Proteomic Analysis of Sea Cucumber (Holothuria leucospilota) from the South China Sea
Le WANG Lizi SHEN Jiao HE Zuliang WU Chen CHEN Bingmiao GAO


Abstract [Objectives] Proteomics methods were used to analyze the total proteome of Holothuria leucospilota in the South China Sea, so as to provide a theoretical basis for elucidating the medicinal material basis of sea cucumber proteins and peptides.
[Methods] H. leucospilota was collected from the South China Sea. Label-free proteomics methods were used to detect the protein profile of enzymatically hydrolyzed H. leucospilota total protein, and identify sea cucumber protein and peptide sequences, and gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) bioinformatics analysis software were used to analyze significantly enriched proteins. The String database was used to construct a protein-protein interaction network of significantly enriched H. leucospilota proteins (PPI network).
[Results] Proteomics identified 1 200 sea cucumber proteins and 2 967 peptides. GO analysis and KEGG analysis showed that the four significantly-enriched sea cucumber proteins hsc70, COX, CYTB and rbbp4 participated in 8 metabolic pathways, and COX1 and CYTB among them were significantly related to cardiac muscle contraction, oxidative phosphorylation and metabolic pathways. The PPI network analysis showed that the 13 significantly-enriched sea cucumber proteins had close protein-protein interaction relationships.
[Conclusions]This study used proteomics to conduct preliminary identification and research on the proteins and peptides of H. leucospilota , providing a scientific basis for the application of H. leucospilota proteins and peptides in the fields of functional health food and drug research and development.
Key words Holothuria leucospilota ; Sea cucumber protein; Sea cucumber peptide; Proteomics
Received: September 2, 2021 Accepted: November 3, 2021
Supported by Scientific Research Program of Higher Education Institutions in Hainan Province (Hnky2019-110).
Le WANG (1987-), female, P. R. China, lecturer, master, devoted to research about functional food, nutrition and health.
*Corresponding author. E-mail: gaobingmiao@qq.com.
Holothuria leucospilota belongs to Holothuriae of Aspidochirotida in Holothuroidea of Echinodermata. It is mainly distributed in Fujian, Taiwan, Guangdong, Guangxi, Hainan Island and Paracel Islands of China, and is the most common edible sea cucumbers in Hainan[1]. Sea cucumbers have similar medicinal properties to Panax ginseng and have become a valuable ingredient in China, Japan and other Asian countries. The nutritional and medicinal value of sea cucumbers is extremely high. Sea cucumbers are not only rich in proteins, essential amino acids, various trace elements, vitamins, etc. , but also contain many biologically active substances[2-3]. Modern pharmacological studies have shown that sea cucumbers have good effects such as anti-oxidation, ACE inhibitory activity, immune regulation, anti-fatigue, lowering blood pressure, lowering blood lipids, anti-tumor, and anti-aging[4-7].
At present, the impact of biologically active peptides on human health has attracted much attention, and its extraction in organisms and its application in humans have become the focus of attention of researchers[8]. Total sea cucumber protein can produce small peptides composed of 2-12 amino acids or peptides with larger molecular weights through enzymatic hydrolysis, and has good solubility and stability and low viscosity, thus having great application potential[9]. With the continuous improvement of people’s material living standards, sea cucumber peptide health-care functional foods with high nutritional value have attracted more and more attention. Studies have found that Holothuria leucospilota peptides have many functions such as lowering blood pressure, anti-oxidation, anti-tumor and anti-fatigue, and its physiological activity may have a certain relationship with the molecular weight distribution of sea cucumber peptides[10].
Proteomics is one of the hot research fields in the post-genomic era. At present, the research on sea cucumber samples using proteomics methods mainly focuses on the effects of sea cucumber fingerling raising, breeding, and processing technology on the tissue structure and quality of sea cucumbers, as well as basic research on disease prevention and control, while there are few reports about the research of sea cucumber proteins and peptides using proteomics[11]. Therefore, in this study, with H. leucospilota as the research object, preliminary identification and research were conducted on proteins and peptides of H. leucospilota by proteomics, aiming to provide a scientific basis for the application of proteins and peptides from H. leucospilota in the fields of functional health food and drug research and development.
Materials and Methods
Materials
Sea cucumber samples were collected from Wenchang City, Hainan Province, and were identified as H. leucospilota by Professor Feng of Hainan University.
Extraction and analysis of total sea cucumber protein
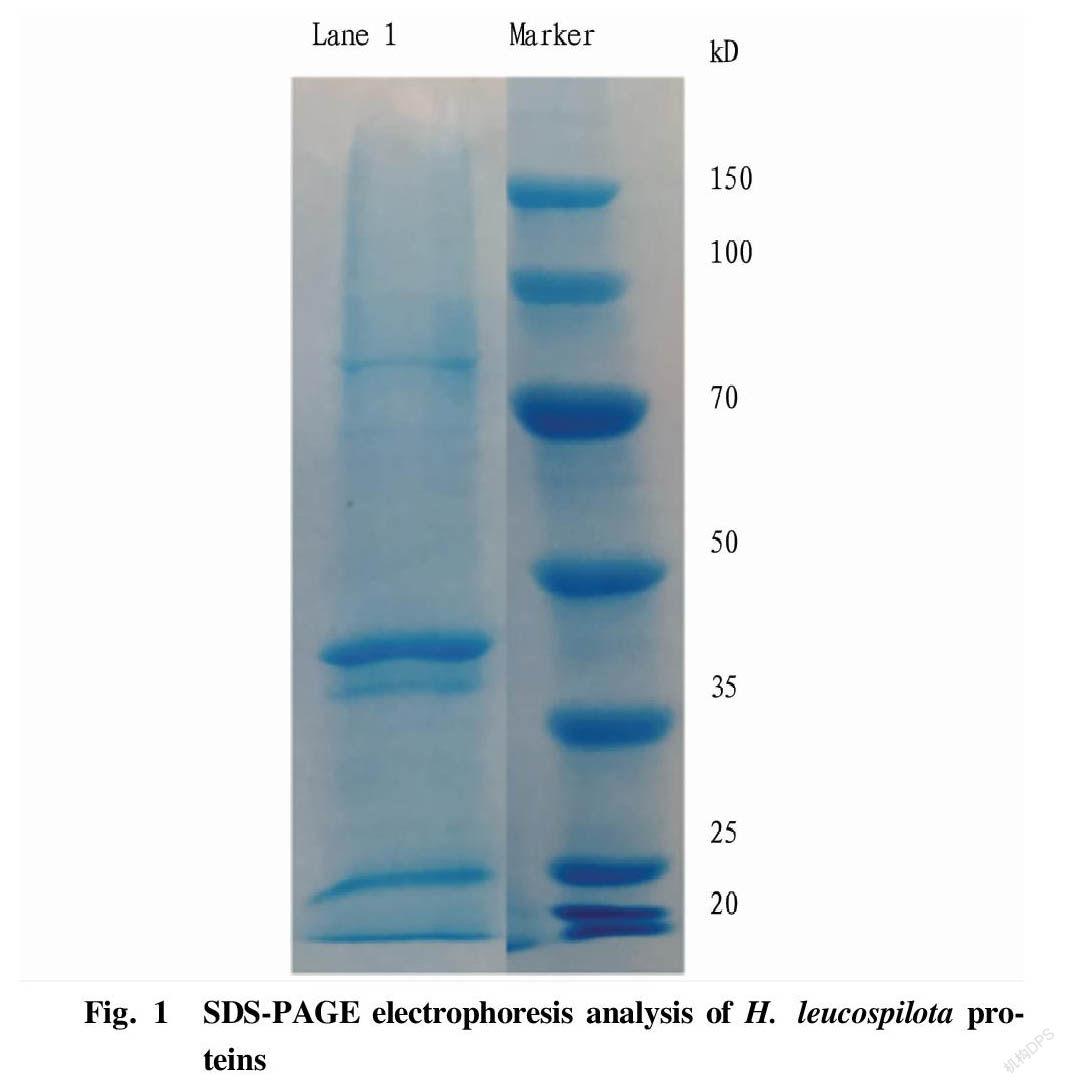
An appropriate amount of sea cucumber sample of H. leucospilota was weighed into a new EP tube, then added with an appropriate amount of RIPA working solution and mixed well. The obtained mixture was ground at a low temperature for 5 min, and ultrasonically treated in an ice bath for 5 min to fully lyse the sample. Next, the sample was rotated and incubated at 4 ℃ for 10 min to fully dissolve the proteins. After centrifugation at 12 000 rpm at 4 ℃ for 15 min, the supernatant was transferred to a new EP tube. SDS-PAGE electrophoresis was used to analyze the total protein of sea cucumbers, and the protein concentration was measured by the BCA kit method.
Nano LC-MS/MS analysis
For each sample, 1 μg of total peptides were separated and analyzed with a nano UPLC (EASY nLC1200) coupled to a Q Exactive HFX Orbitrap instrument (Thermo Fisher Scientific) with a nano electrospray ion source. Separation was performed using a reversed phase column (100 μm ID×15 cm, Reprosil Pur 120 C18 AQ, 1.9 μm, Dr. Maisch). Mobile phases were H2O with 0.1% FA, 2% ACN (phase A) and 80% ACN, 0.1% FA (phase B). Separation of samples was executed with a 120 min gradient at 300 nl/min flow rate (gradient B: 2%-5% for 2 min, 5%-22% for 88 min, 22%-45% for 26 min, 45%-95% for 2 min, 95% for 2 min).
Data dependent acquisition (DDA) was performed in profile and positive mode with Orbitrap analyzer at a resolution of 120 000 (@200 m/z) and m/z range of 350-1 600 for MS1. For MS2, the resolution was set to 15 000 with a dynamic first mass. The automatic gain control (AGC) target for MS1 was set to 3E6 with max IT 50 ms, and 1E5 for MS2 with max IT 110 ms. The top 20 most intense ions were fragmented by HCD with normalized collision energy (NCE) of 27%, and isolation window of 1.2 m/z. The dynamic exclusion time window was 45 s, and single charged peaks and peaks with charge exceeding 6 were excluded from the DDA procedure.
Proteome Discoverer database search
Vendor’s raw MS files were processed using Proteome Discoverer (PD) software (Version 2.4.0.305) and the built-in Sequest HT search engine. MS spectra lists were searched against their species-level UniProt FASTA databases, Carbamidomethyl[C] as a fixed modification, Oxidation (M) and Acetyl (Protein N-term) as variable modifications. Trypsin was used as proteases. A maximum of 2 missed cleavage(s) was allowed. The false discovery rate (FDR) was set to 0.01 for both PSM and peptide levels. Peptide identification was performed with an initial precursor mass deviation of up to 10 ppm and a fragment mass deviation of 0.02 Da. Unique peptide and Razor peptide were used for protein quantification and total peptide amount for normalization. All the other parameters were reserved as default.
Bioinformatics analysis
GO functional annotation analysis (http://www.geneontology.org) was performed on the identified proteins, and the KEGG software (http://www.genome.jp/kegg/pathway.html/) was used for pathway analysis and protein interaction (protein- to-protein interaction, PPI) analysis. The GO system was divided into three parts: cellular component, molecular function and biological process. The pathway analysis through the KEGG software determined the main biochemical metabolic pathways and signal transduction pathways in which the proteins involved. PPI analysis used the String database.
Statistical analysis
All data were processed with Graph Prism 6.0 software. The peak area of each protein was expressed by the relative intensity value, and the average protein abundance of various groups was calculated. Difference analysis adopted the t test, with P <0.05 indicating that the difference was statistically significant.
Results and Analysis
Protein extraction and enzymatic hydrolysis
H. leucospilota proteins were extracted by RIPA lysis method, and the integrity of the protein was evaluated by SDS-PAGE combined with Coomassie brilliant blue staining. The electrophoresis results showed that the sea cucumber proteins had clear and distinct bands and the protein molecular weight distribution was uniform (Fig. 1). The concentration of H. leucospilota proteins was determined by the BCA method to be 0.31 mg/ml.
Identification of sea cucumber proteins and peptides
After trypsin digestion and LC-MS/MS detection, the proteomics research of H. leucospilota proteins identified a total of 1 200 proteins and 2 967 peptides. The number of proteins and peptides identified is shown in Fig. 2-A, and the distribution of protein molecular weights is shown in Fig. 2-B. The proteins with a molecular weight of 20-40 kD were distributed the most. The identified high-abundance proteins were mainly of five family types, that is, actin, myosin, tubulin, filamin-B, and spectrin, of which actin was the most abundant, because actin is one of the most abundant proteins in eukaryotes. In addition, the polypeptide length distribution obtained by enzymatic hydrolysis of sea cucumber proteins is shown in Fig. 2-C, and the distribution of peptide coverage of the identified proteins is shown in Fig. 2-D. Among them, peptides with 7 to 13 amino acids were the most abundant, laying an important material foundation for the research on the pharmacological activity of sea cucumber peptides.
GO analysis
The GO functional classification system was used to analyze the distribution of the identified sea cucumber proteins in cellular component (CC), molecular function (MF), and biological process (BP) (Fig. 3). Among them, the top 10 items that were significantly enriched in the CC term were cell, cell part, intracellular, intracellular part, organelle, intracellular organelle, cytoplasm, protein-containing complex, cytoplasmic part and ribosome; the top 10 items that were significantly enriched in the MF term were small molecule binding, anion binding, guanyl ribonucleotide binding, purine nucleoside binding, GTP binding, purine ribonucleoside binding, guanyl nucleotide binding, structural molecule activity, GTPase activity and structural constituent of ribosome; and the top 10 items that were significantly enriched in the BP term were small molecule metabolic process, cellular amide metabolic process, peptide metabolic process, amide biosynthetic process, translation, peptide biosynthetic process, carboxylic acid metabolic process, oxoacid metabolic process, organic acid metabolic process and drug metabolic process.
KEGG pathway analysis
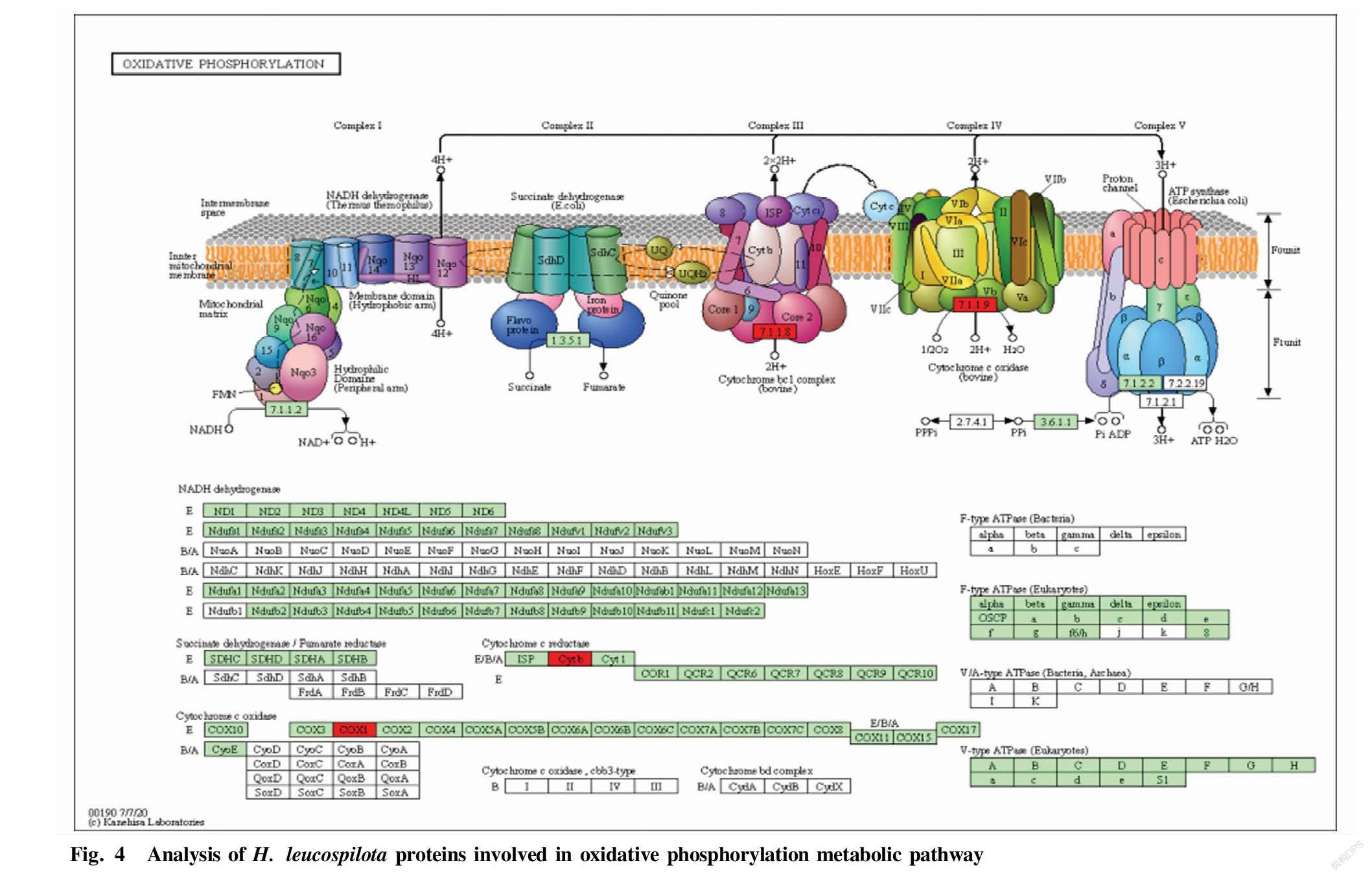
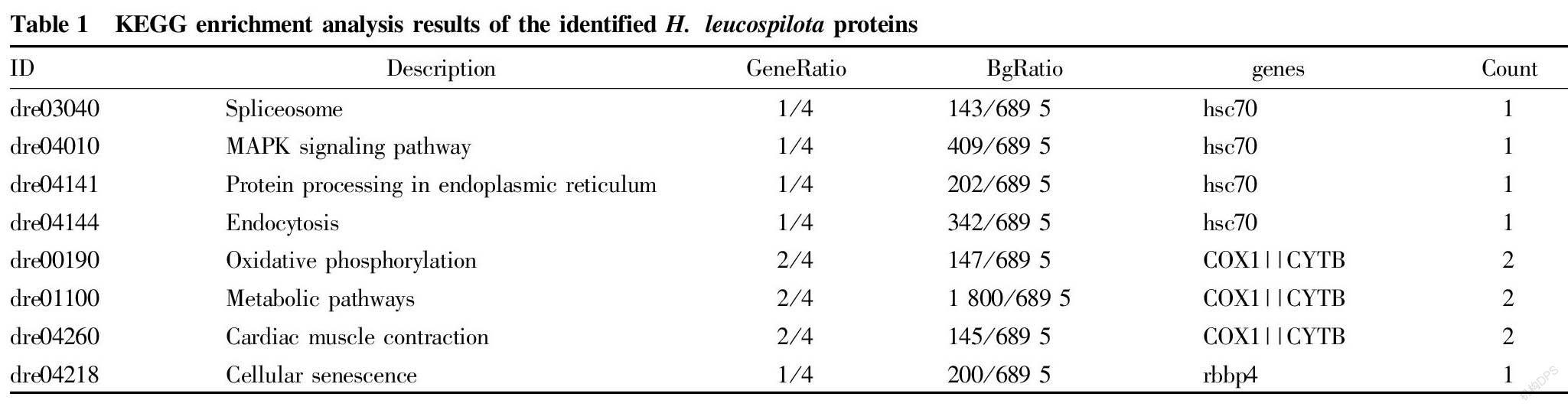
The KEGG online tool was used to analyze the metabolic pathways of the identified sea cucumber proteins to determine which types of biochemical metabolic pathways they were involved in. KEGG analysis showed that the four identified proteins hsc70, COX, CYTB and rbbp4 were involved in eight metabolic pathways, namely spliceosome, MAPK signaling pathway, protein processing in endoplasmic reticulum, endocytosis, oxidative phosphorylation, metabolic pathways, cardiac muscle contraction and cellular senescence (Table 1). The metabolic pathways in which the proteins were significantly enriched were oxidative phosphorylation, cardiac muscle contraction, and metabolic pathways. These three signaling pathways all involved two proteins COX1 and CYTB. In particular, proteins COX1 and CYTB were involved in the oxidative phosphorylation metabolic pathway, and their expression levels were both up-regulated, indicating that the regulation of these two proteins can enhance the oxidative phosphorylation process, suggesting that sea cucumbers have anti-aging pharmacological effects (Fig. 4).
Interaction network of differential proteins
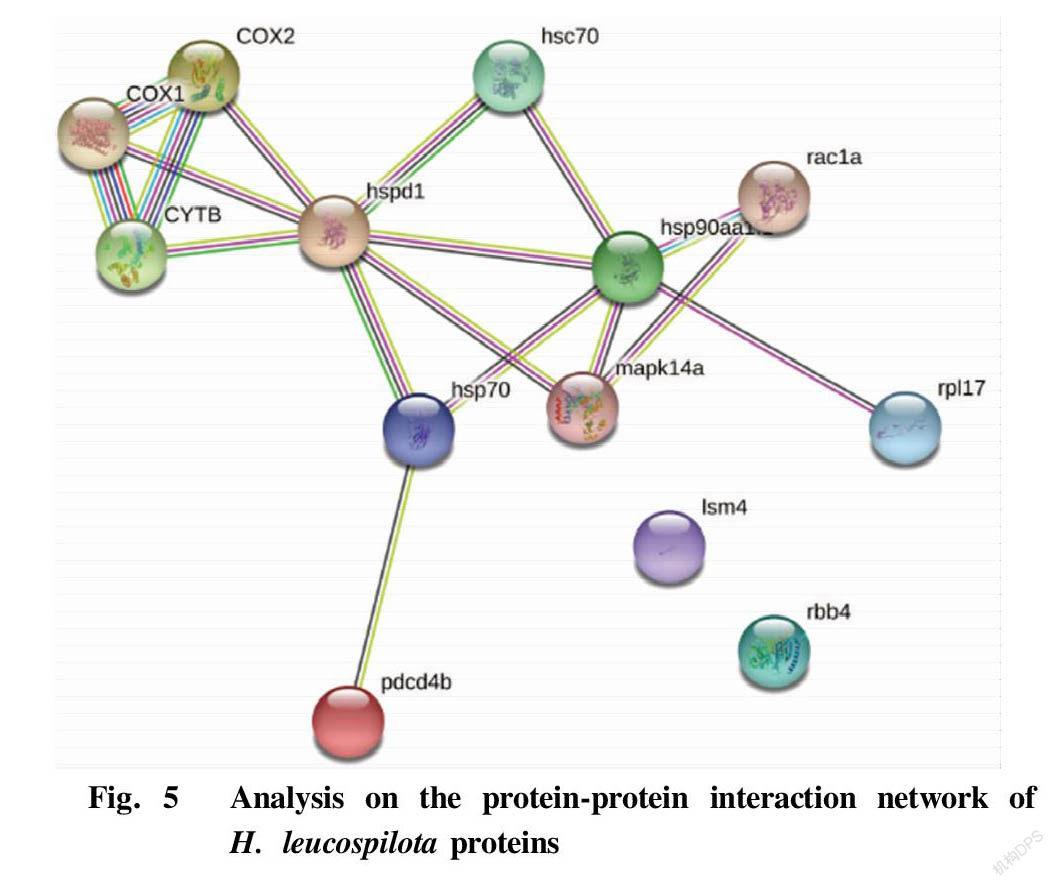
In this study, the analysis on the protein interaction information of the protein-protein interaction network (PPI network) constructed by the STRING database showed that the 13 significantly-enriched sea cucumber proteins had close protein-protein interaction relationship, thereby constructing a diagram of the interaction network of the expressed H. leucospilota proteins with complex structure (Fig. 5). Analysis of the results showed that the 13 significantly-enriched proteins were COX1, COX2, CYTB, hspd1, hsc70, hsp70, hsp90aa, rac1a, mapk14a, pdcd4b, rpl17, Ism4 and rbb4. Among them, the two proteins Ism4 and rbb4 were independent of each other and did not participate in the PPI network. In addition, 11 proteins including COX1, COX2, CYTB, hspd1, hsc70, hsp70, hsp90aa, rac1a, mapk14a, pdcd4b, and rpl17 interacted with each other, which was mainly reflected in the strongest interaction relationship between COX1, COX2 and CYTB, followed by hspd1, hsc70, hsp70, hsp90aa, rac1a, mapk14a.
Discussion and Conclusions
Since ancient times, sea cucumbers have been regarded as a high-quality nutritional and health food, which has the effects of strengthening muscle strength, regulating immunity, preventing arthritis, anemia and impotence, and regulating blood pressure and resisting fatigue, so sea cucumbers are also widely applied in functional foods[12]. In recent years, research on the effects of sea cucumber peptides on human health has attracted the attention of scientific researchers and has become a hot research topic[13]. Liu et al. [14] found that sea cucumber peptides with molecular masses between 1 000 and 3 000 Da have a strong scavenging ability on DPPH free radicals. Li et al. [15] treated Acaudina molpadioides with sodium sulfite with the assistance of twin-screw extrusion, which increased the extraction rate of peptides and enhanced the antioxidant activity of A. molpadioides peptides. The crude peptide was purified to obtain purified component I (1 000-3 000 Da), which had the strongest superoxide anion free radical scavenging rate. Wang et al. [16] studied the antioxidant properties of sea cucumber peptides and found that the antioxidant properties of sea cucumber peptides were better than those of natural antioxidant vitamin C. They also found that the antioxidant properties of sea cucumber peptides were closely related to molecular weight, and sea cucumber peptides with different molecular weight ranges had different ability to scavenge oxygen free radicals. Yu et al. [17] found that sea cucumber peptides could improve the oxidative stress state of skeletal muscle in exercise-induced fatigue rats, reduce the swelling and expansion of skeletal muscle mitochondria and mitochondrial membrane permeability, enhance the respiratory function of skeletal muscle mitochondria, and up-regulate the expression of mitochondrial transcription factors PGC-1α and ERR.
There are more and more researches on sea cucumber peptides, while peptides are prepared by enzymatic hydrolysis of sea cucumber proteins, so it is particularly necessary to use proteomics methods to study sea cucumber proteins. Chen et al. [18] first used isobaric tag based TMT labeling and IMAC enrichment strategy for quantitative phosphorylation proteomics analysis of sea cucumber summer dormancy. Sun et al. [19] used relative and absolute quantitative isobaric markers (iTRAQ) to investigate the changes in the protein content of sea cucumbers in the intestinal regeneration process, and there were significant differences in the expression of 2 321 of the 4 073 identified proteins. However, sea cucumber proteomics analysis and research on sea cucumber proteins and peptides to clarify their pharmacological targets and metabolic pathways has not yet been carried out.
In this study, label-free proteomics methods were used to conduct a preliminary proteomic analysis on H. leucospilota from the South China Sea. A total of 1 200 sea cucumber proteins and 2 967 peptides were identified, and the peptides with a length of 7-13 amino acids had molecular weights of about 700-1 300 Da, which is consistent with the results of Liu et al. [14] and Li et al. [15] studying the molecular weight range of sea cucumber peptides. GO analysis and KEGG analysis showed that the four significantly-enriched sea cucumber proteins hsc70, COX, CYTB and rbbp4 participated in 8 metabolic pathways, and COX1 and CYTB among them were significantly related to cardiac muscle contraction, oxidative phosphorylation and metabolic pathways. It is worth noting that the oxidative phosphorylation pathway reflects the enhancement of mitochondrial metabolism. It is currently believed that a variety of late-onset human diseases and the aging process itself involve impaired oxidative phosphorylation[20]. The two proteins COX1 and CYTB identified in sea cucumbers from H. leucospilota were involved in the oxidative phosphorylation metabolic pathway, and their expression levels were all up-regulated, indicating that the regulation of these two proteins can enhance the oxidative
phosphorylation process, thereby having anti-aging pharmacological effects. The involvement of H. leucospilota proteins in oxidative phosphorylation metabolic pathway is consistent with the results of Yu et al. [17] that sea cucumber peptides enhanced the respiratory function of skeletal muscle mitochondria and improved the oxidative stress state of skeletal muscle in exercise-induced fatigue rats. In addition, this study also clarified that there was close protein interaction and regulatory networks among the 13 sea cucumber proteins that were significantly enriched. Therefore, we used proteomics to conduct preliminary identification and research on the proteins and peptides of H. leucospilota , so as to provide a scientific basis for the application of H. leucospilota proteins and peptides in the fields of functional health food and drug research and development.
References
[1] YU ZH, HUANG W, MA WG, et al. The effects of Chaetoceros moulinii and marine red yeast on the development, growth and survival rate of Holothuria leucospilota larvae[J]. Journal of Fishries China, 2021: 1-8. (in Chinese)
[2] BORDBAR S, ANWAR F, SAARI N. High-value components and bioactives from sea cucumbers for functional foods: A review[J]. Mar Drugs, 2011, 9(10): 1761-805.
[3] ZHAO L, MA HW, CAO R, et al. Analysis of nutritional component in 10 kinds of sea cucumbers[J]. Journal of Food Safety & Quality, 2016, 7(7): 2867-2872. (in Chinese)
[4] Khotimchenko Y. Pharmacological potential of sea cucumbers[J]. Int J Mol Sci., 2018, 19(5): 1342.
[5] JANAKIRAM NB, MOHAMMED A, RAO CV. Sea cucumbers metabolites as potent anti-cancer agents[J]. Mar Drugs, 2015, 13(5): 2909-2923.
[6] MALAIWONG N, CHALORAK P, JATTUJAN P, et al. Anti-Parkinson activity of bioactive substances extracted from Holothuria leucospilota [J]. Biomed Pharmacother, 2019(109): 1967-1977.
[7] ZHAO F, LIU Q, CAO J, et al. A sea cucumber ( Holothuria leucospilota ) polysaccharide improves the gut microbiome to alleviate the symptoms of type 2 diabetes mellitus in Goto-Kakizaki rats[J]. Food Chem Toxicol, 2020(135): 110886.
[8] TONG JJ, ZHANG YB, YE ZZ, et al. Research progress of sea cucumber polypeptides[J]. Science and Technology of Food Industry, 2013, 34(11): 356-360. (in Chinese)
[9] CHIEU HD, SUWANSA-ARD S, WANG T, et al. Identification of neuropeptides in the sea cucumber Holothuria leucospilota [J]. Gen Comp Endocrinol, 2019(283): 113229.
[10] ZHANG X, LI H, WANG L, et al. Anti-inflammatory peptides and metabolomics-driven biomarkers discovery from sea cucumber protein hydrolysates[J]. J Food Sci. 2021, 86(8): 3540-3549.
[11] CHEN X, LIN TY, XUE WF, et al. Study on sea cucumber samples based on proteomics[J]. Journal of Food Safety & Quality, 2019, 10(23): 7869-7874. (in Chinese)
[12] YU YH. Research on the anti-fatigue effect and mechanism of sea cucumber peptides[D]. Wuxi: Jiangnan University, 2021. (in Chinese)
[13] LE QQ, LIAO JJ, TANG GQ, et al. Evaluation of the efficacy of sea cucumber peptides in improving immunity[J]. Modern Food, 2021(10): 111-114. (in Chinese)
[14] LIU CH, ZHU BW, DONG XP, et al. Study on the separation and antioxidant activity of enzymatic hydrolysates from sea cucumber[J]. Food and Fermentation Industries, 2007(9): 50-53. (in Chinese)
[15] LI WC, WU GB, CHEN FH. Effect of twin-screw extrusion on the extraction rate and antioxidant activity of low-value sea cucumber polypeptides[J]. Progress in Fishery Sciences, 2021(6): 142-151. (in Chinese)
[16] WANG J, ZHANG JL, WANG DX, et al. Antioxidant activity of peptide fractions from sea cucumber protein hydrolysates[J]. Food & Machinery, 2010, 26(2): 67-71. (in Chinese)
[17] YU YF, WANG T, WANG GF, et al. Effect of sea cucumber peptide on mitochondrial function of skeletal muscle in exercise-induced fatigue rats[J]. Shandong Medical Journal, 2019, 59(20): 31-34. (in Chinese)
[18] CHEN M, ZHU A, STOREY KB. Comparative phosphoproteomic analysis of intestinal phosphorylated proteins in active versus aestivating sea cucumbers[J]. J Proteomics, 2016(135): 141-150.
[19] SUN L, XU D, XU Q, et al. iTRAQ reveals proteomic changes during intestine regeneration in the sea cucumber Apostichopus japonicus [J]. Comp Biochem Physiol Part D Genomics Proteomics, 2017(22): 39-49.
[20] WEN YL, LIU X, WANG JQ, et al. Research progress of mitochondrial dynamics and oxidative phosphorylation in mammal[J]. Acta Veterinaria Et Zootechnica Sinica, 2021, 52(2): 273-285. (in Chinese)
- 农业生物技术(英文版)的其它文章
- Rice Blast Resistance-associated Genes Based on Different RNA-seq Resources
- Research Progresses on QTLs for Main Grain Shape Genes in Rice
- Effects of Raising Chickens Under Moringa oleifera
- Preliminary Research on Radiation Breeding of Pteroceltis tatarinowii Maxim
- Comparison of Spring Radish Varieties with Entire Leaves
- Occurrence and Chemical Control Techniques of Rice Black-streaked Dwarf Disease in Rongshui County

