DNA Barcode Screening and Preliminary Study on Phylogeny of Sea Snake Based on Genes COI and cytb
Xiangjun WANG Wen CHEN Haiying SU





Abstract [Objectives] The most common gene fragment used in animal DNA barcode technology is COI , but it is not necessarily suitable for all species. This study was conducted to screen genes suitable for the DNA barcode of sea snakes.
[Methods]All COI and cytb gene sequences on GenBank were searched and downloaded. After the comparison with Mega software, clustering trees of MrBayes system were established.
[Results] Interspecies distances were greater than intraspecies distances for the two genes. The topological structures of their molecular hierarchical clustering trees were clear, and the support rates were high.
[Conclusions]Therefore, it is concluded that not the DNA barcode of each species must be gene COI . Cytb is more suitable in terms of the mitochondrial gene of sea snakes.
Key words Sea snake; DNA barcode; Clustering tree; COI gene; cytb gene
Received: August 23, 2021 Accepted: October 26, 2021
Supported by Hainan Provincial Natural Science Foundation of China, High-level Talent Project (321RC587), Classification of sea snakes in the South Sea China based on molecular Systematics, morphology and climate model; Special Scientific Research Trial Production Project of Sanya City (2016KS05), Identification of sea snake species and construction of DNA barcoding based on molecular systematics.
Xiangjun WANG (1982-), female, associate professor, PhD, devoted to research about ecology and zoology.
*Corresponding author. E-mail: qilong_chen@163.com.
As a fast, effective, accurate and simple method of species identification, DNA barcode technology greatly compensates for the shortcomings of traditional morphological classification methods. It represents the future development trend of species identification research, and is worthy of increasing promotion and application in customs, catering, and identification of Chinese medicinal materials and further in-depth research[1].
At present, the most commonly used gene fragment for animal DNA barcode technology is gene COI . The interspecies variation of this sequence can better distinguish species except Cnidaria, and COI has the advantage of being easy to be amplified by universal primers while ensuring sufficient variation. The DNA sequence itself rarely contains insertions and deletions, and is recognized as a standard DNA barcode gene in the animal kingdom[2-3].
The COI sequence has become the basic fragment of animal DNA barcodes, and has been verified to be effective in identifying wild animals and their fakes[4-6]. In addition, the Cytb gene is also one of the genes commonly used as DNA barcodes. Song et al. [7] used gene Cytb to sequence and analyze Agkistrodon medicinal materials and samples collected in the market, and the results showed that the difference rate of Cytb gene sequence between genuine individuals of the species is far less than that between common adulterants.
Liao et al. [8] pointed out that the smallest interspecies difference in the cytb sequences of various sea snakes is greater than the largest intraspecies difference, indicating that the cytb barcode can be used as a DNA barcode for identifying sea snake species. The COI sequence can also be used as a method for species identification of sun-dried sea snakes, but the number of species mentioned in articles is too small, only 5 species of sea snakes[9].
In this study, a preliminary screening of sea snake genes was conducted by downloading the COI and cytb gene sequences and establishing a phylogenetic tree, and the above two genes were compared for whether they are suitable to be used as DNA barcodes.
Materials and Methods
Materials
All the COI and cytb genes of sea snakes were searched from GenBank, and the information of 46 species of sea snakes with these two genes were downloaded, including a total of 216 sequences. If the sequence number of a certain gene was less than 5, they would be kept all; and if the sequence number of a certain gene was more than 5, 5 sequences would be kept at most. It is necessary to try to choose the longest sequences from different numbering sources. The results are shown in Table 1 and Table 2.
Methods
The downloaded sequences were aligned and cut with the clustal function of the Mega6.0 software. After cutting, COI was retained about 705 bp and cytb was retained about 1 080 bp. fas format was converted to nex format using the SequenceMatrix1.7.8 software, and the MrBayes3.2.6 software was used for calculation. For COI sequences, a tree was constructed with Naja naja as the outer group, on a total of 4 species, about 500 000 generations, and saved about 3 h later. For the Cytb sequence, a tree was constructed with Naja naja as the outer group, on a total of 46 species, about 3.5 million generations, and saved about 36 h later. The FigTree software was adopted for editing of coefficients.
Results and Analysis
Bayesian system clustering tree based on gene COI
Through the Bayesian hierarchical clustering tree (Fig.1), it can be seen that the sequences of the same genus of 2 genera including 4 species in Laticaudinae and Hdrophiinae were clustered together, and individuals of the same species had a significant aggregation trend. The support rates of the corresponding branches of the same species were all higher than 90%, specifically, 95% for one branch and 100% for another branch.
The overall tree had a clear topological structure, and there were no unclear points. However, there were too few species and the overall credibility was not high.
Bayesian system clustering tree based on gene cytb
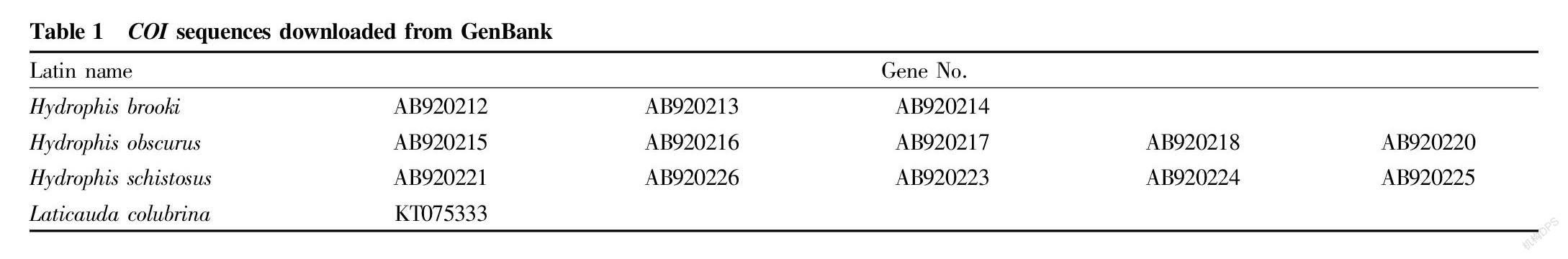
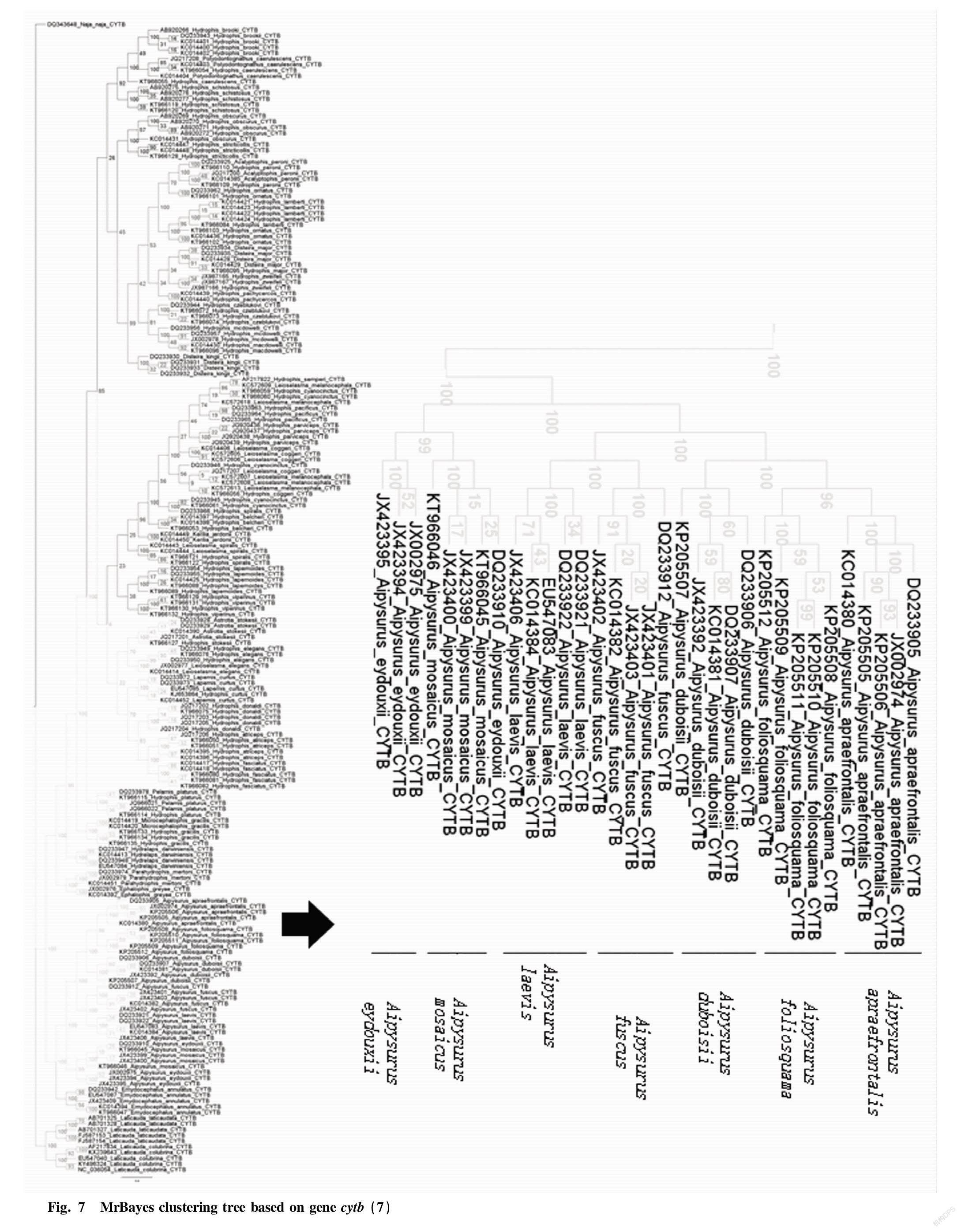
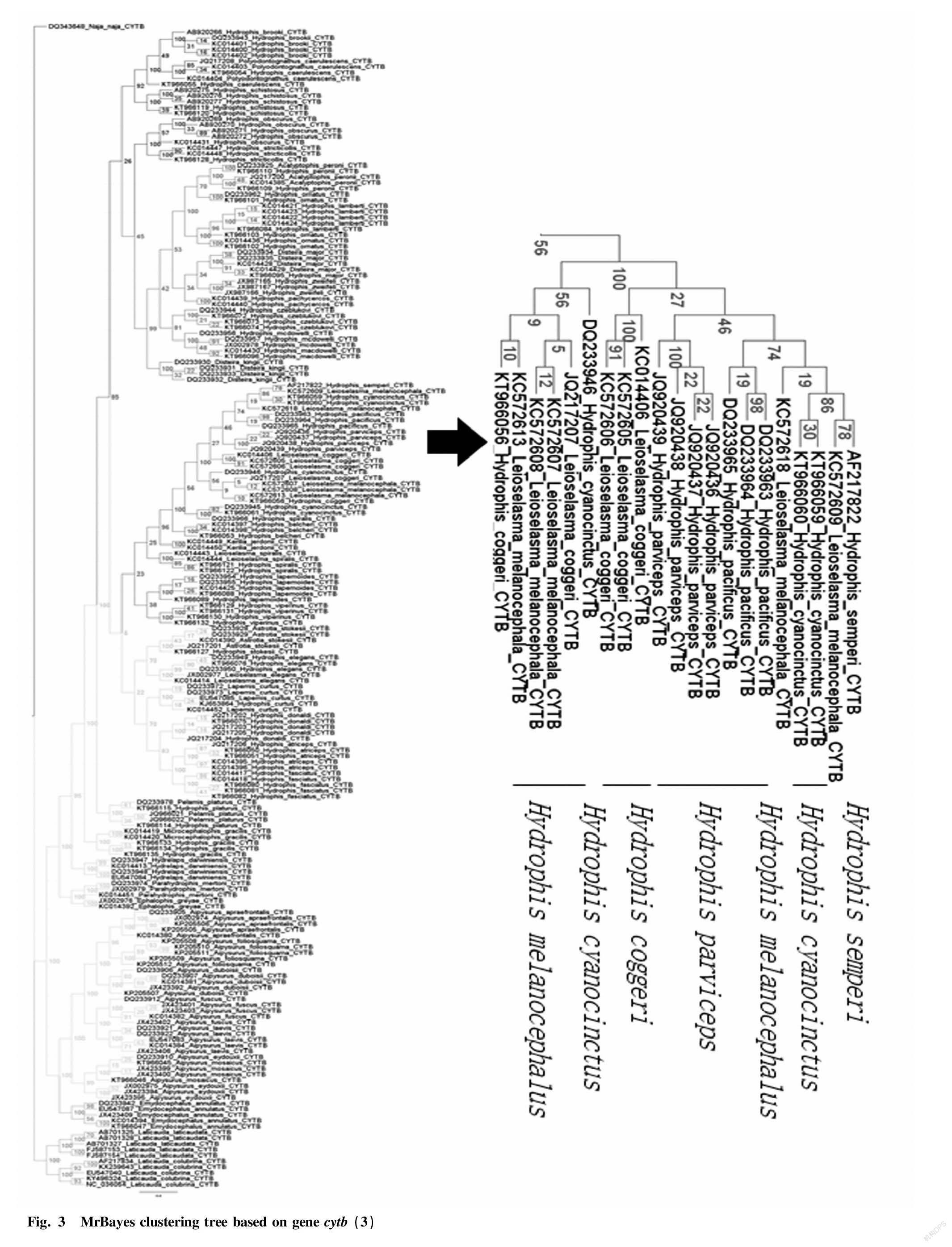
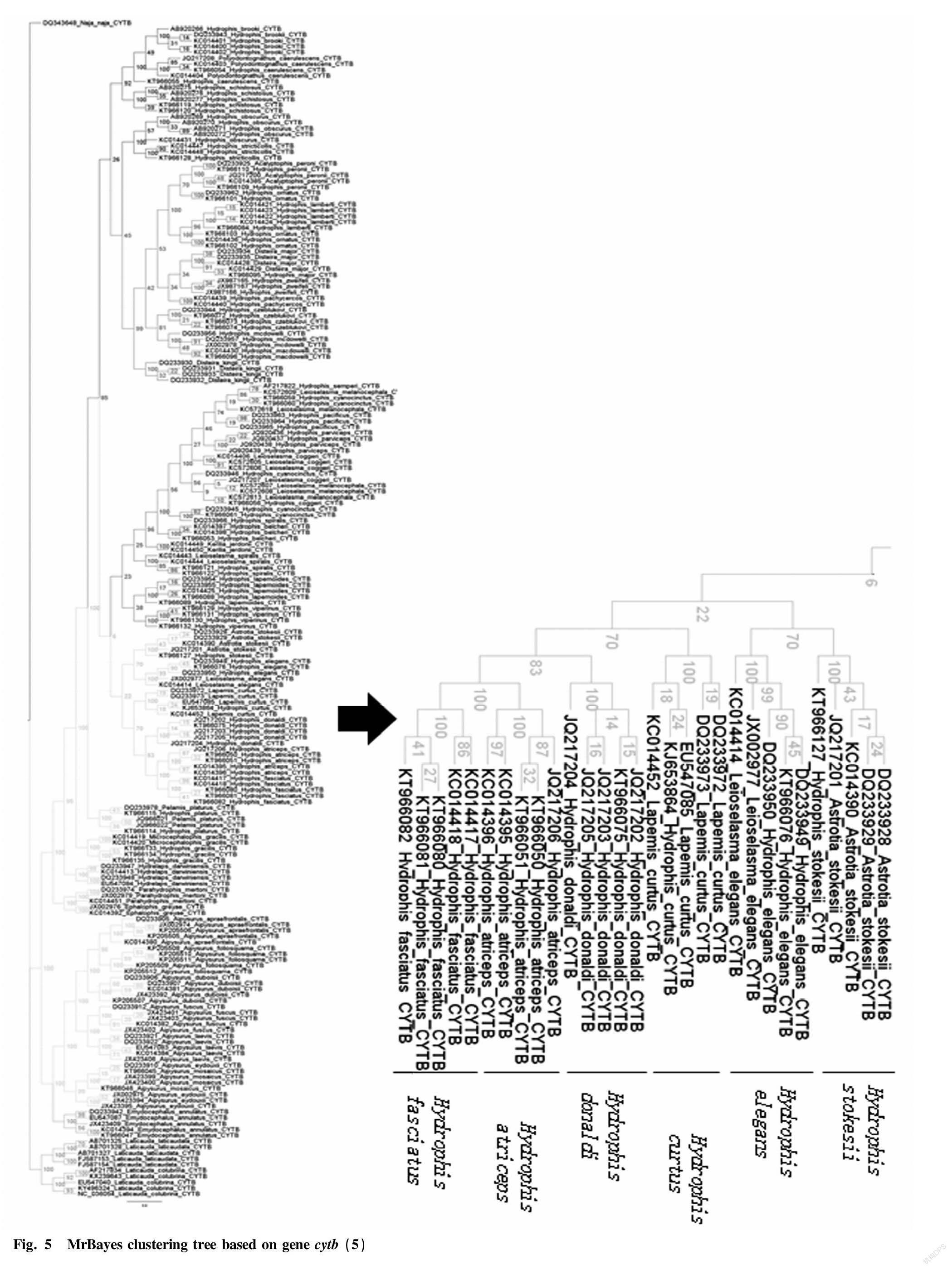
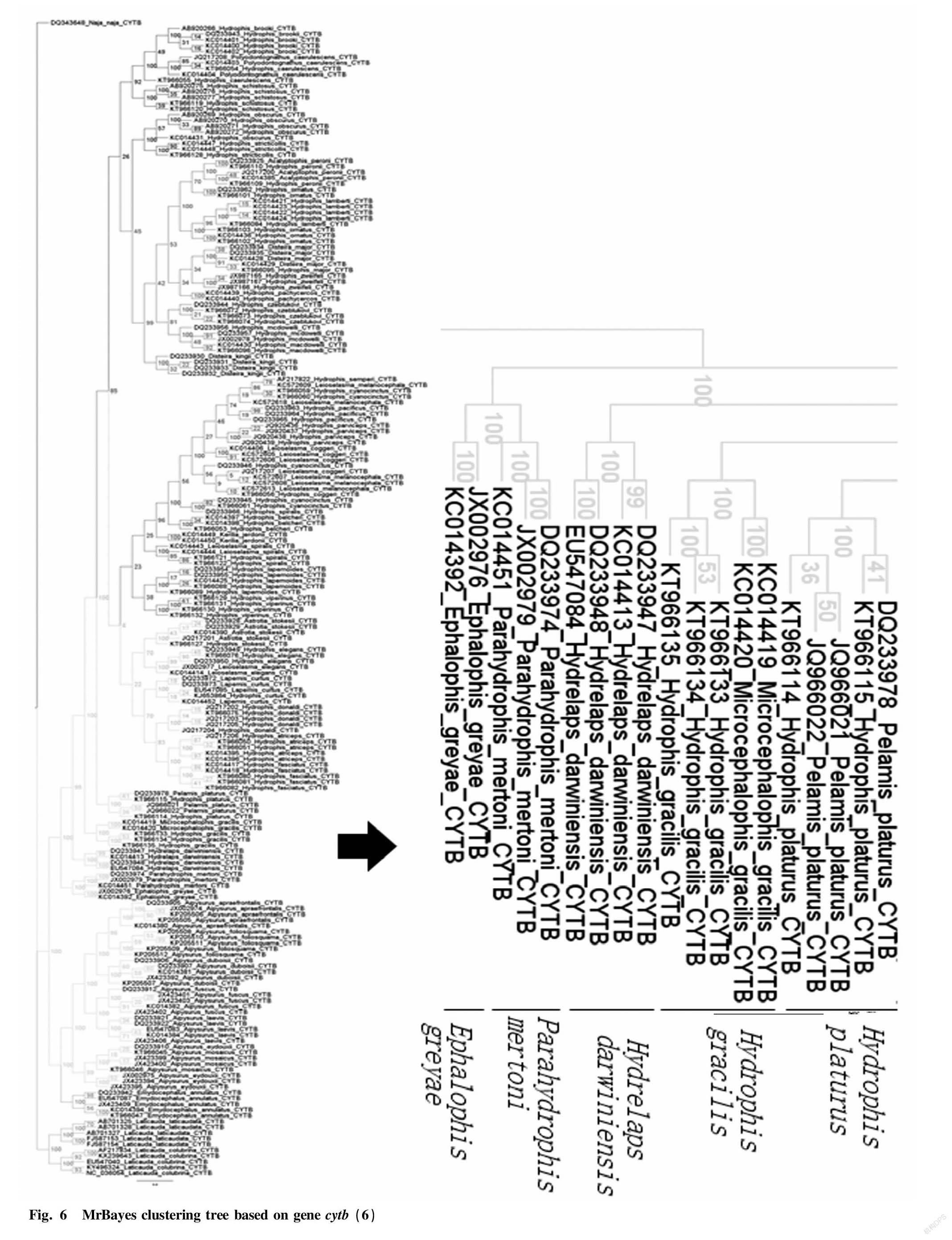
Through the Bayesian hierarchical clustering tree (Fig. 2 to Fig. 8), it can be seen that the sequences of the same genus of 9 genera including 46 species in Laticaudinae and Hdrophiinae were clustered together, and individuals of the same species had a significant tendency to cluster together. The support rates of the corresponding branches of the same species were all 100%, but the support rates among individual branches were not high, and that of one branch was less than 10%.
The overall tree topology was clear, but some nodes were not clear. The names of following species had changed: the present name of Polyodontognathus caerulescens is Hydrophis caerulescens ; the present name of Acalyptophis peronii is Hydrophis peronii ; the present name of Disteira major is Hydrophis major ; Leioselasma coggeri is now Hydrophis coggeri ; the present name of Astrotia stokesii is Hydrophis stokesii ; the present name of Leioselasma elegans is Hydrophis elegans ; the present name of Pelamis platurus is Hydrophis platurus ; and the present name of Microcephalophis gracilis is Hydrophis gracilis .
Individual species were cross-clustered: Hydrophis cyanocinctus , Leioselasma melanocephala and Hydrophis coggeri , the above 3 species were cross-clustered, and the support rates were low.
The genus names of individual species were not consistent, so the classification was not clear. However, the overall tree had high credibility and was suitable for identifying sea snake species.
Xiangjun WANG et al. DNA Barcode Screening and Preliminary Study on Phylogeny of Sea Snake Based on Genes COI and cytb
Discussion
Barcode gene screening
At present, among mitochondrial genes, gene COI is most commonly used for DNA barcoding[4-6]. In the research literature of snake DNA barcoding, except for a few about 16S and cytb genes[8-10], the rest are also about gene COI . For the study of sea snake DNA barcode, CNKI included two articles[9-10], which are based on genes cytb and COI , respectively.
Because both genes COI and cytb have a moderate rate of change, and because the primers used to amplify the above two genes in snakes are highly universal and the amplification conditions are mild, they are often used as DNA molecular markers[5-9,11-12], and interspecies genetic distances are greater than intraspecies genetic distances[8-9]. Therefore, we used both as screening materials for sea snake DNA barcodes.
We searched for all the COI sequences of sea snakes on GenBank, and there were only 17 sequences, which were not suitable for DNA barcodes. The COI gene species of sea snakes were also too few. Although the phylogenetic tree had clear branches and high support rates, but its operability was not great in actual identification of unknown samples.
As for gene cytb , there were as many as 46 species on GenBank (among which 8 species of sea snakes produced in the South China Sea, except for H. melanocephalus , all had sequences) and most species had more than 5 sequences, which were submitted from different sources; and its hierarchical clustering tree did not lose the characteristics of clear branches and high support rates, so gene cytb as a sea snake DNA barcode is more realistic.
To build a clustering tree of DNA barcodes, the MEGA software NJ method is commonly used[1,8-9,13], because of its fast construction speed and high support rates. We tried to use MrBayes software to build a molecular clustering tree, which had a clear topological structure and higher support rates, and most nodes reached more than 90%. In terms of calculation time, single-core calculation took about 1-2 d, but if multi-core calculation was performed, the time could be greatly shortened and the identification result could be obtained quickly.
Phylogeny
From the cytb gene phylogenetic tree, it can be seen that genus Laticauda , genus Aipysurus and genus Emydocephalus were relatively distantly related to other sea snakes, and the species within each genus were obviously clustered into one group, exhibiting clear systematic relationship; and genus Aipysurus was closely related to genus Emydocephalus .
Genus Acalyptophis , genus Astrotia , genus Lapemis , genus Pelamis and genus Praescutata recorded in Chinese Zoology were all included in genus Hydrophis . Therefore, the validity of these five genera needs to be further studied.
Five species of genus Hydrophis, H. caerulescens, H. cyanocinctus, H. fasciatus, H. gracilis , and H. Ornatus , which are distributed in the seas of China, were relatively distant in the phylogeny.
In Chinese Zoology, A. peronii and H. Ornatus , which belong to different genera, were closely related to each other.
Conclusions
Gene COI is the most commonly used gene for DNA barcodes, but it is not necessarily suitable for all species. For genes COI and cytb of sea snakes, cytb on GenBank included many species and sequences, and cytb is more suitable for DNA barcodes compared with gene COI . And the topological structure was clear and the support rates were high when using MrBayes for building a molecular tree.
The validity of some sea snake genera recorded in China Zoology needs to be further studied.
References
[1] HUANG Y, ZHANG YY, ZHAO CJ, et al. DNA barcoding and its utility in commonly-used medicinal snakes[J]. China Journal of Chinese Materia Medica, 2015, 40(5): 868-874. (in Chinese)
[2] COSTA FO, DE WAARD JR, BOUTILLIER J, et al. Biological identifications through DNA barcodes: The case of the Crustacea[J]. Canadian Journal of Fisheries and Aquatic Sciences. 2007, 64(2): 272-295.
[3] HEBERT PD, RATNASINGHAM S, DEWAARD JR. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species[J]. Proceedings: Biological Sciences, 2003, 270(S1): S96-S99.
[4] CHEN N. Sequence analysis of 16SrRNA gene of 14 crude snake venoms sold in the market[C]//Guangdong Pharmaceutical Association. Proceedings of the 2009 Annual Conference of Guangdong Pharmaceutical Association. Guangdong Pharmaceutical Association: Guangdong Pharmaceutical Association, 2010. (in Chinese)
[5] QIN Q. There will be a reliable basis for authenticity identification of medicinal animals: Research on DNA barcoding of medicinal animals starts[J]. Journal of Traditional Chinese Medicine Management, 2010, 18(9): 819-819. (in Chinese)
[6] PENG JL, WANG XZ, WANG D, et al. Application of DNA barcoding based on mitochondrial COI gene sequence in species identification of Culter in Cyprinidae[J]. Acta Hydrobiologica Sinica, 2009, 33(2): 271-276. (in Chinese)
[7] SONG WC, SONG SW, LIU DF, et al. Sequence and analysis of Cytb gene of Agkistrodon and its adulterants sold in market[J] Chinese Traditional and Herbal Drugs, 2006, 20(12): 1862-1865. (in Chinese)
[8] LIAO LX, ZENG KW, TU PF. Hydrophidae identification through analysis on Cytb gene barcode[J]. China Journal of Chinese Materia Medica, 2015, 40(16): 3179-3182. (in Chinese)
[9] LIAO LX, ZENG KW, TU PF. Hydrophidae identification through analysis on cytochrome c oxydase I( COI ) and ribosome 16S rDNA gene barcode[J]. China Journal of Chinese Materia Medica, 2016, 41(10): 1792-1796. (in Chinese)
[10] DENG LB, TANG XL, SHAO JM, et al. Initially study of the 16 S rRNA gene in DNA barcoding for snakes from Jiangxi[J]. Journal of Nanchang University: Natural Science, 2011, 35(1): 95-102. (in Chinese)
[11] AN XY, HUANG Y, DING L, et al. Relationships between Elaphe perlacea and Elaphe mandarinus based on morphological characters and mitochondrial 12S, Cytb and ND4 gene sequences[J]. Journal of Sichuan Forestry Science and Technology, 2017, 38(2): 85-90. (in Chinese)
[12] SHEN X, ZHOU KY, WANG YQ. 12S rRNA and Cytb gene fragments sequence of some species of the genus Agkistrodon [J]. Chinese Journal of Zoology, 2000, 44(1): 18-21. (in Chinese)
[13] CHEN SL, PANG XH, YAO H, et al. Identification system and perspective for DNA barcoding traditional Chinese materia medica[J]. Modernization of Traditional Chinese Medicine and Materia Medica: World Science and Technology, 2011, 13(5): 747-754. (in Chinese)
- 农业生物技术(英文版)的其它文章
- Rice Blast Resistance-associated Genes Based on Different RNA-seq Resources
- Research Progresses on QTLs for Main Grain Shape Genes in Rice
- Effects of Raising Chickens Under Moringa oleifera
- Preliminary Research on Radiation Breeding of Pteroceltis tatarinowii Maxim
- Comparison of Spring Radish Varieties with Entire Leaves
- Occurrence and Chemical Control Techniques of Rice Black-streaked Dwarf Disease in Rongshui County

