal tissue (P=0.021).Moreover the presence ofFusobacterium nucleatumis associated with significantly shorter survival time in patients with ESCC (17).Prophyromonas gingivalishas also been found to be associated with ESCC (6,31) but we did not observe a significant difference in its relative abundance in cases of malignant esophageal lesions and their matched controls in our study population.One possible explanation is that these studies assessed for the presence ofProphyromonas gingivalisrather than its relative abundance.A recent report from a study also conducted in Henan,China supported our findings (32).This study showed that tumor tissues had a greater abundance ofFusobacteriumthan paired non-tumor tissues (67 pairs),but no significant difference in the abundance ofPorphyromonaswas observed.Altogether,certain oral bacterial species,includingFusobacterium nucleatum,might contribute to and predict the carcinogenesis of malignant esophageal lesions.Identification and manipulation of carcinogenic oral bacteria may offer actionable strategies for prevention of this highly fatal disease.
Sensitivity analysis of the performance of this predictive model showed that after excluding cases diagnosed within 1 year of collection of the baseline specimen and their matched controls,the AUC remained high.More importantly,this model performed well for both endoscopically screened and clinically diagnosed casecontrol triads.Identification of early esophageal lesions is a primary concern of endoscopic screening,as early lesions are of greater clinical and public-health importance than clinically diagnosed SDA cases.Most of the latter are advanced lesions which are less likely to benefit from treatment (33).When analysis was limited to the baseline data of the AECCS cohort,the final prediction model stably yielded good discriminatory results.Additionally,we found that the overall CV across all specimens from all individuals with multiple sampling was about 2.3 times higher than the intra-individual CV (251.2%vs.107.9%),indicating the time-stability of these species.Consistent with our findings,previous studies have also suggested that the abundance of core members of the oral microbiome is fairly stable over time,although more precise microbiome estimates can be obtained by measurement at multiple time points (34,35).These findings indicate our model may also be generalized in settings without intensive sampling,where single-time only specimen collection is employed.Altogether,the oral microbiome holds promise as a noninvasive early warning biomarker for risk stratification for esophageal cancer screening programs.
Due to the high incidence of ESCC in China,endoscopic surveillance of esophageal cancer has come to be viewed as an important national undertaking.Since 2006,more than 1 million endoscopies sponsored by the Chinese government have been carried out in several regions of high ESCC incidence (36,37).The cost of endoscopic examination is high,and endoscopy is an invasive procedure.Therefore,identification of high-risk subjects in the general population is a strategy which is cost-effective.We previously established an easy-to-use risk prediction model for ESCC using demographic and lifestyle factors (37).Use of the model in screening could have allowed 27% of subjects 60 years or younger and 9%of subjects older than 60 years to avoid endoscopy without missing SDAs,which means that approximately 16.6% of endoscopies in total could have been avoided.Oral microbiome markers could be combined in the future with demographic/lifestyle factors to construct a more comprehensive and accurate prediction model for malignant esophageal lesions.Incorporation of modelbased risk assessment into large-scale screening programs for esophageal cancer with endoscopic examination of only high-risk individuals identified by the model may render screening programs safer and more cost-effective.
Although this is the first population-based nested casecontrol study with multiple sampling,its limitations should be noted.First,due to the prospective matched casecontrol design,the added predictive value of matching factors such as age could not be evaluated.Second,despite access to a repository of >40,000 oral specimens from the AECCS cohort,the absolute number of case-control triads included in this study with multiple specimens was still relatively small due to the low incidence of SDA lesions which rendered some temporal analysis inaccurate.Third,an independent external cohort is needed to validate the results of this study.
Conclusions
This prospective study shows that specific members of the oral microbiome are associated with the subsequent risk of malignant esophageal lesions,and a model based upon a panel of 11 lesion-associated oral species achieves excellent classification performance.This lends support to the hypothesis that the oral microbiome may play a causative and predictive role in the aetiopathogenesis of esophageal cancer,and raises the possibility that the non-invasive microbiome biomarkers,alone or in combination with other factors,may enable risk-stratification of esophageal cancer screening programs in the future.Our findings have implications for a personalized approach to primary and secondary prevention of esophageal cancer.Further studies are needed to validate our findings and to elucidate mechanisms of the causal relationship.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No.30930102,82073626,81502855,81773501),the National Key R&D program of China (No.2016YFC0901404),the National Special Programme of Scientific and Technological Resources Investigation (No.2019FY101102), the Digestive Medical Coordinated Development Center of Beijing Hospitals Authority (No.XXZ0204),the Beijing Natural Science Foundation (No.7182033), the Beijing Municipal Administration of Hospital’s Youth Programme (No.QML20171101),and the Science Foundation of Peking University Cancer Hospital (No.2020-7).
Footnote
Conflicts of Interest:The authors have no conflicts of interest to declare.

Table S1 Number of filtered sequence reads per specimen*
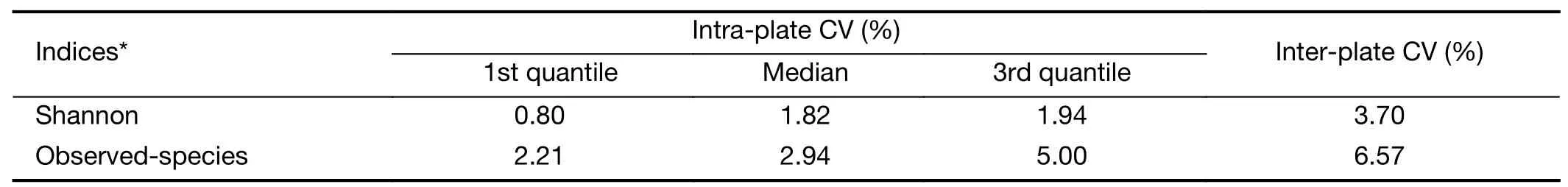
Table S2 Coefficients of variation for Shannon diversity index and number of species observed among quality control samples
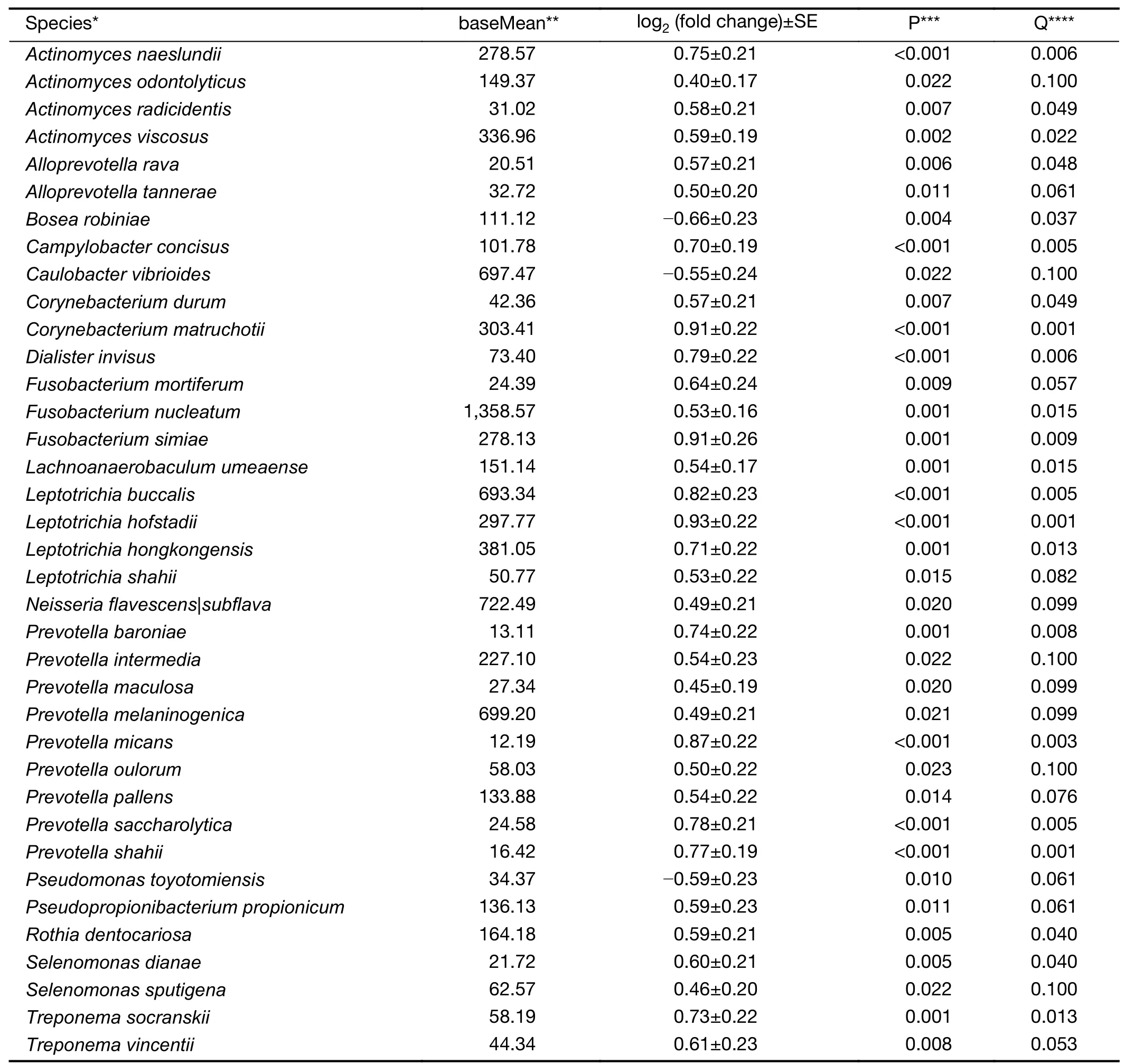
Table S3 Species with differential abundance in cases of malignant esophageal lesions and controls based on DESeq analysis in Anyang,China,2006-2017

Table S4 Temporal stability of relative abundance of predictive oral species within an individual with three or more specimens from Anyang China,2006-2017*
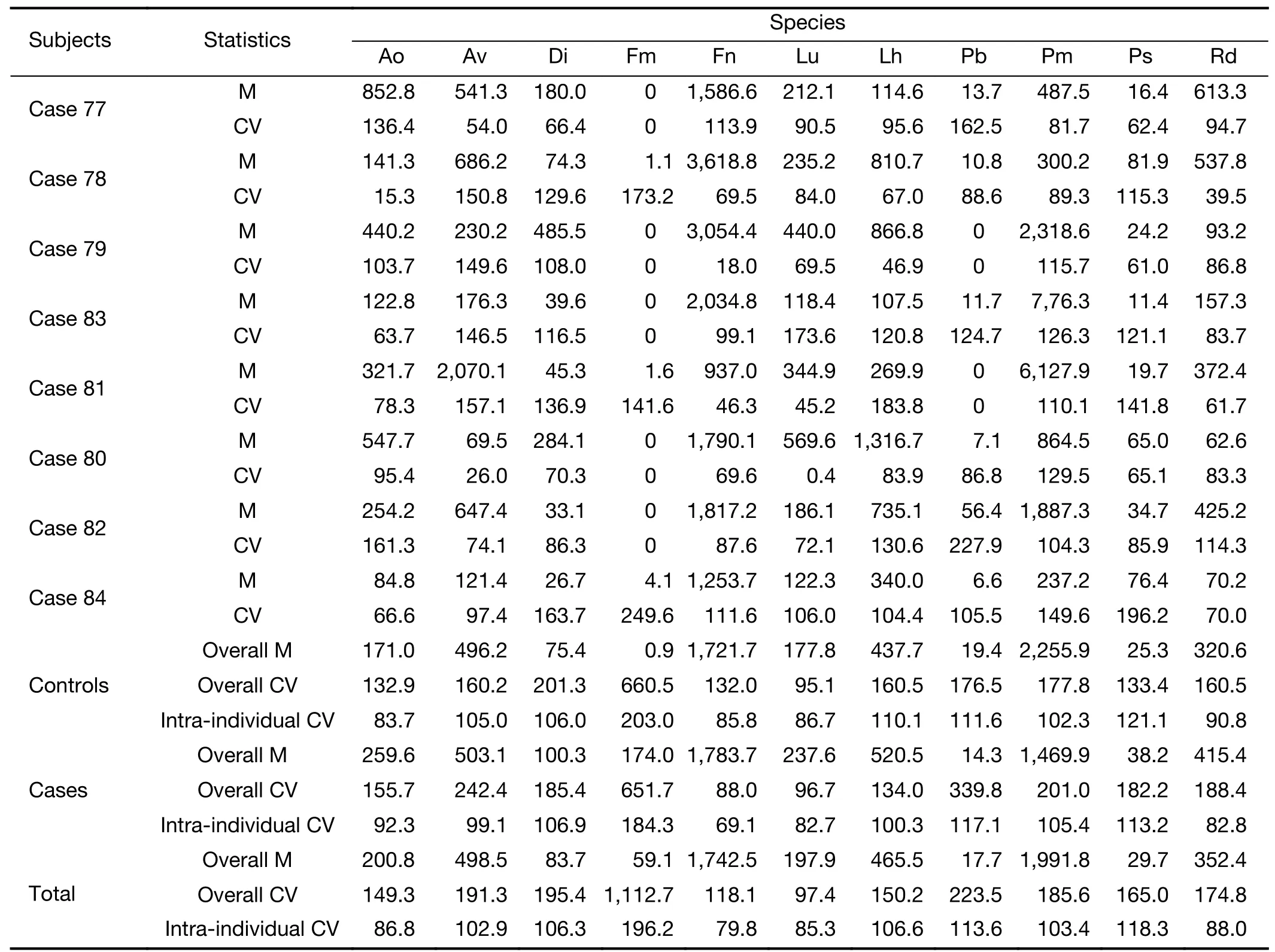
Table S4 (continued)
 Chinese Journal of Cancer Research2020年6期
Chinese Journal of Cancer Research2020年6期
- Chinese Journal of Cancer Research的其它文章
- Development and validation of prognostic nomogram based on log odds of positive lymph nodes for patients with gastric signet ring cell carcinoma
- Separate lateral parametrial lymph node dissection improves detection rate of parametrial lymph node metastasis in early-stage cervical cancer:10-year clinical evaluation in a single center in China
- Prognostic factors affecting long-term outcomes in patients with brain metastasis from esophageal carcinoma
- Precision screening for esophageal squamous cell carcinoma in China
- Trends and risk factors of lung cancer in China
- Current epidemiology of pancreatic cancer:Challenges and opportunities
