Identification of Mulberry Endophytic Fungus Macrophomina phaseolina Strain
Xue’e DOU Jingrui HAN

Abstract An endophytic fungus MOD-1 was isolated from the roots of mulberry trees, and was identified as Macrophomina phaseolina by morphology and molecular biology. The 5.8 S rDNA/ITS region sequence of the strain has been registered in GeneBank with the accession number of EU250575.
Key words Macrophomina phaseolina ; Morphological identification; Molecular biological identification
Received: October 7, 2021 Accepted: November 9, 2021
Supported by National Cocoon and Silk Development Risk Fund (GJXBH 200732); Natural Science Foundation of Shandong Province (2007ZRB01872).
Xue’e DOU (1969-), female, P. R. China, master, livestock enginee, devoted to research about livestock breeding technology and management.
*Corresponding author. E-mail: 121780313@qq.com.
Mulberry trees are a medicinal and edible plant, which is treasured throughout the body. In agricultural production, the main use of mulberry trees comes from the breeding of silkworms with mulberry leaves and the production of silkworm cocoons for the development of various uses. With the successful breeding of high-protein feed mulberry trees, research on feed mulberry leaves as additives for various livestock and poultry feed has become increasingly common. Research and development personnel at all levels have conducted various studies on the potential utilization value of mulberry trees from different channels. In this study, based on the isolation and identification of endophytic fungi in mulberry trees and its diversity analysis[1] and the combination of morphological observation and Sudan III staining method, a MOD-1 strain[2] with high oil production and good stability was screened and identified by morphological and molecular biology methods as Macrophomina phaseolina . The identification of mulberry endophytic fungus M. phaseolina has not been reported at home and abroad.
Materials and Methods
Experimental materials
Experimental strain
The MOD-1 strain was isolated and preserved from mulberry trees by the Sericulture Pathology and Molecular Biology Laboratory of Shandong Agricultural University.
Experimental media
Solid PDA medium; TWA+WS medium[3]; liquid PDA medium.
Main reagents and instruments
Taq DNA polymerase, purchased from Sangon Biotech (Shanghai) Co., Ltd.; PCR primers, synthesized by Shanghai Boya Biology Co., Ltd.; agarose; Takara DV807 DNA gel recovery kit; phenol; chloroform; isoamyl alcohol; isopropanol; reagents such as anhydrous ethanol, all domestically produced.
HQL-150A constant temperature shaker; WDP biochemical multipurpose incubator; DHG-9023A electric constant temperature drying oven; RE-52AA rotary evaporator; TU-1810 ultraviolet-visible spectrophotometer; TGL-20G high-speed refrigerated centrifuge; 8002 type electric-heated thermostatic water bath; DYY-III type electrophoresis instrument; GDS-8000 type gel imaging analysis system; PTC-200 PCR instrument.
Experimental methods
Morphological identification of mulberry endophytic fungus MOD-1 strain
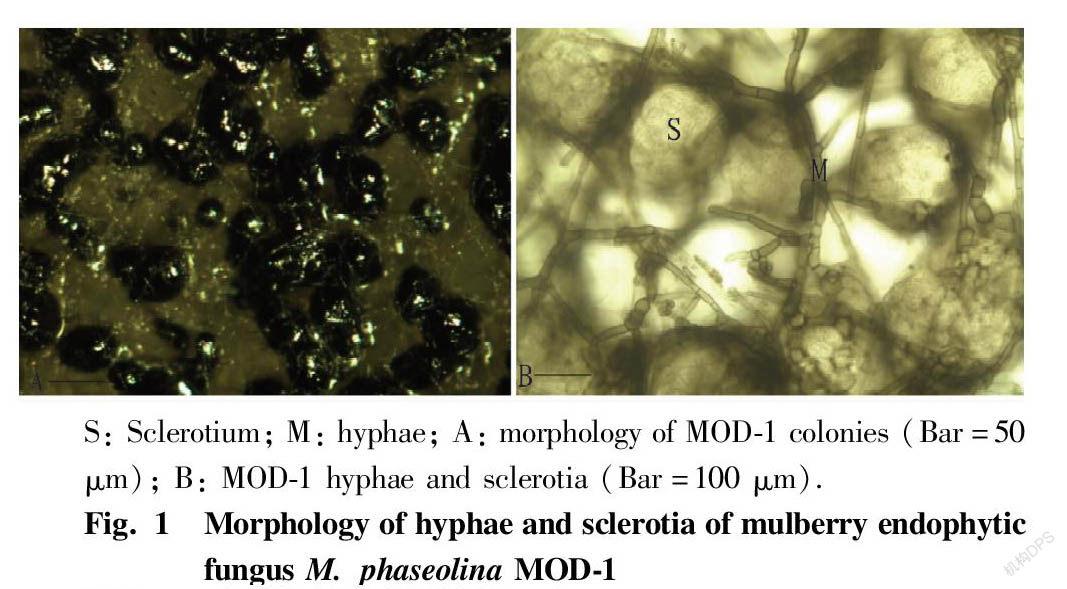
Spot planting method and fungal slide culture method are adopted to make glass slides[4], which were observed for hypha morphology, conidial structure and spore morphology under an optical microscope, and strain identification was carried out referring to the fungal morphology identification literature[5-7]. For the MOD-1 strain that did not produce spores under artificial culture conditions, methods such as colony wounding, near-ultraviolet radiation, TWA+WS medium culture, and short-term refrigeration in refrigerators were adopted to induce sporulation[4]. A digital camera was used to take pictures of fungal colony morphology in time and record relevant shapes at any time; and observation was performed under a Motic digital microscope and fields of view with typical characteristics were selected to take pictures and directly measure related data for description and recording.
Molecular biology identification of mulberry endophytic fungus MOD-1 strain
The genomic DNA of mulberry endophytic fungus MOD-1 strain was extracted by the improved CTAB method[8]. First, clean mycelia cultured in a PDA liquid medium at 26 ℃ on a shaker for 3 d were collected, and dried at 65 ℃ for 12 h. The mycelia were ground into powder in liquid nitrogen, and 20-25 mg was added into a 1.5 ml centrifuge tube containing 500 μl extraction solution (150 mM NaCl, 100 mM EDTA, 50 mM Tris·HCl, pH 8.0), obtaining a mixture after mixing well. The obtained mixture was then added with 50 μl 10% SDS solution, mixed gently again and incubated at 37 ℃ for 1 h. Next, it was added with 75 μl of 5 M NaCl solution, mixed gently, then added with 65 μl of CTAB/NaCl (10% CTAB, 0.7 M NaCl) solution, and mixed well, and the resultant system was kept at 65 ℃ for 10-20 min. After cooling to room temperature, it was added with an equal volume (700 μl) of chloroform: isoamyl alcohol (24∶1), and mixed by inversion for 5 min. Centrifugation (10 000 r/min, 12 min, room temperature) was performed, and the supernatant was transferred to another centrifuge tube. Next, 0.6 times of cold isopropanol (450 μl) was added to the supernatant, which was then gently inverted to mix evenly, and frozen at -20 ℃ for at least 1 h. After centrifugation (10 000 r/min, 10 min, 4 ℃), the precipitate was washed twice with 100 μl of 70% ethanol, and dried after absorbing ethanol at 37 ℃ for 20 min. It was then dissolved with 200 μl 1×TE (10 mM Tris·HCl; 1 M EDTA). The product was mixed with 6×Loading Buffer and detected by 0.8% agarose gel electrophoresis.
5.8 S rDNA/ITS gene fragment were amplified using universal primers ITS5 5’-GGAAGTAAAAGTCGT- AACAAGG-3’, ITS4: 5’-TCCTCCGCTTATTGATATGC-3’[9]. The 25 μl reaction system included 10×buffer 2.5 μl, 25 mmol/L Mg 2+ 3 μl, 10 mmol/L dNTP 2.0 μl, 10 μmol primers each 0.5 μl, Taq enzyme 0.15 μl, DNA template 2 μl, and sterile deionized water added to 25 μl. The amplification procedure started with pre-denaturation at 94 ℃ for 5 min, followed by 35 cycles of denaturation at 94 ℃ for 1 min, annealing at 56 ℃ for 1 min, and 72 ℃ for 1 min, and finally completed by extension at 72 ℃ for 10 min. The PCR amplification products were detected by 1.25% agarose gel electrophoresis, and observed by EB staining. Sequencing was completed by Sangon Biotech (Shanghai) Co., Ltd. The sequencing results were compared in GenBank with BLAST software. The ClustalX software was used for multiple sequence alignment, and the NJplot program was used for phylogenetic analysis.
Results and Analysis
Classification and identification of MOD-1 strains
Morphological identification of MOD-1 strain
The fungus grew rapidly on PDA medium at 28 ℃, and its colony diameter could reach 9 cm in 2 d. The hyphae were gray-white at first, separated, and not constricted at the separation positions; the apex hyphae had many branches, which were acute or nearly right-angled, and had a diameter of 15.60-31.87 μm. After 3 d, black particles began to appear near the center of colonies. After 5 d, the particles were densely distributed in the whole dish. The hyphae on the surface gradually became sparse, gray-black and shiny. The dark-brown particles were microsclerotia under microscopic examination, and no sporulation mechanism was found. The microsclerotia were scattered or adherent, mostly spherical, oval, and had a diameter of 156.00-202.82 μm, and a few of them were irregular. When the sclerotia were crushed, a large number of oil balls of different sizes overflowed, and thick dark-brown hyphae often grew on the sclerotia (Fig. 1). The colony dorsal view medium was blue-black. With reference to related literature[5], the fungus was identified as M. phaseolina .
Molecular biology identification of MOD-1 strain
A fragment with a molecular weight of about 600 bp was amplified from the MOD-1 strain. According to the alignment results of the 5.8 S rDNA/ITS region sequences of most fungi included in Genbank, the actual length of the entire 5.8 S rDNA/ITS was between 480 and 522 bp, and after aligning MOD-1 strain and 10 sequences downloaded from Genbank, a full-length sequence of 581 bp was taken for analysis. The GenBank sequence number of the 5.8 S rDNA/ITS region sequence of strain MOD-1 is EU250575. A phylogenetic tree was generated using the NJ (Neighbour-Joining) method in ClastalX, and the phylogenetic tree was tested with Bootstrap with 1 000 times of repetition. The phylogenetic tree of the MOD-1 strain constructed with Botryosphaeria iberica as an outgroup is shown in Fig. 2. Through phylogenetic analysis of the 5.8 S rDNA/ITS region of the MOD-1 strain, it was found that it was clustered with M. phaseolina (AF132795) on a branch, and combined with morphology, it was identified as M. phaseolina in mulberry trees.
Conclusions and Discussion
The traditional identification method of fungi is mainly based on the morphological characteristics of fungal hyphae, spore-forming structure, and spore morphology. Due to the complex nutritional requirements of many microorganisms in nature, some strains cannot grow on artificial media or can grow but do not produce sexual or asexual spores, while non-spore producing strains cannot be identified by traditional morphological methods, and have certain limitations. The development of molecular biology technology has made DNA molecules an effective means to distinguish species, varieties, and geographic strains, and the 5.8 S rDNA/ITS region of fungi is widely used in the study of fungal molecular systems due to its multiple copies in the primary structure and the high conservation of hypervariable regions. In this study, it was found that the mulberry endophytic fungus M. phaseolina did not produce spores under artificial culture conditions, which brought certain difficulties to its morphological identification. The identification of this fungus using the 5.8 S rDNA/ITS region is of greater significance. The position of the mulberry endophytic fungus M. phaseolina MOD-1 in the phylogenetic tree established in this study (Fig. 2) is highly consistent with the pathogenic fungus M. phaseolina [10] of soybean blight isolated from the northern United States, and they shared a homology of 100% and had a genetic similarity of 85%. M. phaseolina is mostly studied at home and abroad as a plant pathogen. On ginkgo, pine, China fir[11] and other diseased seedlings, it generally does not produce conidia, but only produces small sclerotia, which are black brown, smooth in surface, oblate or oval, small as powder; the conidia produced on sesame, jute[11], cantaloupe[12] and other hosts are buried in the tissues, with the ostiole opening outside the epidermis, and they are oval, colorless, and single-celled; and under artificial culture conditions, no spores are produced, and only microsclerotia are produced[7]. However, there has been no report on the pathogenicity of this kind of fungus to mulberry trees at home and abroad, and the endophytic fungus has not been found to be pathogenic to mulberry trees (data not attached). The relationship between this mulberry endophytic fungus and mulberry trees and its infection and colonization mechanism need to be further studied.
References
[1] DOU XE, MOU ZM, HAN JR, et al. Isolation and identification of endophytic fungi in mulberry and its diversity analysis[J]. Science of Sericulture, 2008, 34(1): 6-10. (in Chinese)
[2] DOU XE. Isolation and identification of mulberry endophytic fungi and study on oil-producing fungus Macrophomina phaseolina MOD-1[D]. Tai’an: Shandong Agricultural University, 2008: 31. (in Chinese)
[3] ZHANG TY. Chinese fungi[M]. Beijing: Science Press, 2003: 19. (in Chinese)
[4] FANG ZD. Plant disease research methods[M]. Beijing: China Agriculture Press, 1998: 112-121; 146-149. (in Chinese)
[5] WEI JC. Fungus identification manual[M]. Shanghai: Shanghai Scientific & Technical Publishers, 1979: 408-522. (in Chinese)
[6] SHAO LP, SHEN RX, ZHANG SX, et al. Fungal taxonomy[M]. Beijing: China Forestry Publishing House, 1984: 54-346.
[7] XU ZG. General plant pathology[M]. Beijing: China Agriculture Press, 2000: 95-96. (in Chinese)
[8] HE YQ. Improvement of the method of fungal mycelium culture and DNA extraction[J]. Mycosystema, 2000, 19(3): 434. (in Chinese)
[9] WHITE T, BRUN J, LEE S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetic[M]. New York: American Academia Press, 1990: 315-322.
[10] HARRINGTON TC, STEIMEL JP, WORKNEH F, et al. Molecular identification of fungi associated with vascular discoloration of soybean in the North Central United States[J].Plant disease, 2000, 84(1): 83-89.
[11] FANG ZD, YUAN XR, LI CD, et al. Control experiment of ginkgo stem rot[J]. Acta Phytopathologica Sinica, 1956, 2(1): 43-54. (in Chinese)
[12] WANG XD, LI GY. Identification of the pathogen of Hami melon charcoal rot and its biological characteristics[J]. Journal of Shihezi University: Natural Science, 2004, 22(Sup.): 98-100. (in Chinese)
- 农业生物技术(英文版)的其它文章
- Rice Blast Resistance-associated Genes Based on Different RNA-seq Resources
- Research Progresses on QTLs for Main Grain Shape Genes in Rice
- Effects of Raising Chickens Under Moringa oleifera
- Preliminary Research on Radiation Breeding of Pteroceltis tatarinowii Maxim
- Comparison of Spring Radish Varieties with Entire Leaves
- Occurrence and Chemical Control Techniques of Rice Black-streaked Dwarf Disease in Rongshui County

