Rice Blast Resistance-associated Genes Based on Different RNA-seq Resources
Tao ZHOU Yalong ZHANG Dewei TAI Xiaozhong ZHANG Xueshi LIU Wei ZHANG



Abstract Rice is the main food crop in Asian countries, and rice blast causes an annual loss of rice yield of 30%-35%. Cultivating rice blast resistant varieties and discovering resistance genes are important means to prevent rice blast. By analyzing the changes in rice transcriptome expression levels before and after infection by Magnaporthe oryzae of different physiological races, we found that 5 transcripts were up-regulated together after infection, Os07t0125500-01, Os07t0129300-00, Os07t0126301-01 and Os07t0124900-01 belongs to the CAP superfamily, and Os10t0409400-01 encodes a BURP domain containing protein.
Key words Rice blast; BURP; CAP; RNA-seq
Received: October 6, 2021 Accepted: November 9, 2021
Supported by Construction of a China-Angola Joint research Center for Crop Breeding (KY202001010); Key Research and Development Project of Anhui Province (202004a06020027).
Tao ZHOU (1995-), male, P. R. China, master, devoted to research about genetic breeding of rice.
* Corresponding author. E-mail: wzhangahas@163.com.
Rice ( Oryza sativa L.) is one of the staple food that feeds more than half of the population in the world[1]. It is hard to satisfy people’s demand for rice in the next 30 years, unless some of the risk factors which harm to rice yield are removed[2-3]. Rice disease is one of the important factors of rice yield reduction, and rice blast is the main disease in rice growth and development. It is caused by the fungus Magnaporthe oryzae [4]. The relative yield loss caused by rice blast in different production areas each year ranges from 30% to 50%[5-6]. Therefore, the use of molecular genetics to discover rice blast resistance genes/quantitative trait locus (QTLs) and uncover the resistance mechanism is particularly important for breeding rice-blast-resistant varieties.
Rice blast resistance is divided into complete resistance and partial resistance[7] or field resistance. Complete resistance is controlled by a single gene and most of them are race-specific resistance genes, while partial resistance is determined by QTLs and has non-specific and durable resistance[8]. Among the major genes that have been cloned, Pi-b [9] is the first rice blast resistance gene successfully cloned. It is located near the end of the long arm of chromosome 2 in rice and resistant to most of the races of rice blast in Japan. The broad-spectrum resistance genes Pi9 and Pi2 located on chromosome 6 showed resistance to 43 blast isolates in different countries[10], and Chen et al. [11] found that Pi2 showed resistance to most of the 792 Pyricularia grisea race in China. Pik locus is the main locus for controlling rice blast disease, located on chromosome 11, containing 4 cloned rice blast resistance genes Pi-1, Pik-h, Pik-m, Pik-p , among which Pik-p is the major allele. Studies have also shown that most of the genes at the Pik locus had broad spectrum resistance. The resistance mediated by the blast resistance gene Pb1 from the indica rice Modan is classified as a field resistance gene. Pi-37 is located on the first chromosome of rice. It shows complete resistance to rice blast isolates "CHL1405", "CHL1340" and "CHL1159" in southern China[12]. Pi63 [13] shows strong resistance to blast isolates Kyu9439013 and H10-168-1. It has moderate susceptibility to Kyu89-246 and H09-132-1, and its disease resistance is closely related to its expression.
To date, more than 277 rice blast resistance genes/quantitative trait locus (QTLs) have been identified (https://www.ricedata.cn/ontology/ontology.aspx?ta=TO:0000074), of which 84 are major genes for blast resistance, and 24 major genes have been successfully cloned (http://www.ricedata.cn/gene/gene_pi.htm). Most of the cloned resistance genes encode a nucleotide binding site (NBS) at the N-terminal, and a leucine-rich repeats (LRR) domain at the C-terminal, such as Pi1, Pi2, Pi5, Pi9, Pi25, Pi36, Pi37, Pi56, Pia, Pita, Pib, Piz, Pizt, Pik-m, Pid3, Pik-p, etc. Although Pb1 [14] also encodes the NBS-LRR protein, it lacks the P-loop structure necessary for nucleotide binding in the NBS region. Pi21 [15] the first invisible rice blast resistance gene to be located, is different from common disease resistance genes. The protein product it encodes is rich in proline and contains heavy metal binding domains and protein interaction domains.
Most of the above genes related to rice blast resistance were identified and cloned through map-based cloning, such as Pib, Pit, Pia, etc. With the development of bioinformatics, the application of some new bioinformatics resources and tools will help the identification, location and cloning of genes will help to understand the resistance mechanism of rice to rice blast, which in turn contributes to disease-resistant breeding. With the development of "next-generation" sequencing technology, researchers can quickly obtain a large amount of genome and transcriptome sequencing data at a relatively low cost. The current RNA-seq technology has high accuracy, good technical reproducibility and better gene estimation. Absolute expression status makes it the main method of gene expression analysis[16]. Lu et al. [17] obtained three candidate genes related to M. oryzae S182 resistance through RNA-Seq and Genome-wide association studies (GWAS) analysis, among which the phenylalanine ammonia-lyase gene family members LOC_Os02g41630 and OsPAL4 are broad-spectrum resistance-related genes. Yoshihiro et al. [18] analyzed the gene expression of mixed samples of host plants and pathogenic bacteria in rice leaves infected with rice blast 24 h after infection to understand the attack and defense mechanisms of rice and rice blast, all of the 240 up-regulated fungal transcripts encoding putative secreted proteins, indicating that these candidate rice blast resistance genes may play an important role in the initial stage of infection.
The distinctive feature of BURP domain containing protein is that the C-terminus contains a BURP-domain consisting of 230 amino acids, the N-terminus of the domain has two conserved phenylalanine (FF), and the C-terminus is highly conserved amino acids such as valine (V), aspartic acid (D), threonine (T), proline (P), glycine (G) and four repeated cysteine-histidine (CH) Motif. BURP domain was named for the four member of group BNM2, USP, RD22 and PG1[19]. Proteins containing BURP domains are ubiquitous in plants and have important functions in plant growth, development and stress response. The BURP genes of Phaseolus vulgaris were expressed differently in those with drought stress[20]. The expression of RD22-like subfamily induced by salicylic acid and abscisic acid in different tissues of Gossypium hirsutum [21]. In rice, OsRAFTIN1 was an anther specific BURP protein, and essential in the late stage of rice pollen development[22]. Analysis of the expression patterns of 17 BURP family genes ( OsBURP01-17 ) from rice showed that most of the genes were induced by stress (drought, salt, cold, and abscisic acid treatment, etc .)[23]. Although BURP domain-containing proteins have been identified in several plant species, few studies have been done on the expression of rice BURP domain-containing gene under rice blast infection.
CAP domain exists in widely organisms, and it has a superfamily consisting of cysteine-rich secretory proteins (CRISP), Antigen 5 (Ag5), and pathogenesis-related 1 (PR-1) proteins[24]. They are known to play significant roles in immune defense in mammals and plants[25]. PR-1 is the first member of the known CAP superfamily in 1970[26], and overexpression of the PR-1 gene leads to increased plant resistance to fungi[27], oomycetes[28] and bacteria[29], but not to viruses[30]. Ag5 protein is widely found in insect venom and saliva, such as venoms of vespids and fire ants[31]. The third and last founding member of the CAP superfamily, CRISP, it has an N-terminal CAP domain and a C-terminal cysteine-rich domain (CRD), and its Cysteine-rich domain may modulate the activity of K+ ion channels[32]. Collectively, CAP superfamily is closely related to the infection of germs.
There are few researches on the common defense mechanism and gene expression of rice infected with different rice blast physiological races[33]. Obtaining some transcriptome sequencing data of rice after blast infection through public gene databases will help us to use RNA-seq technology to explore the response mechanism of rice after blast infection. In this study, we obtained the RNA sequencing data of rice infected by different rice blast races and the RNA sequencing data of rice after the same race of rice blast infected with different time lengths, combined with transcriptome analysis. We obtained a total of 5 up-regulated genes in rice after rice blast infection. One of them is BURP domain-containing gene, and the other is CAP superfamily genes. In order to further understand the functions of their gene families, we performed GO and KEGG analysis on these families to study their functions and enrichment.
Materials and Methods
Materials
RNA-seq data composed of two parts, one of which is ZH11 and ostga5 infected by M. Oryzae for 0, 12, 24, and 48 h, the other is infected by M. Oryzae 248, 235 and 162 for 0, 8, 24, 48, 72, 96 h of Nipponbare. All the RNA data were sequenced by Illumina HiSeq 2000 sequencing instrument. RNA sequence of Nipponbare, ZH11 and ostga5 mutant were obtained from NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA739552 and PRJNA741871. Nipponbare genome and annotation were obtained from ensemble plants (http://plants.ensembl.org/Oryza_sativa/Info/Index).
Data processing
We transformed the SRA to FASTQ using the sratoolkit software, and mapped the RNA reads to Nipponbare genome by Hisat2. Then we generated the bam files by Samtools. Finally, the read counts of gene expression were estimated by the Stringtie. RNA-seq data of rice cultivar ZH11 and ostga5 mutant was different with nipponbare. Due to the lack of reference genome, we have to de novo assembly the transcripts by Trinity, and the gene expression was analyzed by RSEM. Then, the transcripts were mapped to Nipponbare genome by GMAP and compared with Nipponbare genome bedtools.
We used the DEseq2 to analyze the differential expression levels of genes and use transcripts per million (TPM) as the normalization standard for read counts. The groups were considered as differential expression between condition at adject p-value (padj)<0.01 and log2 foldchange>2.
Furthermore, in order to further study the expression, function and evolution of those genes, we annotated the genes by Pfamscan and eggong-mapper, then obtained the GO, KEGG and found homologous gene families and conserved domains.
Results and Analysis
De novo assembly and mapping
After assembly of the transcriptome data of zh11 and ostga5, the mean length of the transcripts was 1 089 nt and 1 104 nt, region from 184 nt and 191 nt to 23 092 nt and 15 426 nt, the transcripts longer than 1 000 nt was 30.07% and 35.44%, longer than 2 000 nt was 16.96% and 17.45%. Long sequences with high quality enable us to gain more information on gene. Then the assembled transcripts of ZH11 and ostga5 mutant were mapped to Nipponbare geneme with overlap at 95%.
Screening of differentially expressed genes
According to the differential expression analysis of the gene expression in different samples, it was found that Nipponbare, ZH11 and ostga5 had 5 transcripts that were co-up-regulated in infection by Magnaporthe grisea , namely Os07t0125500-01, Os10t0409400-01, Os07t0129300-00, Os07t0126301-01 and Os07t0124900-01 , but no co-down-regulated genes were found under the same conditions. The annotation shows that Os07t0125500-01, Os07t0129300-00, Os07t0126301-01 and Os07t0124900-01 have CAP domain belongs to CAP superfamily (PF00188), and Os10t0409400-01 encodes BURP domain containing protein (PF03181).
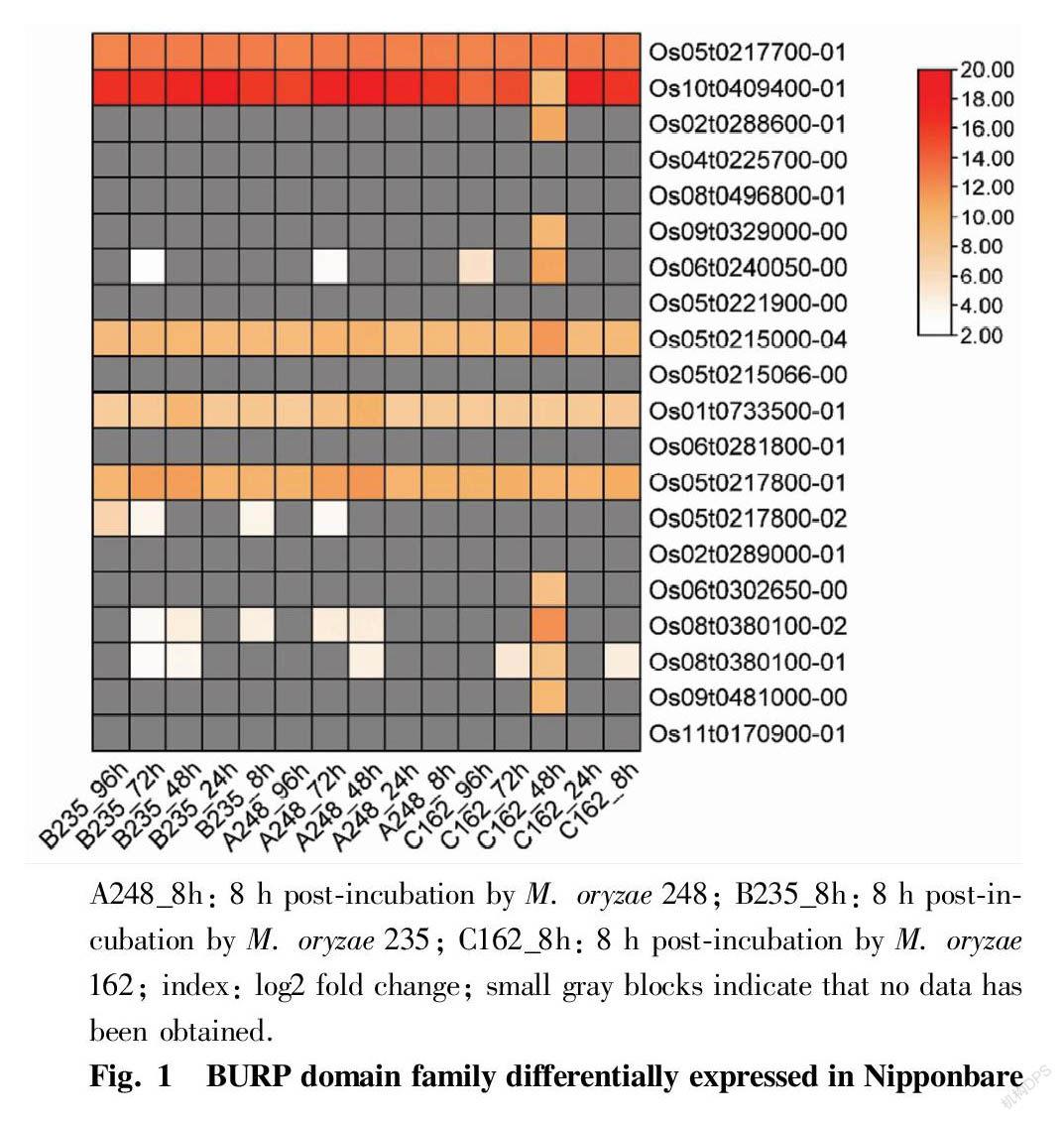
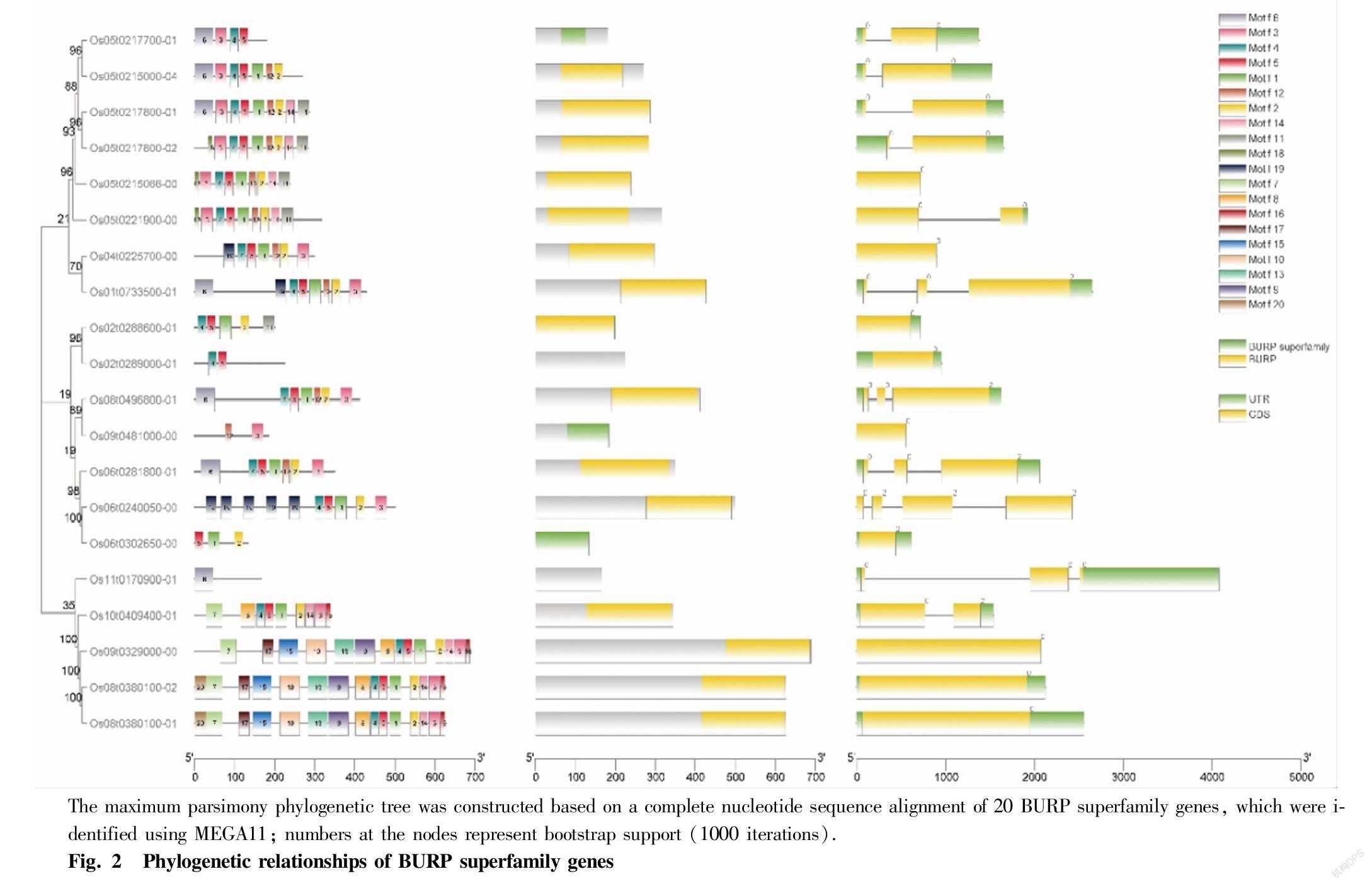
All of the 20 BURP domain containing family genes were searched with pfamscan software and annotated with eggnog-mapper. In those BURP domain family in Nipponbare, Os10t0409400-01, Os05t0217700-01, Os05t0215000-04, Os01t0733500-01 and Os05t0217800-01 were all expressed, and in addition, Os10t0409400-01 and Os05t0217700-01 had significant differences in expression with infection of before and after (Fig. 1). Consistent with previous studies[34], phylogenetic shows that BURP family genes are divided into 8 groups, Os10t0409400-01, Os08t0380100-01, Os08t0380100-02 and Os09t0329000-00 belong to PG1 β-like proteins, which are divided into a group due to their close distance (Fig. 2). Eight of them are annotated in the GO database (Table 1). The 5 genes which annotated in the KEGG database are all involved in endocytosis (ko04144).
CAP superfamily
We identified 35 genes that carried CAP superfamily, and most of the genes were up-regulated expression after being treated with different rice blast fungus for different time (Fig. 3). Then we constructed the maximum parsimony phylogenetic tree, the result show that the CAP superfamily can be divided into 8 groups. Among these groups, Os07t0125500-01, Os07t0129300-00, Os07t0126301-01, Os07t0124900-01 and the cloned genes OsPR1a (Os07t0129200-01), OsPR1b (Os01t0382000-01) are a group, indicating that their functions are similar (Fig. 4). Only two genes were annotated in the GO database, involved in biological processes (GO:0008150), multicellular organismal process (GO:0032501), response to stimulus (GO:0050896), biological regulation (GO:0065007), regulation of biological quality (GO:0065008). 26 genes had annotated in the KEGG database, including MAPK signaling pathway-plant (ko04016), plant hormone signal transduction (ko04075) and Plant-pathogen interaction (ko04626).
Conclusions and Discussion
To study the co-expressed genes of rice after inoculating different race of rice blast fungus and reveal the defense mechanism of rice resistance to rice blast, we used the transcriptome profiling of nipponbare, zh11 and ostga5 that infected with rice blast fungus. And co-up-regulated expression of five genes plays an important role in rice defense and resistance in blast fungus. One of those Os10t0409400-01 was named as OsBURP16 which is the β-subunit of polygalacturonase 1 in rice, and OsBURP16 overexpression reduces the pectin content and cell adhesion of rice, while increasing the resistance to abiotic stress[35-37]. Moreover, OsBURP16 had induced maximally expression in cold sensitive genotype, and had an increased transcript level[38]. There was a lot of research in OsBURP16 with abiotic stress and we have not foundreports on rice blast. What was interesting was that OsBURP16 was up-regulated expression in all samples of our study. We can reasonably inference that OsBURP16 was an important part of the rice blast response mechanism. GO shows that BURP domain protein family is significantly concentrated in pollen development and response to stress in biological processes, and Golgi apparatus, extracellular matrix part in cellular component. KEGG shows that some of those genes has involved in endocytosis. Previous study has also supported that BURP domain protein family has an important function in response of stress conditions in plants. BURP domain protein 4 ( OsBURP04 ) is up-regulated under all stress conditions, such as dry and cold[39]. CaBDP1 ( C. arabica BURP domain-containing proteins) expression can be highly induced in coffee plants subjected to drought, cold, salt, or ABA. It can inhibit cotyledon greening and seedling growth while the up-regulation of ABA[40]. Analysis of the transcription level of MtBURP ( Medicago truncatula BURP domain-containing proteins) genes in response to drought stress indicated that all 39 BURP genes are regulated by drought stress[41]. These all indicate the important role of the burp domain gene family in abiotic resistance.
Cladogram indicated that Os07t0125500-01, Os07t0129300-00, Os07t0126301-01 and Os07t0124900-01 was similar to the cloned gene OsPR1a and OsPR1b were searched in CAP superfamily (Fig. 4). PR-1 proteins were found ubiquitously distributed among plants and play an important role in stress response[42-44]. WjAMP-2 from leaves of Wasabia japonica L. and homology with PR-1 in most plant, it was induced by inoculation with fungal pathogens, and overexpression of WjAMP-2 will enhance resistance against Botrytis cinereal [27]. Exogenous ethylene and jasmonic acid treatment of rice shoots could induce a strong systemic resistance response in the roots, and the expression of pathogenesis-related gene OsPR1a and OsPR1b was significantly up-regulated[45], and the expression of OsPR1a was significantly induced in water stress and salt stress[46]. An OsPR1 expression/regulation model was proposed in 2001 which revealed the important role of OsPR1 in rice defense or stress response, and OsPR1 was highly responsive to the rice blast[47]. Protein Elicitor PemG1 from M. grisea improved resistant of rice, induced transient expression of PR-1 in rice[48]. Thus, it is reasonably speculated that these four co-up-regulated genes were related the response of rice blast.
References
[1] ZHANG Q. Purple tomatoes, black rice and food security[J]. Nat Rev Genet, 2021, 22(7): 414.
[2] HICKEY LT, HAFEEZ AN, ROBINSON H, et al. Breeding crops to feed 10 billion[J]. Nat Biotechnol, 2019, 37(7): 744-754.
[3] RAY DK, RAMANKUTTY N, MUELLER ND, et al. Recent patterns of crop yield growth and stagnation[J]. Nat Commun, 2012(3): 1293.
[4] WANG JC, CORRELL JC, JIA Y. Characterization of rice blast resistance genes in rice germplasm with monogenic lines and pathogenicity assays[J]. Crop Protection, 2015(72): 132-138.
[5] GREER C, WEBSTER R. Occurrence, distribution, epidemiology, cultivar reaction, and management of rice blast disease in California[J]. Plant Disease, 2001, 85(10): 1096-1102.
[6] SKAMNIOTI P, GURR SJ. Against the grain: Safeguarding rice from rice blast disease[J]. Trends Biotechnol, 2009, 27(3): 141-150.
[7] PARLEVLIET JE. Components of resistance that reduce the rate of epidemic development[J]. Annual review of phytopathology, 1979, 17(1): 203-222.
[8] ASHKANI S, RAFII M, SHABANIMOFRAD M, et al. Molecular progress on the mapping and cloning of functional genes for blast disease in rice ( Oryza sativa L.): current status and future considerations[J]. Crit Rev Biotechnol, 2016. 36(2): 353-367.
[9] WANG ZX, YANO M, YAMANOUCHI U, et al. The Pib gene for rice blast resistance belongs to the nucleotide binding and leucine-rich repeat class of plant disease resistance genes[J]. The Plant Journal, 1999, 19(1): 55-64.
[10] LIU G, LU G, ZENG L, et al. Two broad-spectrum blast resistance genes, Pi9 (t) and Pi2 (t), are physically linked on rice chromosome 6[J]. Mol Genet Genomics, 2002, 267(4): 472-480.
[11] CHEN HL, CHEN BT, ZHANG DP, et al. Pathotypes of Pyricularia grisea in rice fields of central and southern China[J]. Plant Disease, 2001, 85(8): 843-850.
[12] CHEN S, WANG L, QUE ZQ, et al. Genetic and physical mapping of Pi37(t), a new gene conferring resistance to rice blast in the famous cultivar St. No. 1[J]. Theoretical and Applied Genetics, 2005, 111(8): 1563-1570.
[13] XU X, HAYASHI N, WANG CT, et al. Rice blast resistance gene Pikahei-1(t), a member of a resistance gene cluster on chromosome 4, encodes a nucleotide-binding site and leucine-rich repeat protein[J]. Molecular Breeding, 2014, 34(2): 691-700.
[14] NAGAO HAYASHI, HARUHIKO INOUE, TAKAHIRO KATO, et al. Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication[J]. Plant J, 2010, 64(3): 498-510.
[15] FUKUOKA S, OKUNO K. QTL analysis and mapping of pi21, a recessive gene for field resistance to rice blast in Japanese upland rice[J]. Theoretical and Applied Genetics, 2001, 103(2-3): 185-190.
[16] WANG Z, GERSTEIN M, SNYDER M. RNA-Seq: A revolutionary tool for transcriptomics[J]. Nature reviews genetics, 2009, 10(1): 57-63.
[17] LU Q, WANG C, NIU X, et al. Detecting novel loci underlying rice blast resistance by integrating a genome-wide association study and RNA sequencing[J]. Molecular Breeding, 2019, 39(6): 1-10.
[18] KAWAHARA Y, OONO Y, KANAMORI H, et al. Simultaneous RNA-seq analysis of a mixed transcriptome of rice and blast fungus interaction[J]. PLoS One, 2012, 7(11): e49423.
[19] HATTORI J, BOUTILIER KA, LOOKEREN CAMPAGNE MM, et al. A conserved BURP domain defines a novel group of plant proteins with unusual primary structures[J]. Molecular & General Genetics Mgg, 1998, 259(4): 424-428.
[20] KAVAS M, YLDRM K, SEGIN Z, et al. Genome-wide identification of the BURP domain-containing genes in Phaseolus vulgaris [J]. Physiology and Molecular Biology of Plants, 2021, 27(9): 1885-1902.
[21] SUN H, WEI H, WANG H, et al. Genome-wide identification and expression analysis of the BURP domain-containing genes in Gossypium hirsutum [J]. BMC Genomics, 2019, 20(1): 558.
[22] WANG AM, XIA Q, XIE WS, et al. The classical Ubisch bodies carry a sporophytically produced structural protein (RAFTIN) that is essential for pollen development[J]. Proceedings of the National Academy of Sciences, 2003, 100(24): 14487-14492.
[23] DING XP, HOU X, XIE KB, et al. Genome-wide identification of BURP domain-containing genes in rice reveals a gene family with diverse structures and responses to abiotic stresses[J]. Planta, 2009, 230(1): 149-163.
[24] TAKASHI TADOKORO, CASSANDRA M MODAHL, KATSUMI MAENAKA, et al. Cysteine-rich secretory proteins (CRISPs) from venomous snakes: An overview of the functional diversity in a large and underappreciated superfamily[J]. Toxins (Basel), 2020, 12(3): 175.
[25] GAIKWAD AS, HU J, CHAPPLE DG, et al. The functions of CAP superfamily proteins in mammalian fertility and disease[J]. Hum Reprod Update, 2020, 26(5): 689-723.
[26] LOON LCV, KAMMEN AV. Polyacrylamide disc electrophoresis of the soluble leaf proteins from Nicotiana tabacum var. ‘Samsun’and ‘Samsun NN’: II. Changes in protein constitution after infection with tobacco mosaic virus[J]. Virology, 1970, 40(2): 199-211.
[27] KIBA A, NISHIHARA M, NAKATSUKA T, et al. Pathogenesis-related protein 1 homologue is an antifungal protein in Wasabia japonica leaves and confers resistance to Botrytis cinerea in transgenic tobacco[J]. Plant Biotechnology, 2007, 24(2): 247-253.
[28] ALEXANDER D, GOODMAN RM, GUT-RELLA M, et al. Increased tolerance to two oomycete pathogens in transgenic tobacco expressing pathogenesis-related protein 1a[J]. Proceedings of the National Academy of Sciences, 1993, 90(15): 7327-7331.
[29] SANG HS, PAK JH, MI JK, et al. An acidic pathogenesis-related1 gene of Oryza grandiglumis is involved in disease resistance response against bacterial infection[J]. The plant pathology journal, 2014, 30(2): 208.
[30] CARR JP, BEACHY RN, KLESSIG DF. Are the PR1 proteins of tobacco involved in genetically engineered resistance to TMV[J]. Virology, 1989, 169(2): 470-473.
[31] KING TP, SPANGFORT MD. Structure and Biology of Stinging Insect Venom Allergens[J]. International Archives of Allergy and Immunology, 2000, 123(2): 99-106.
[32] GUO M, TENG M, NIU L, et al. Crystal structure of the cysteine-rich secretory protein stecrisp reveals that the cysteine-rich domain has a K+ channel inhibitor-like fold[J]. J Biol Chem, 2005, 280(13): 12405-12412.
[33] PRIYANKA J, VINAY S, HIMANSHU D, et al. Identification of long non-coding RNA in rice lines resistant to Rice blast pathogen Maganaporthe oryzae [J]. Bioinformation, 2017, 13(8): 249-255.
[34] DING XP, HOU X, XIE KB, et al. Genome-wide identification of BURP domain-containing genes in rice reveals a gene family with diverse structures and responses to abiotic stresses[J]. Planta, 2009, 230(1): 149-163.
[35] LIU H, MA Y, CHEN N, et al. Overexpression of stress-inducible OsBURP16, the beta subunit of polygalacturonase 1, decreases pectin content and cell adhesion and increases abiotic stress sensitivity in rice[J]. Plant Cell Environ, 2014, 37(5): 1144-1158.
[36] JIN J, DUAN J, SHAN C, et al. Ethylene insensitive3-like2 (OsEIL2) confers stress sensitivity by regulating OsBURP16, the β subunit of polygalacturonase (PG1β-like) subfamily gene in rice[J]. Plant Science, 2020(292): 110353.
[37] XU Y, HU D, HOU X, et al. OsTMF attenuates cold tolerance by affecting cell wall properties in rice[J]. New Phytologist, 2020, 227(2): 498-512.
[38] DE FREITAS GM, THOMAS J, LIYANAGE R, et al. Cold tolerance response mechanisms revealed through comparative analysis of gene and protein expression in multiple rice genotypes[J]. PloS one, 2019, 14(6): e0218019.
[39] TIEN VQ, DUONG N H, NHAN DT, et al. In Silico Analysis of Osa-miR164 Gene Family in Rice ( Oryza Sativa )[J]. VNU Journal of Science: Natural Sciences and Technology, 2021, 37(3).
[40] DINH SN, KANG H. An endoplasmic reticulum-localized Coffea arabica BURP domain-containing protein affects the response of transgenic Arabidopsis plants to diverse abiotic stresses[J]. Plant Cell Rep, 2017, 36(11): 1829-1839.
[41] LI Y, CHEN X, CHEN Z, et al. Identification and expression analysis of BURP domain-containing genes in Medicago truncatula [J]. Front Plant Sci, 2016(7): 485.
[42] HON WC, GRIFFITH M, MLYNARZ A, et al. Antifreeze proteins in winter rye are similar to pathogenesis-related proteins[J]. Plant Physiology, 1995, 109(3): 879-889.
[43] ZEIER J, PINK B, MUELLER MJ, et al. Light conditions influence specific defence responses in incompatible plant-pathogen interactions: Uncoupling systemic resistance from salicylic acid and PR-1 accumulation[J]. Planta, 2004, 219(4): 673-83.
[44] STINTZI A, HEITZ T, PRASAD V, et al. Plant ‘pathogenesis-related’ proteins and their role in defense against pathogens[J]. Biochimie, 1993, 75(8): 687-706.
[45] NAHAR K, KYNDT T, VLEESSCHAUWER D DE, et al. The jasmonate pathway is a key player in systemically induced defense against root knot nematodes in rice[J]. Plant Physiol, 2011, 157(1): 305-316.
[46] KOTHARI KS, DANSANA PK, JITENDER G, et al. Rice stress associated protein 1 (OsSAP1) interacts with aminotransferase (OsAMTR1) and pathogenesis-related 1a protein (OsSCP) and regulates abiotic stress responses[J]. Frontiers in plant science, 2016(7): 1057.
[47] AGRAWAL GK, RAKWAL R, JWA NS, et al. Signalling molecules and blast pathogen attack activates rice OsPR1a and OsPR1b genes: A model illustrating components participating during defence/stress response[J]. Plant Physiology and Biochemistry, 2001, 39(12): 1095-1103.
[48] PENG DH, QIU DW, RUAN LF, et al. Protein elicitor PemG1 from Magnaporthe grisea induces systemic acquired resistance (SAR) in plants[J]. Molecular plant-microbe interactions, 2011, 24(10): 1239-1246.
- 农业生物技术(英文版)的其它文章
- Research Progresses on QTLs for Main Grain Shape Genes in Rice
- Effects of Raising Chickens Under Moringa oleifera
- Preliminary Research on Radiation Breeding of Pteroceltis tatarinowii Maxim
- Comparison of Spring Radish Varieties with Entire Leaves
- Occurrence and Chemical Control Techniques of Rice Black-streaked Dwarf Disease in Rongshui County
- Overview of Research on Tissue Culture and Rapid Propagation Technology of Pholidota

