奶牛育种中的长寿性状
张海亮,刘澳星,米思远,李想,罗汉鹏,鄢新义,王雅春
奶牛育种中的长寿性状
张海亮,刘澳星,米思远,李想,罗汉鹏,鄢新义,王雅春
(中国农业大学动物科学技术学院,北京 100193)
长寿性状是奶牛育种中最重要的功能性状之一,除产量性状之外,长寿性状具有最大的经济价值。由于遗传力低、数据分布特殊和性状表现晚等特点,长寿性状也是选育难度最大的性状之一。自上世纪50年代起,长寿性状就进入了各国奶牛育种家的视野,针对长寿性状的研究持续进行;上世纪90年代后,各奶业发达国家陆续将长寿性状纳入其奶牛综合选择指数。目前,我国奶牛综合选择指数(CPI)中尚不包含长寿性状,对长寿性状的研究也处于起步阶段。文章通过整理分析奶牛长寿性状的有关研究,从性状定义、遗传评估方法、与其他性状的关系、遗传标记、性状选育策略等方面系统介绍了长寿性状的研究和选育情况。通过汇总有关长寿性状与其他性状遗传相关的研究,阐述了长寿性状与其他性状之间的复杂关系;通过收集各国奶牛选育方案中的相关信息,重点介绍了各主要奶业发达国家对长寿性状的选育策略。此外,本文还通过收集在我国奶牛群体中针对长寿性状开展的研究,概述了奶牛长寿性状在我国的研究现状。长寿性状有许多不同的定义,可使用不同的模型对其进行遗传评估,包括线性模型、阈模型、生存分析模型和随机回归模型等。长寿性状与产量、体型、繁殖、健康和管理类性状等存在低到中等的遗传相关,线性体型性状中,与泌乳系统有关的体型性状与长寿性状的遗传相关较高;繁殖和健康性状表现更好的奶牛,其长寿性状通常表现更好;长寿性状与其他性状之间的遗传关系受牛群选育方向的影响较大,在不同群体中,长寿性状与同一性状的遗传相关不尽相同。奶业发达国家的奶牛综合选择指数中均包含长寿性状,这些国家除了使用直接长寿进行选择之外,部分国家还同时采用间接选择的方法对长寿性状进行选育,常用于间接选择长寿性的其他性状主要包括泌乳系统、腰强度、尻角度、肢蹄和乳房炎抗性等。在不同群体中,均发现了大量与长寿性状相关的遗传标记,其中大多数标记都定位在已报道的与繁殖、疾病和体型等性状相关的遗传区域内。文章还提出了从数据收集、遗传参数估计、遗传标记挖掘、评估模型和选择策略探讨等方面针对我国奶牛群体进行长寿性状研究的必要性。
奶牛;长寿性;遗传评估;遗传相关;遗传标记;遗传选育
长寿是奶牛生产盈利的基础要素,除了产量性状之外,长寿性状具有最大的经济价值,是奶牛育种中最重要的功能性状之一。每头奶牛只有弥补了其在后备牛阶段的培育成本之后,才有可能为牧场带来收益。一般认为奶牛在第二个泌乳期能够保持收支平衡,只有达到第三个泌乳期及以上,才能够获得较高的利润率[1]。提高奶牛长寿性,可以延长奶牛盈利期的长度,获得更高的收益;可以减少后备牛的饲养头数,降低牛群更新的成本;在牛群规模不变的情况下,可以允许牧场进行更高比例的主动淘汰,增加选择强度,进而加快牛群的遗传进展;牛群更加长寿意味着群体的健康水平更高,牧场的兽药和兽医投入也会更少[2-4]。
影响奶牛长寿性的因素有很多,所有能够影响奶牛淘汰的因素均会对奶牛长寿性产生影响。奶牛淘汰时所记录的淘汰原因多种多样,按照牧场管理者淘汰时的意愿,可以分为主动淘汰和被动淘汰[5]。主动淘汰的原因主要有年老、低产或育种计划等,被动淘汰则包括繁殖障碍、产后疾病、乳房疾病、肢蹄疾病、消化代谢疾病、外伤等,这些疾病和外界因素将导致奶牛生产能力下降或消失,甚至个体死亡,从而影响奶牛的长寿性。由于长寿性影响因素复杂、性状定义多样、数据分布特殊、性状表现晚、遗传力低(大多在0.02—0.15[6-8])等特点,长寿性状是奶牛育种中最复杂、选育最困难的性状之一。本文将从长寿性状的定义、遗传评估、与其他性状的关系、遗传标记、选育方法等方面对长寿性状进行概述,以期能帮助我国相关研究人员系统地了解奶牛长寿性状,对我国奶牛长寿性状的研究和选育提供参考和借鉴。
1 长寿性状的定义及遗传评估
1.1 长寿性状定义
根据观察值的数据类型,可将各种长寿性状的定义分为两类。第一类定义以奶牛在牧场中整个生存周期或某一(几)个生存阶段中的生存时间(单位为天、月、胎次等)作为观察值[9-10],包括在群寿命(herd life)、生产寿命(productive life)、产奶寿命(milking life)和产犊次数(胎次数)等。在群寿命代表了一头奶牛从出生到离群的整个过程中,在牧场中度过的时间;生产寿命代表了一头奶牛从头胎产犊到离群,在牧场中度过的时间;在生产寿命的基础上,去除奶牛各胎次的干奶期长度,即为产奶寿命。第二类定义以个体在某一(几)个生存阶段内生存或死亡的状态(survival)作为观察值,作为二分类阈性状(0或1)[11-13],可将此类性状称为某阶段(观测时间点)存活率。为了能与以时间长度定义的第一类长寿性状保持一致,通常将存活定义为1,淘汰或死亡定义为0。在计算个体的表型时,不同定义的性状所需要的信息不同;在选育中,不同定义的长寿性状对奶牛的侧重点也不尽相同。
此外,对长寿性状进行遗传评估时,根据是否校正个体产量水平,可将长寿性状分为功能长寿(functional longevity,校正产量)和真实长寿(true longevity,不校正产量)[11]。功能长寿代表了奶牛抗被动淘汰的能力,而真实长寿则同时代表了奶牛抗被动淘汰和抗主动淘汰的能力。根据长寿性状的信息组成,可将长寿性状分为直接长寿和间接长寿。直接长寿利用奶牛的生存信息定义性状,直接描述了一头奶牛的存活状态或存活时间;而间接长寿则利用各种能够早期测定且与直接长寿遗传相关较高的其他性状,间接预测了一头奶牛的生存能力[13]。
1.2 长寿性状遗传评估
针对不同定义的各种长寿性状,不同国家和研究单位使用不同的方法对长寿性状进行遗传评估,常用的模型有线性模型、阈模型[14]、随机回归模型[15-16]和生存分析模型等[17-18]。线性模型可以对以时间长度定义的寿命类或以存活状态定义的生存率类长寿性状进行遗传评估,阈模型可以对存活率类性状进行遗传评估。这两种模型的数据准备过程简单,可选用的评估软件较多,各种软件的计算速度通常较快,其评估结果一般能够满足公牛排队中对育种值估计可靠性和参数估计稳定性的要求。对存活率类长寿性状进行遗传评估时,线性模型和阈模型评估结果之间的秩相关高达0.90以上[14],这两种评估模型对公牛排队的影响较小;使用相同软件时,线性模型的计算速度通常更快。
生存分析是研究生存数据及其统计规律的一种分析方法,生存数据描述了从事件起始到事件结束之间的时间长度,长寿性状的观测值是一类典型的生存数据。根据模型特点,可将生存分析模型划分为参数模型、半参数模型和非参数模型,常用的参数模型有指数分布模型、对数逻辑斯谛分布模型、威布尔分布模型、伽玛分布模型、对数正态分布模型等。根据比例风险模型中基础风险函数的分布,可划分为基础风险函数为常数的指数比例风险模型、基础风险函数服从Weibull 分布的Weibull比例风险模型、基础风险函数任意的Cox比例风险模型等。生存分析能更好地拟合长寿性状的数据分布,且能利用存活个体的数据(删失数据),其评估结果的可靠性通常优于其他方法[7];但该方法也存在计算速度慢、表型数据完整度要求较高、数据准备复杂等限制因素。此外,受软件计算能力限制,使用参数模型的生存分析目前无法使用动物模型进行评估。评估生产寿命性状时,线性模型与生存分析模型评估结果的差异主要来自于所用数据的差异;数据相同时(例如,生存分析中不考虑删失数据),两种方法评估结果的秩相关可达0.90以上[19-20]。
2 长寿性状与其他性状的关系
出于对长寿性状进行间接选择的考虑,长寿性状与其他性状之间的关系一直是长寿性状的研究重点。通过不同的方法,长寿性状与体型、产量、繁殖、健康、管理等五大类性状之间的关系被大量报道。例如,采用多性状遗传分析的方法,探究长寿性状与其他性状之间的遗传相关;采用生存分析的方法,研究各性状的变异对奶牛淘汰风险的影响,确定各性状能够解释长寿性状变异的相对大小;利用单性状模型分别评估获得的育种值,计算长寿性状与其他性状之间的近似遗传相关;利用回归的方法确定长寿性状与其他性状的关系等。
2.1 长寿性状与其他性状的遗传关系
在荷斯坦牛[21-22]、西门塔尔牛[23]、更赛牛[24]和瑞士褐牛[25]等常见乳用品种中,长寿性状与部分线性体型性状之间均存在低到中等的遗传相关。本文收集分析了各国奶牛群体中长寿性状与主要线性体型性状之间的遗传相关,长寿性状与各体型性状的遗传相关范围如图1所示。在不同群体中,泌乳系统相关的线性体型性状与长寿性状存在较高的遗传相关,如泌乳系统得分、乳房深度、前乳头位置、后乳房高度、后乳房宽度等[26-28];与运动相关的体型性状与长寿性之间的遗传相关较低,甚至接近于零,如肢蹄、骨质地、蹄角度、后肢后视等[29];与体躯大小相关的线性体型性状与长寿性状之间多呈低到中等的负遗传相关,如体躯容量、体高、体深等。同一群体中,随着牛只出生年份的变化,荷斯坦牛线性体型性状与长寿性状之间的遗传相关也在不断变化[21, 28]。
本文收集分析了各国奶牛群体中常见的产量、繁殖、健康和管理性状与长寿性状之间的遗传相关,长寿性状与各性状的遗传相关范围如图2所示。在各国群体中,产量性状与长寿性状间的遗传相关范围较大。在一些群体中,基于头胎产量水平进行的大量主动淘汰造成长寿性状和产量性状呈现较高的正遗传相关[26, 30];当使用校正头胎产量的功能长寿时,长寿性状与产量性状之间的遗传相关明显降低。随着对牛群产量的高强度选育,高产给予奶牛对抗主动淘汰的优势不再明显。例如,在1979—1993年间出生的美国荷斯坦牛中,产量性状与生产寿命之间的遗传相关逐渐减小[28]。与低产奶牛相比,高产奶牛抗被动淘汰的能力更差;在部分群体中,产量性状与功能长寿之间存在负遗传相关[25]。在各国群体中,繁殖性状与长寿性状存在低到中等的遗传相关,繁殖性能表现更好的母牛,其长寿性表现更好。疾病是引起奶牛淘汰的重要因素,奶牛乳房炎和部分繁殖疾病(繁殖障碍、卵巢囊肿)与长寿性状存在低到中等的负遗传相关[30-33],奶牛疾病抗性越强则长寿性表现越好。此外,与体细胞评分相比,临床乳房炎与长寿性之间的遗传相关更高[32- 34]。
2.2 长寿性状与其他性状之间的表型关系
采用生存分析的方法可以分析各性状的变异对长寿性的影响,尤其适用于各种分类性状,如线性体型性状、疾病性状和部分繁殖性状等。通过比较模型中纳入各性状时模型似然率的变化,可以估计各性状能够解释长寿性状变异的相对大小;通过计算各性状不同水平的相对淘汰风险(OR),可以反应各性状的变异对长寿性的具体影响。
在各线性体型性状中,泌乳系统相关的性状对长寿性有最大的影响,如乳房深度、前乳房附着、后乳房高度、中央悬韧带等性状[35-38];除乳头长度外,泌乳系统相关性状表现更好的奶牛,其相对淘汰风险越低[35]。对于体躯容量性状,荷斯坦牛中,拥有中等体型的奶牛,其相对淘汰风险更低[35, 38];而在娟姗牛和爱尔夏等体型较小的品种中,体型更大的奶牛,其淘汰风险更低[39]。体躯容量相关的性状受牛群遗传改良方向的影响较大,在不同群体间,同一种性状对长寿性的影响差异较大。此外,乳用特征、泌乳速度等体型性状对长寿性也有较大的影响。

以存活状态定义的长寿性状统一转换为存活率(将存活定义为1),其估计育种值的有利方向与以时间长度定义的长寿性状一致;不返情率、临床乳房炎和繁殖疾病分别以不返情、临床乳房炎发病、繁殖疾病发病定义为1;图中“”“”“”分别表示绘图时参考的遗传相关研究数目<5条、5—10条、>10条;遗传相关数值的参考文献略
在荷斯坦牛、娟姗牛和爱尔夏牛中,产犊难易、犊牛大小、授精次数、产后首配日龄、首配至妊娠间隔及空怀天数等繁殖或产犊性状对功能生产寿命有显著影响,繁殖性状表现越好的奶牛,其相对淘汰风险越低[40]。例如,产犊间隔、空怀天数和首配至妊娠间隔越短,奶牛淘汰风险越低。体细胞评分对功能生产寿命有显著的影响,母牛的淘汰风险随体细胞评分的升高而增加,高体细胞评分奶牛的淘汰风险可达到牛群平均体细胞评分个体的4—6倍[41];与高体细胞数牛场相比,个体体细胞数相同幅度的增加,低体细胞数牛场的个体淘汰风险的增加幅度更大[42];与健康牛相比,泌乳期内任何时间患乳房炎均会造成奶牛淘汰风险的增加,泌乳高峰期时淘汰风险的增加幅度最大[34]。此外,通过生存分析的方法,发现泌乳性情[43]、泌乳速度[43]、出生时母亲年龄[44]、牛奶尿素氮含量[45]、乳糖率[45]、近交率[46]等性状的变异对荷斯坦牛、爱尔夏牛和娟姗牛的长寿性存在显著影响。
3 长寿性状的相关基因及分子标记
3.1 长寿性状候选基因关联分析
筛查前人通过QTL定位的结果或根据基因已知的生理功能初步筛选长寿性状潜在的候选基因,通过候选基因关联分析的方法,可以评估目标基因与长寿性状的关系,进而发现长寿性状的候选基因。在长寿性状的候选基因关联分析中,通常使用生产寿命育种值作为关联表型,使用线性混合模型或生存分析模型进行关联分析,各研究中选择进行关联分析的基因大多为已报道的影响奶牛产奶性能、健康状况和繁殖性能的基因。在不同群体中,α酪蛋白s1(casein alpha s1,)催乳素受体(prolactin receptor,)奶脂肪球-EGF因子蛋白8(milk fat globule-EGF factor 8 protein,)非受体酪氨酸激酶,SRC原癌基因(SRC proto-oncogene, non-receptor tyrosine kinase,)酰基甘油O-酰基转移酶1(diacylglycerol O-acyltransferase 1,)β酪蛋白(casein beta)[47]瘦素(leptin,)[48]硬脂酰辅酶A去饱和酶(stearoyl-CoA desaturase,)[49]丝氨酸蛋白酶抑制因子,分支A(α-1抗蛋白酶,抗胰蛋白酶),成员14(serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin),member 14,别称)[50]钙抑素(calpastatin,)[51]和serpin家族A成员1(serpin family A member 1,)[52]等基因存在与长寿性状显著关联的SNP或单倍型,这些基因可作为长寿性状的候选基因。
在部分发达国家的奶牛群体中,因低产而进行的主动淘汰是影响奶牛长寿性的重要原因;由于对产量性状的影响,可能造成了[53]和[54-55]等基因在特定群体中间接影响了长寿性状;在波兰群体中,当校正个体产量水平的影响后,则对长寿性状没有显著的影响[48]。酪蛋白家族基因在奶牛的泌乳和健康中扮演着主要的角色[53-56],和与长寿性状间的关系进一步证明了这一点。在不同群体中,与奶牛的适应性、代谢和健康之间的具体关系存在争议,其中一些研究报道了与疾病风险[57]、死产[58]和犊牛存活率[58]等性状之间存在显著的关系,而这些性状导致的奶牛淘汰均是长寿性状的重要组成部分。
3.2 长寿性状全基因组关联分析
全基因组关联分析(genome-wide association analysis,GWAS)是一种鉴定目标性状相关分子标记的有效方法,各国研究人员采用不同的GWAS方法鉴定了大量与长寿性状显著相关的分子标记(表1),这些信息为长寿性状的标记辅助选择和基因组选择提供了有用信息,同时也为阐释长寿性状的分子遗传基础奠定了基础。在长寿性状的GWAS中,以存活时间长度为单位的生产寿命和在群寿命是最常用的性状定义方法,单标记回归是最常用的关联分析方法,逆回归育种值则是最常用的关联分析伪表型。
在各关联分析中,在6、14、16、18和X染色体上发现的显著SNP最多,这些SNP大多定位在已知的与奶牛临床乳房炎、体细胞数(评分)、产犊性状(产犊难易、犊牛大小、死产)、繁殖性状、体型性状相关的QTL区间内。如,Cole等[59]利用1 654头荷斯坦母牛进行长寿性GWAS时,分别在7号染色体15.8 Mb处和X染色体106 Mb处发现了两个与体细胞评分、女儿妊娠率共同相关的遗传区域;Nayeri等[60]利用6 116头公牛进行长寿性GWAS时,在13号染色体59 Mb和18号染色体42 — 65 Mb处定位到的遗传区域均与前人发现的产犊性状QTL区域重合。在各国群体中,健康和繁殖问题均是引起奶牛淘汰的主要原因,各种长寿性状实际上度量了奶牛受产量、健康和繁殖等因素影响的存活时间(能力)。因此,对奶牛长寿性状进行的GWAS,大多发现了与上述性状共同相关的遗传标记。
4 长寿性状的选育
4.1 各国奶牛育种体系中的长寿性状
上世纪50年代以来,对长寿性状的大量研究已经证明长寿性是可遗传的,对牛群的长寿性进行选育提高是可能的。上世纪90年代之后,各国奶牛育种协会陆续将长寿性状纳入遗传评估计划,随着对长寿性状的持续研究,长寿性状的定义和遗传评估方法也在不断发展[66]。目前,世界各主要奶业发达国家对长寿性状的选育情况如表2所示。大多数国家的奶牛综合选育指数中都包含长寿性状,其权重多在5%—10%,德国尤其重视奶牛的长寿性选育,其总性能指数(RZG,2018)中长寿性状的权重高达20%。各国对长寿性状的定义和评估方法不尽相同,将长寿性状定义为奶牛在某几个生命阶段的存活状态是最常见的定义方法;大多数国家采用线性模型对各种长寿性状进行遗传评估,以法国为代表的欧洲联合遗传评估组织使用生存分析的方法进行遗传评估,荷兰则使用随机回归的方法对长寿性状进行遗传评估。
4.2 长寿性状的间接选择
利用线性模型评估不同定义的直接长寿,需等到公牛的部分后裔淘汰或死亡后,才能获得表型信息;能够利用删失数据的生存分析方法,其估计育种值的可靠性也仅取决于公牛后裔中非删失的女儿数[67],这使得长寿性状的选育十分困难,长寿性状成为了奶牛育种中世代间隔最长的性状。因此,在利用直接长寿进行选择的同时,许多国家也考虑了一些与长寿性状遗传相关较高、并能早期测定的性状进行间接选择,如美国、加拿大、法国和新西兰等(表2)。
在性状的选择上,早期测定、遗传力较高的线性体型性状成为了长寿性状间接选择的首选性状[68],其中体型总分、泌乳系统、腰强度、尻角度、肢蹄等最为常用;在各国群体中,体细胞评分(数)和临床乳房炎也是间接选择长寿性的常用性状;在美国群体中,因产量水平进行的主动淘汰比例较高,产奶量、乳脂量和乳蛋白量也是长寿性状间接选择的重要性状;此外,泌乳速度和部分繁殖性状也比较常用。
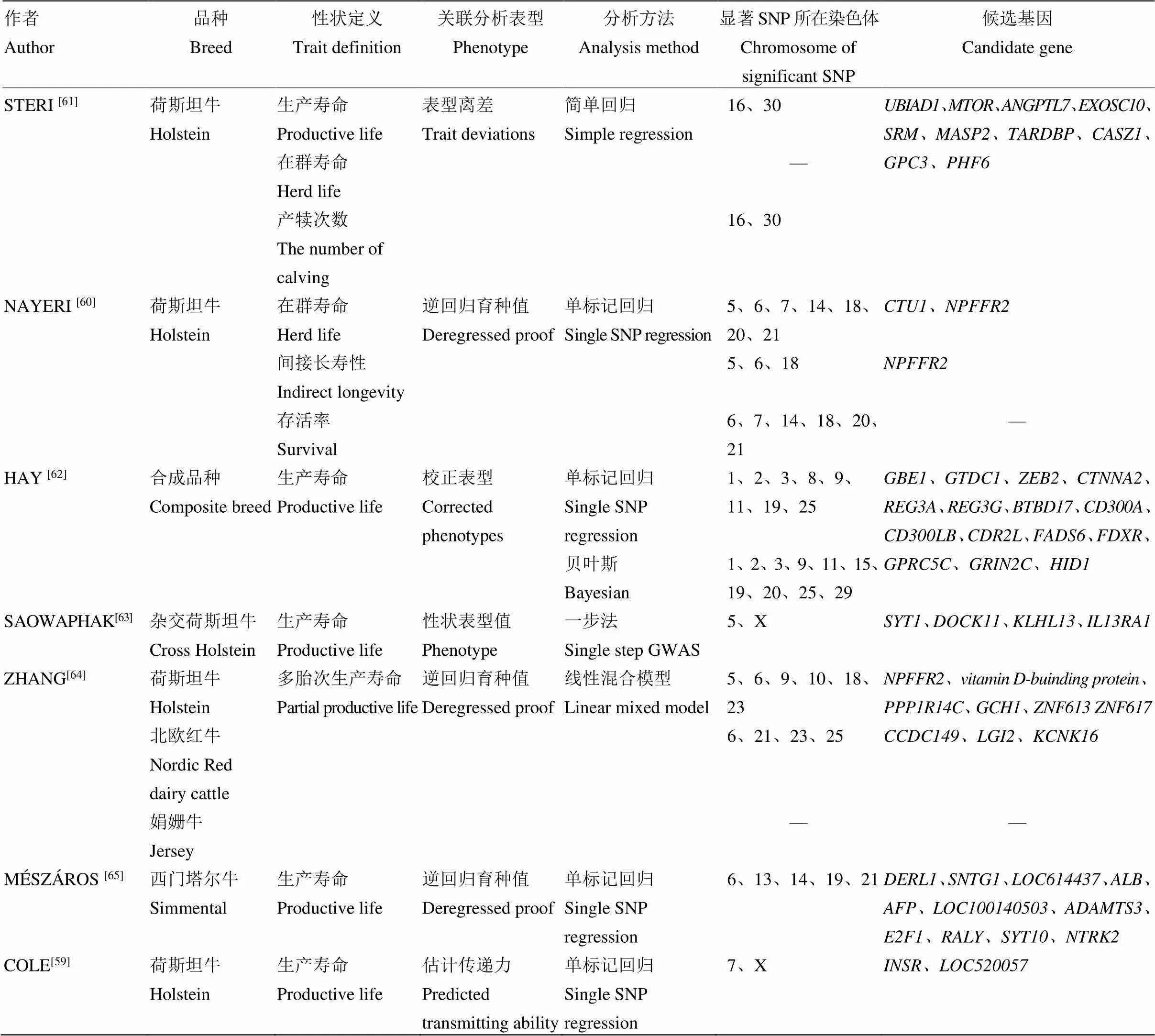
表1 长寿性状全基因组关联分析研究
4.3 长寿性状的基因组选择
由于能够缩短世代间隔、提高选择的准确性,加快遗传进展,基因组选择方法已经在越来越多国家的奶牛育种中得以应用[69]。对于表现时间晚、遗传力低的长寿性状,基因组选择的应用潜力巨大。通过建立长寿性状基因组选择的参考群,使长寿性的选育不再依赖被选择个体的后裔淘汰信息,能够大幅缩短长寿性状的世代间隔。目前,许多国家都已经开展了长寿性状基因组选择的研究和应用,奶业发达国家的成功应用证明对长寿性状进行基因组选择是可行的。据报道,德国荷斯坦公牛长寿性状的基因组育种值可靠性可以达到51%,比系谱指数提高了17%[70];美国荷斯坦公牛长寿性状的基因组育种值可靠性可以达到45%,比系谱指数提高了18%[71]。
5 展望
据调查,我国规模化奶牛场中荷斯坦牛的平均利用胎次不足三胎[72],远没有达到奶牛生产力最高的阶段,严重影响着奶牛养殖的效益。目前,在奶牛育种中,兼顾高产和健康长寿的平衡育种理念已经成为各国奶牛育种界的共识。在我国,长寿性状还没有纳入中国奶牛选育指数,对长寿性状的研究处于起步阶段,仅有使用局部地区的牛群数据进行的少量研究。例如,分别针对北京地区[10]和上海地区[73]牛群进行的遗传参数估计、使用微卫星标记定位方法[74]和候选基因关联分析方法[75-78]挖掘长寿性状的遗传标记。目前,我国奶牛群体的长寿性表现较差,不仅严重影响了生产效率,而且也挤压了进行高强度主动淘汰选育其他性状的空间,我国牛群的长寿性状选育刻不容缓。在常规遗传评估中,各国研究人员从模型和性状定义等角度提出了许多对长寿性状进行早期选择的方法;基因组选择技术在奶牛育种中成功应用之后,有效解决了该性状选择世代间隔长的问题;因此,在基因组评估中,长寿性状评估时所使用的定义和模型还需进一步探讨。有研究指出,在基因组选择中,将与目标性状相关的分子标记信息加入到基因组数据中可以提高基因组选择的准确性[79-80];在我国奶牛群体中,挖掘与长寿性状相关的遗传标记可以为该性状的基因组选择和标记辅助选择提供有用的信息。此外,长寿性状的遗传力较低,其受遗传与环境互作效应及气候条件、牧场管理等非遗传因素的影响较大;我国奶牛养殖在全国各地均有分布,不同地区之间生产环境条件和管理模式的差异较大,研究长寿性状中遗传与环境之间的互作具有较大意义。综上,针对我国奶牛群体,有必要统筹全国牛群数据,从长寿性状的遗传参数估计、遗传评估方法论证、遗传标记挖掘、基因组选择方法和选育策略等方面开展系统研究十分必要。

表2 各国长寿性状遗传评估及选育情况
信息节选自Interbull(https://interbull.org/index)、各国品种协会官方网站;表中各项信息均为长寿性状在荷斯坦牛中的选育信息
Adapted from website of Interbull (https://interbull.org/index) and breed associations from various countries. The information in this table is the selection information of longevity traits in Holstein cattle
随着我国奶牛养殖对长寿性状的不断认识,以及育种数据的不断积累,未来一段时间,长寿性状将会成为我国奶牛遗传育种领域的研究热点,将长寿性状纳入奶牛选育指数是我国奶牛育种的必然趋势。最终,通过遗传选育的手段,改善牛群的长寿性,进而提高奶牛的终生效益,不断提高我国奶牛养殖的竞争力。
[1] MURRAY B. Finding the tools to achieve longevity in Canadian dairy cows., 2013, 25: 15-28.
[2] JAIRATH L K, HAYES J F, CUE R I. Multitrait restricted maximum likelihood estimates of genetic and phenotypic parameters of lifetime performance traits for Canadian Holsteins., 1994, 77(1): 303-312. DOI: 10.3168/jds.S0022-0302(94)76955-1.
[3] ESSL A. Longevity in dairy cattle breeding: a review., 1998, 57(1): 79-89. DOI: 10.3168/jds.S0022- 0302(94)76955-1.
[4] BROTHERSTONE S, VEERKAMPeerkamp R F, Hill W G. Genetic parameters for a simple predictor of the lifespan of Holstein- Friesian dairy cattle and its relationship to production., 1997, 65(1): 31-37. DOI: 10.1017/S135772980001626X.
[5] WEIGEL K A, PALMER R W, CARAVIELLO D Z. Investigation of factors affecting voluntary and involuntary culling in expanding dairy herds in Wisconsin using survival analysis., 2003, 86(4): 1482-1486. DOI: 10.3168/jds.S0022-0302(03)73733-3.
[6] SEWALEM A, KISTEMAKER G J, DUCROCQ V. Genetic analysis of herd life in Canadian dairy cattle on a lactation basis using a Weibull proportional hazards model., 2005, 88(1): 368-375. DOI: 10.3168/jds.S0022-0302(05)72696-5.
[7] CARAVIELLO D Z, WEIGEL K A, GIANOLA D. Comparison between a Weibull proportional hazards model and a linear model for predicting the genetic merit of US Jersey sires for daughter longevity., 2004, 87(5): 1469-1476. DOI: 10.3168/ jds.S0022-0302(04)73298-1.
[8] DUCROCQ V, QUAAS R L, POLLAK E J. Length of productive life of dairy cows. 2. variance component estimation and sire evaluation., 1988, 71(11): 3071-3079. DOI: 10.3168/ jds.S0022-0302(88)79907-5.
[9] TSURUTA S, MISZTALisztal I, LAWLOR T J. Changing definition of productive life in US Holsteins: effect on genetic correlations., 2005, 88(3): 1156-1165. DOI: 10.3168/ jds.S0022-0302(05)72782-X.
[10] 李想, 鄢新义, 罗汉鹏, 刘林, 郭刚, 王新宇, 王雅春. 不同模型估计中国荷斯坦牛生产寿命遗传参数. 畜牧兽医学报, 2019, 50(06): 1162-1170. DOI: 10.11843/j.issn.0366-6964.2019.06.006.
LI X, YAN X Y, LUO H P, LIU L, GUO G, WANG X Y, WANG Y C. Genetic parameters for productive life of Chinese Holsteins by different models.2019, 50(06): 1162-1170. DOI: 10.11843/j.issn.0366-6964.2019.06. 006. (in Chinese)
[11] SHORT T H, LAWLOR T J. Genetic parameters of conformation traits, milk yield, and herd life in Holsteins., 1992, 75(7): 1987-1998. DOI: 10.3168/jds.S0022-0302(92)77958-2.
[12] VISSCHER P M, GODDARD M E. Genetic parameters for milk yield, survival, workability, and type traits for Australian dairy cattle., 1995, 78(1): 205-220. DOI: 10.3168/jds. S0022-0302(95)76630-9.
[13] JAIRATH L, DEKKERS J C, SCHAEFFER L R. Genetic evaluation for herd life in Canada., 1998, 81(2): 550-562. DOI: 10.3168/jds.S0022-0302(98)75607-3.
[14] BOETTCHER P J, JAIRATH L K, DEKKERS J C. Comparison of methods for genetic evaluation of sires for survival of their daughters in the first three lactations., 1999, 82(5): 1034. DOI: 10.3168/jds.S0022-0302(99)75324-5.
[15] VEERKAMP R F, BROTHERSTONE S, ENGEL B. Analysis of censored survival data using random regression models., 2001, 72(1): 1-10. DOI: 10.1017/S1357729800055491.
[16] PELT M L VAN, MEUWISSEN T H E, JONG G DE. Genetic analysis of longevity in Dutch dairy cattle using random regression., 2015, 98(6): 4117-4130. DOI: 10.3168/jds. 2014-9090.
[17] IMBAYARWO-CHIKOSI V E, DZAMA K, HALIMANI T E. Genetic prediction models and heritability estimates for functional longevity in dairy cattle., 2015, 45(2): 105-121. DOI: 10.4314/sajas.v45i2.1.
[18] SASAKI O. Estimation of genetic parameters for longevity traits in dairy cattle: A review with focus on the characteristics of analytical models., 2013, 84(6): 449-460. DOI: 10.1111/asj.12066.
[19] VOLLEMA A R, GROEN A F. A comparison of breeding value predictors for longevity using a linear model and survival analysis., 1998, 81(12): 3315-3320. DOI: 10.3168/jds.S0022-0302(98)75897-7.
[20] LUBBERS R, BROTHERSTONE S, DUCROCQ V P. A comparison of a linear and proportional hazards approach to analyse discrete longevity data in dairy cows., 2000, 70(2): 197-206. DOI: 10.1017/S1357729800054667.
[21] VOLLEMA A R, GROEN A F. Genetic correlations between longevity and conformation traits in an upgrading dairy cattle population., 1997, 80(11): 3006-3014. DOI: 10.3168/jds.S0022-0302(97)76267-2.
[22] ZAVADILOA L, STIPKOVA M. Genetic correlations between longevity and conformation traits in the Czech Holstein population., 2012, 57(3): 125-136. DOI: 10.17221/5566-CJAS.
[23] ZAVADILOA L, NEMCOVA E, STIPKOVA M. Relationships between longevity and conformation traits in Czech Fleckvieh cows., 2009, 54(9): 385-394. DOI: 10. 17221/1685-CJAS.
[24] CRUICKSHANK J, WEIGEL K A, DENTINE M R. Indirect prediction of herd life in Guernsey dairy cattle., 2002, 85(5): 1307-1313. DOI: 10.3168/jds.S0022-0302(02) 74195-7.
[25] VUKASINOVIC N, MOLL J, KUNZI N. Genetic relationships among longevity, milk production, and type traits in Swiss Brown cattle., 1995, 41(1): 11-18. DOI: 10.1016/0301-6226(94)00044-8.
[26] WEIGEL K A, LAWLOR J T J, VANRADEN P M. Use of linear type and production data to supplement early predicted transmitting abilities for productive life., 1998, 81(7): 2040-2044. DOI: 10.3168/jds.S0022-0302(98)75778-9.
[27] SETATI M M, NORRIS D, BANGA C B. Relationships between longevity and linear type traits in Holstein cattle population of Southern Africa., 2004, 36(8): 807-814. DOI: 10.1023/B:TROP.0000045965.99974.9c.
[28] TSURUTA S, MISZTAL I, LAWLOR T J. Genetic correlations among production, body Size, udder, and productive life traits over time in Holsteins., 2004, 87(5): 1457-1468. DOI: 10.3168/jds.S0022-0302(04)73297-X.
[29] PEREZ-CABAL M A, GARCIA C, GONZALEZ-RECIO O. Genetic and phenotypic relationships among locomotion type traits, profit, production, longevity, and fertility in Spanish dairy cows., 2006, 89(5): 1776-1783. DOI: 10.3168/jds.S0022-0302 (06)72246-9.
[30] HAILE-MARIAM M, BOWMAN P J, GODDARD M E. Genetic and environmental relationship among calving interval, survival, persistency of milk yield and somatic cell count in dairy cattle., 2003, 80(3): 189-200. DOI: 10.1016/ S0301-6226(02)00188-4.
[31] HOLTSMARK M, HERINGSTAD B, MADSEN P. Genetic relationship between culling, milk production, fertility, and health traits in Norwegian Red cows., 2008, 91(10): 4006-4012. DOI: 10.3168/jds.2007-0816.
[32] PFEIFFER C, FUERST C, DUCROCQ V. Short communication: Genetic relationships between functional longevity and direct health traits in Austrian Fleckvieh cattle., 2015, 98(10): 7380-7383. DOI: 10.3168/jds.2015-9632.
[33] ROGERS G W, BANOS G, NIELSEN U S. Genetic correlations among somatic cell scores, productive life, and type traits from the United States and udder health measures from Denmark and Sweden., 1998, 81(5): 1445-1453. DOI: 10.3168/ jds.S0022-0302(98)75708-X.
[34] NEERHOF H J, MADSEN P, DUCROCQ V P. Relationships between mastitis and functional longevity in Danish Black and White dairy cattle estimated using survival analysis., 2000, 83(5): 1064-1071. DOI: 10.3168/jds.S0022-0302(00) 74970-8.
[35] BUENGER A, DUCROCQ V, SWALVE H H. Analysis of survival in dairy cows with supplementary data on type scores and housing systems from a region of northwest Germany., 2001, 84(6): 1531-1541. DOI: 10.3168/jds.S0022-0302(01) 70187-7.
[36] LARROQUE H, DUCROCQ V. Relationships between type and longevity in the Holstein breed., 2001, 33(1): 39-59. DOI: 10.1186/1297-9686-33-1-39.
[37] SEWALEM A, KISTEMAKER G J, MIGLIOR F. Analysis of the relationship between type traits and functional survival in Canadian Holsteins using a Weibull proportional hazards model., 2004, 87(11): 3938-3946. DOI: 10.3168/jds.S0022- 0302(04)73533-X.
[38] DADPASAND M, MIRAEI-ASHTIANI S R, MORADI SHAHREBABAK M. Impact of conformation traits on functional longevity of Holstein cattle of Iran assessed by a Weibull proportional hazards model., 2008, 118(3): 204-211. DOI: 10.1016/j.livsci.2008. 01.024.
[39] SEWALEM A, KISTEMAKER G J, VAN DOORMAAL B J. Relationship between type traits and longevity in Canadian Jerseys and Ayrshires using a Weibull proportional hazards model., 2005, 88(4):1552-1560. DOI: 10.3168/jds.S0022- 0302(05)72824-1
[40] SEWALEM A, MIGLIOR F, KISTEMAKER G J. Relationship between reproduction traits and functional longevity in Canadian dairy cattle., 2008, 91(4): 1660-1668. DOI: 10.3168/jds.2007-0178.
[41] SEWALEM A, MIGLIOR F, KISTEMAKER G J. Analysis of the relationship between somatic cell score and functional longevity in Canadian dairy cattle., 2006, 89(9): 3609- 3614. DOI: 10.3168/jds.S0022-0302(06)72400-6.
[42] CARAVIELLO D Z, WEIGEL K A, Shook G E. Assessment of the impact of somatic cell count on functional longevity in Holstein and Jersey cattle using survival analysis methodology., 2005, 88(2): 804-811. DOI: 10.3168/jds.S0022-0302(05) 72745-4.
[43] SEWALEM A, MIGLIOR F, KISTEMAKER G J. Analysis of the relationship between workability traits and functional longevity in Canadian dairy breeds., 2010, 93(9): 4359-4365. DOI: 10.3168/jds.2009-2969.
[44] FUERST-WALTL B, REICHL A, FUERST C. Effect of maternal age on milk production traits, fertility, and longevity in cattle., 2004, 87(7): 2293-2298. DOI: 10.3168/jds.S0022- 0302(04)70050-8.
[45] MIGLIOR F, SEWALEM A, JAMROZIK J. Analysis of milk urea nitrogen and lactose and their effect on longevity in Canadian dairy cattle., 2006, 89(12): 4886-4894. DOI: 10.3168/jds.S0022-0302(06)72537-1.
[46] SEWALEM A, KISTEMAKER G J, MIGLIOR F. Analysis of inbreeding and its relationship with functional longevity in Canadian dairy cattle., 2006, 89(6): 2210-2216. DOI: 10.3168/jds.S0022-0302(06)72291-3.
[47] FONTANESI L, CALO D G, GALIMBERTI G. A candidate gene association study for nine economically important traits in Italian Holstein cattle., 2014, 45(4): 576-580. DOI: 10.1111/ age.12164.
[48] SZYDA J, MOREK-KOPEC M, KOMISAREK J. Evaluating markers in selected genes for association with functional longevity of dairy cattle., 2011, 12:30. DOI: 10.1186/1471-2156-12-30.
[49] KOMISAREK J, DORYNEK Z. Effect of,,andgene polymorphism on estimated breeding values for functional and production traits in Polish Holstein-Friesian bulls., 2009, 50(2): 125-132. DOI: 10.1007/ BF03195663.
[50] KHATIB H, SCHUTZKUS V, CHANG Y M. Pattern of expression of the uterine milk protein gene and its association with productive life in dairy cattle., 2007, 90(5): 2427-2433. DOI: 10.3168/jds.2006-722.
[51] GARCIA M D, MICHAL J J, GASKINS C T. Significant association of the calpastatin gene with fertility and longevity in dairy cattle., 2006, 37(3): 304-305. DOI: 10.1111/j.1365-2052. 2006.01443.x.
[52] KHATIB H, HEIFETZ E, DEKKERS J C. Association of the protease inhibitor gene with production traits in Holstein dairy cattle., 2005, 88(3): 1208-1213. DOI: 10.3168/jds.S0022- 0302(05)72787-9.
[53] RUSSO V, FONTANESI L, DOLEZAL M, DOLEZAL M, LIPKIN E, SCOTTI E, ZAMBONELLI P, DALL’OLIO S, BIGI D, DAVOLI R, CANAVESI F, MEDUGORAC I, FOSTER M, SOLKNER J, SCHIAVINI F, BAGNATO A, SOLLER M. A whole genome scan for QTL affecting milk protein percentage in Italian Holstein cattle, applying selective milk DNA pooling and multiple marker mapping in a daughter design., 2012, 43(1): 72. DOI: 10.1111/ j.1365-2052.2012.02353.x.
[54] HILL R, CANAL A, BONDIOLI K, MORELL R, GARCIA M D. Molecular markers located on the DGAT1, CAST, and LEPR genes and their associations with milk production and fertility traits in Holstein cattle., 2016, 15(1). DOI:10.4238/gmr.15017794.
[55] GAUTIER M, CAPITAN A, FRITZ S, EGGEN A, BOICHARD D, DRUET T. Characterization of the DGAT1 K232A and variable number of tandem repeat polymorphisms in French dairy cattle., 2007, 90(6): 2980-2988. DOI:10.3168/jds. 2006-707.
[56] HUANG W, PENAGARICANO F, AHMAD K R, LUCEY J A, WEIGEL K A, KHATIB H. Association between milk protein gene variants and protein composition traits in dairy cattle., 2012, 95(1): 440-449. DOI:10.3168/jds.2011-4757.
[57] CHEBEL R C, SUSCA F, SANTOS J E P. Leptin genotype is associated with lactation performance and health of Holstein cows., 2008, 91(7): 2893-2900. DOI:10.3168/ jds.2007-0891.
[58] BRICKELL J S, POLLOTT G E, CLEMPSON A M, OTTER N, WATHES D C. Polymorphisms in the bovine leptin gene associated with perinatal mortality in Holstein-Friesian heifers., 2010, 93(1): 340-347. DOI:10.3168/jds.2009-2457.
[59] COLE J B, WIGGANS G R, MA L. Genome-wide association analysis of thirty one production, health, reproduction and body conformation traits in contemporary U.S. Holstein cows., 2011, 12(1): 408.
[60] NAYERI S, SARGOLZAEI M, ABO-ISMAIL M K. Genome-wide association study for lactation persistency, female fertility, longevity, and lifetime profit index traits in Holstein dairy cattle., 2017, 100(2): 1246-1258. DOI: 10.3168/jds.2016- 11770.
[61] STERI R, MOIOLI B, CATILLO G. Genome-wide association study for longevity in the Holstein cattle population., 2019, 13(7): 1350-1357. DOI: 10.1017/S1751731118003191.
[62] HAY E H, ROBERTS A. Genomic prediction and genome-wide association analysis of female longevity in a composite beef cattle breed., 2017, 95(4): 1467-1471. DOI: 10.2527/jas2016.1355.
[63] SAOWAPHAK P, DUANGJINDA M, PLAENGKAEO S. Genetic correlation and genome-wide association study (GWAS) of the length of productive life, days open, and 305-days milk yield in crossbred Holstein dairy cattle., 2017, 16(2). DOI: 10.4238/gmr16029091.
[64] ZHANG Q, GULDBRANDTSEN B, THOMASEN J R. Genome- wide association study for longevity with whole-genome sequencing in 3 cattle breeds., 2016, 99(9): 7289-7298. DOI: 10.3168/jds.2015-10697.
[65] MESZAROS G, EAGLEN S, WALDMANN P. A genome wide association study for longevity in cattle., 2014, 4(1): 46-55. DOI: 10.4236/ojgen.2014.41007.
[66] MIGLIOR F, FLEMING A, MALCHIODI F. A 100-year review: identification and genetic selection of economically important traits in dairy cattle., 2017, 100(12): 10251-10271. DOI: 10.3168/jds.2017-12968.
[67] VUKASINOVIC N, SCHLEPPI Y, KUNZI N. Using conformation traits to improve reliability of genetic evaluation for herd life based on survival analysis., 2002, 85(6): 1556-1562. DOI: 10.3168/jds.S0022-0302(02)74225-2.
[68] SEWALEM A, MIGLIOR F, KISTEMAKER G J. Short communication: Modification of genetic evaluation of herd life from a three-trait to a five-trait model in Canadian dairy cattle., 2007, 90(4): 2025-2028. DOI: 10.3168/jds.2006-719.
[69] VANRADEN P M. Efficient methods to compute genomic predictions., 2008, 91(11): 4414-4423. DOI: 10.3168/ jds.2007-0980.
[70] LIU Z, SEEFRIED F R, REINHARDT F. Impacts of both reference population size and inclusion of a residual polygenic effect on the accuracy of genomic prediction., 2011, 43:19. DOI: 10.1186/1297-9686-43-19.
[71] VANRADEN P M, VAN TASSELL C P, WIGGANS G R. Invited review: reliability of genomic predictions for North American Holstein bulls., 2009, 92(1): 16-24. DOI: 10.3168/jds.2008-1514.
[72] 鄢新义, 刘澳星, 董刚辉, 郭刚, 王新宇, 刘林, 张胜利, 王雅春. 北京地区中国荷斯坦牛长寿性及其影响因素分析. 中国畜牧杂志, 2016, 52(23):1-6.
YAN X Y, LIU A X, DONG G H, GUO G, WANG X Y, LIU L, ZHANG S L, WANG Y C. Analysis of longevity and its influencing factors in Chinese Holstein population in Beijing.2016, 52(23): 1-6.(in Chinese)
[73] 毛杰. 上海地区荷斯坦牛体型性状与产奶性状、SCS和寿命性状的遗传分析[D]. 南京: 南京农业大学, 2015.
MAO J. Genetic analysis between type traits, milk production traits, SCS and longevity traits of Holstein cattle in Shanghai[D]. Nanjing: Nanjing Agricultural University, 2015.(in Chinese)
[74] 储明星, 叶素成, 陈国宏. 微卫星标记与奶牛数量性状QTL定位. 遗传, 2003(3): 337-340.
CHU M X, YE S C, CHEN G H. Mapping quantitative trait loci for quantitative traits in dairy cattle microsatellite markers.. 2003(3): 337-340.(in Chinese)
[75] 王梦琦, 倪炜, 张慧敏, 杨章平, 王西朴, 蒋彦森, 毛永江. 中国荷斯坦牛基因编码区SNP多态与临床乳房炎和生产寿命的关联分析. 中国农业科学, 2017, 50(12): 2359-2370. DOI:10.3864/j.issn.0578-1752.2017.12.016.
WANG M Q, NI W, ZHANG H M, YANG Z P, WANG X P, JIANG Y S, MAO Y J. Association between SNPs in the CDS regions ofgene and the clinical mastitis and lifetime for Chinese Holstein., 2017, 50(12): 2359-2370. DOI:10.3864/j.issn.0578-1752.2017.12.016. (in Chinese)
[76] 王梦琦, 倪炜, 郭译蔚, 张慧敏, 杨章平, 毛永江. 中国荷斯坦牛基因SNP突变与奶牛疾病及泌乳性能的相关性分析. 中国兽医学报, 2017, 37(12): 2418-2424. DOI: 10.16303/j.cnki.1005- 4545.2017.12.30.
WANG M Q, NI W, GUO Y W, ZHANG H M, YANG Z P, MAO Y J. Correlation analysis between diseases and milking traits and SNP offor Chinese Holstein cow., 2017, 37(12): 2418-2424. DOI: 10.16303/j.cnki.1005-4545. 2017.12.30. (in Chinese)
[77] 赵佳强. 中国荷斯坦奶牛生产寿命、体型线性性状分子标记的研究[D]. 武汉: 华中农业大学, 2013.
ZHAO J Q. Research on molecular markers and productive life, liner type traits of Holstein cattle in China[D]. Wuhan: Huazhong Agricultural University, 2013.(in Chinese)
[78] 陈星. 奶牛乳房炎相关天然免疫基因的筛选及其功能研究[D]. 武汉: 华中农业大学, 2016.
CHEN X. Dairy cow mastitis related innate immune genes screening and function study[D]. Wuhan: Huazhong Agricultural University, 2016.(in Chinese)
[79] LIU A, LUND M S, BOICHARDARD D, KARAMAN E, FRITZ S, AAMAND G P, NIELSEN U S, WANG Y, SU G. Improvement of genomic prediction by integrating additional single nucleotide polymorphisms selected from imputed whole genome sequencing data., 2020, 124:37-49. DOI:10.1038/s41437-019-0246-7.
[80] BRONDUM R F, SU G, JANSS L, SAHANA G, GULDBRANDTSEN B, BOICHARD D, Lund M S. Quantitative trait loci markers derived from whole genome sequence data increases the reliability of genomic prediction., 2015, 98(6): 4107–4116. DOI:10.3168/jds.2014-9005.
A Review on Longevity Trait in Dairy Cattle Breeding
ZHANG HaiLiang, LIU AoXing, MI SiYuan, LI Xiang, LUO HanPeng, YAN XinYi, WANG YaChun
(College of Animal Science and Technology, China Agricultural University, Beijing 100193)
Longevity is an important functional trait in dairy cattle. In addition to yield traits, longevity is more economically important than other traits in dairy breeding. The characteristics of longevity include low heritability, unfollowing normal distribution and performance later, hence longevity is the most difficult trait to select in dairy breeding. Since the 1950s, dairy breeders have begun to pay attention to longevity traits. Now, researches on longevity traits have continued. After the 1990s, longevity has been included in total selection index in many countries. However, longevity traits currently are not included in China Dairy Performance Index (CPI), and the study on longevity traits is also in its infancy in China. This review presented the trait definitions, methods of genetic evaluation, relationships between other traits, genetic makers, and selection strategies on longevity traits to systematically introduce its research and application in dairy industry. The complex relationships between longevity and other traits were described by summarizing the genetic correlation coefficients reported by many studies. The selection strategies for longevity traits in the various countries with developed dairy industry were highlighted by summarizing information collected from their dairy breeding program. Furthermore, this review also summarized the studies on longevity traits in dairy cattle in China. Longevity traits had various definitions, and there were various models used to preform genetic analysis, including linear model, threshold model, survival model and random regression model. There were low to moderate genetic correlations between longevity and other traits, including yield, linear type, fertility, health and workability traits, in which the higher genetic correlations were found between longevity and linear type traits in udder system. Generally, dairy cows with better performance on fertility and health traits were better on longevity. Among different populations, there were some differences on relationships between longevity and other traits, which were largely influenced by selection goals of current population. The total selection indexes in many countries included longevity. In addition to using direct longevity to select it, the indirect selection methods were also used in many countries. Udder system, strength, rump angle, feet and leg system, and mastitis resistance were comment traits used to indirect selection. Many genetic markers associated with longevity have been found in various populations, most of which were in genetic regions that have been reported to be associated with traits such as reproduction, disease and body type. Finally, this review also proposed the necessities of collecting longevity data, estimating genetic parameters, locating genetic markers, exploring evaluation model and selection strategy in China dairy cattle population.
longevity; genetic evaluation; genetic correlation; genetic maker; genetic selection; dairy cattle
2019-11-08;
2020-01-13
现代农业(奶牛)产业技术体系建设专项资金(CARS-36)、长江学者和创新团队发展计划(IRT_15R62)、现代农业产业技术体系北京市奶牛创新团队(BAIC06-2019)、研究生国际化培养提升项目(31051521)
张海亮,Tel:18813071851;E-mail:18813071851@163.com。通信作者王雅春,E-mail:wangyachun@cau.edu.cn
(责任编辑 林鉴非)

