Molecular mechanism prediction analysis of compound Kushen injection in the treatment of COVID-19 based on network pharmacology and molecular docking
Wan-Ying Zhang,Ying Chen,Miao-Miao Zhang,Guo-Wei Zhang
1College of Chinese Medicine,Hebei University,Baoding 071000,China.
Abstract
Background:As one of the eight effective traditional Chinese medicines for the treatment of atypical pneumonia,compound Kushen injection (CKI) played an important role in combating pneumonia caused by severe acute respiratory syndrome coronavirus 2 virus in China in 2003.CKI is known to inhibit inflammation,and its main chemical components,namely matrine and oxymatrine,can promote Th cells to recognize and eliminate viruses.In this study,network pharmacology and molecular docking were used to explore the mechanisms of CKI for treating coronavirus disease 2019. Methods: The Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform and other related literature were used to screen CKI’s active ingredients in the blood.Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform,Swiss Target Prediction and STITCH were used to search for potential targets of the active ingredients.The “ingredient-target” network was constructed using the Cytoscape software.The STRING online database was used to construct a target protein-protein interaction network that can be visualized and analyzed using the Cytoscape software to obtain key targets.Results:Sophocarpine,sophoridine,matrine,(+)-allomatrine,AIDS211310,and sophranol were the six active ingredients.After docking the active ingredients with severe acute respiratory syndrome coronavirus 2 3CL hydrolase and angiotensin-converting enzyme 2 (ACE2),they displayed suitable affinity,which could block viral replication and its binding to ACE2.The key targets mainly involved inflammatory factors,such as interleukin-6 (IL-6)and tumor necrosis factor (TNF).Gene Ontology enrichment analysis mainly indicated the IL-6 cytokine-mediated signaling pathway and cytokine-mediated signaling pathway.The Kyoto Encyclopedia of Genes and Genome pathway enrichment analysis mainly indicated steroid hormone biosynthesis and the TNF signaling pathway. Conclusion:The alkaloids in CKI can block viral replication and its binding to severe acute respiratory syndrome coronavirus 2 and ACE2 receptors.They regulate the IL-6-mediated signaling pathway,TNF signaling pathway,and steroid hormone biosynthesis,thereby initiating therapeutic responses against coronavirus disease 2019.
Keywords: Compound Kushen injection,Novel coronavirus,Molecular docking,Mechanism of action,Severe acute respiratory syndrome coronavirus 2 3CL hydrolase,Angiotensin-converting enzyme 2
Background
In December 2019,multiple cases of unexplained pneumonia were diagnosed in Wuhan,Hubei,China,and its pathogen was subsequently confirmed as a novel coronavirus.On January 20,2020,academician Zhong Nanshan affirmed the human-to-human coronavirus disease 2019 (COVID-19) transmission,by which time 217 cases of COVID-19 have been diagnosed in China.As the epidemic worsened,all provinces across the country launched a level-1 emergency response to prevent and control the COVID-19 epidemic.At the press conference of the Joint Defense and Control Mechanism of the State Council last February 8,2020,pneumonia from novel coronavirus infection was collectively referred to as the “novel coronavirus pneumonia” [1].On February 11,2020,the World Health Organization named this disease as coronavirus disease “COVID-19”,and the International Committee on Taxonomy of Viruses officially named the corresponding virus “severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)”.Related studies [2] have reported that both SARS-CoV-2 and SARS-CoV bind to the angiotensin-converting enzyme 2 (ACE2) receptor in the human body through spiked protein,resulting in viral invasion.SARS-CoV-2 3CL hydrolase is a key protein involved in viral translation and replication in human cells.After binding to the ACE2 receptor,SARS-CoV-2 can activate the classical renin-angiotensin regulatory pathways to act on the lungs and other target organs,eventually leading to multiple organ damage [3].Therefore,SARS-CoV-2 3CL hydrolase and ACE2 were selected for molecular docking in this study.
The combination of traditional Chinese and Western medicine has been shown to be effective in COVID-19 treatment.The traditional Chinese medicine compound Kushen injection(CKI)was launched in China in 1995 with the approval number of State Food and Drug Administration of China Z14021230,which is composed of Kushen (Sophorae Flavescentis Radix)and Baituling (Rhizoma Heterosmilacis Japonicae).It can effectively clear damp heat,and cool and detoxify blood,and these are similar to the anti-inflammatory effects of Western medicine.CKI,one of the eight effective Chinese medicines for atypical pneumonia treatment,played an important role in combating SARS-CoV-related pneumonia in China in 2003.Relevant research has indicated that CKI can effectively protect SARS-infected patients from multiple organ damage,such as injury to the heart,liver,kidneys,and other organs,and enhance immune functions in humans[4].Sun et al.[5]have shown that CKI’s anti-inflammatory effect is mediated by inhibiting the excessive activation of nuclear factor kappa-B in macrophages.The clinical trial conducted by Yu et al.[6] demonstrated that CKI inhibited inflammatory pathways by reducing tumor necrosis factor alpha (TNF-α) expression,transforming growth factor beta synthesis,and cytokine production to prevent and treat radiation pneumonitis.Matrine and oxymatrine can regulate immune functions and enhance Th lymphocyte activity,which contribute to the ability of immune cells to recognize and neutralize viruses [7-8].CKI is now used in COVID-19 clinical treatment,but its molecular mechanism remains unclear and warrants further investigation.
This study explores CKI’s mechanism in COVID-19 treatment using network pharmacology and molecular docking to provide a scientific basis for clinical applications.
Materials and Methods
Screening of active ingredients
CKI’s active ingredients were screened in the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP) database(http://lsp.nwu.edu.cn/tcmsp.php) using the search terms “Sophorae Flavescentis Radix” and “Rhizoma HeterosmilacisJaponicae”.According to pharmacokinetic absorption,distribution,metabolism,and excretion parameters,oral bioavailability (OB ≥30%) and drug-likeness (DL ≥0.18) were set as the screening conditions for the active ingredients.Relevant literature was cross-referenced to further determine the active ingredients in the blood infused with CKI.
Screening of active ingredient targets
The TCMSP,Swiss Target Prediction(http://www.swisstargetprediction.ch/),and STITCH(http://stitch.embl.de/) databases were used to search for the targets of CKI’s active ingredients.In the TCMSP database,the names of the active ingredients were used as the search terms to select the corresponding targets.The Canonical SMILES format of the active ingredients obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) were searched in the Swiss Target Prediction database with the species set to “Homo sapiens”,and the top ten targets were selected from the prediction results.In addition,the active ingredients in the canonical SMILES format that were not queried were input into the database as predictive 2-D structures under the same filtering conditions.Similarly,the names of the active ingredients were searched in the STITCH database,and the top ten targets were selected from the prediction results.The target gene names and the UniProt IDs of the active ingredients were obtained from the UniProt (https://www.uniprot.org/) database with the species set to “Homo sapiens” for subsequent analyses.
Construction of ingredient-target network
CKI’s active ingredients and targets that were obtained from the aforementioned databases were input into the Cytoscape 3.7.2 software to visualize and construct the ingredient-target network.
Construction of protein-protein interaction(PPI)network
Several known or predicted PPIs were collected from the STRING database (https://string-db.org/) [9].The aforementioned genes that were obtained were input into the STRING database with the species setting as“Homo sapiens”.The obtained PPI network was saved as a.tsv file and visualized.Network topology analysis was performed by importing the.tsv file into the Cytoscape 3.7.2 software.
Gene Ontology (GO) functional enrichment analysis and Kyoto Encyclopedia of Genes and Genome(KEGG)pathway enrichment analysis
DAVID (http://www.david.niaid.nih.gov) is an online tool for enriching large-scale genetic biological processes and pathways [10].In this study,DAVID was used to perform CKI-based GO and KEGG pathway enrichment analyses.The obtained target UniProt IDs were input into DAVID with the species set to “Homo sapiens”,the GOTERM_BP function in GO was used to enrich target biological processes.KEGG pathway enrichment analysis was used for channel enrichment,and the key CKI signal pathways withPvalues less than 0.05 were selected.
Molecular docking
The ChemBioDraw plug-in in ChemOffice was used to draw the 2-D structures of the active ingredients,which were imported into the ChemBioDraw3D program to obtain the 3-D structures that were saved as a.mol2 file to reduce its size.The 3-D crystal structures of ACE2 (PDB ID:1R42) and SARS-CoV-2 3CL hydrolase (PDB ID:6LU7)were downloaded from the PDB database (https://www.rcsb.org/) and saved in the pdb format.The PyMOL software was used to separate the protein from the primary ligand,which was dehydrated and hydrogenated.The protein and primary ligand were saved in the pdb format.Using AutoDockTools 1.5.6,the active ingredients and proteins in the pdb format were converted to the pdbqt format.The active pocket parameters were set,and Vina was administered for docking.When the affinity was equal to or less than -5.0 kJ/mol [11],the active ingredient was considered to have good target-binding activity.
Results
Active ingredients
From the TCMSP database,113 active ingredients were obtained for Kushen (Sophorae FlavescentisRadix),whereas there was no active ingredient related to Baituling (Rhizoma Heterosmilacis Japonicae).A total of 66 chemical ingredients in Baituling (Rhizoma Heterosmilacis Japonicae) were selected after cross-referencing with related literature [12-16].The ingredients of Baituling (Rhizoma Heterosmilacis Japonicae) were derived from the internal data of the Beijing Zhendong Guangming Pharmaceutical Research Institute;however,several data have not been published.The obtained ingredients were screened with OB ≥30%and DL ≥0.18,and compared with the CKI ingredients reported by Gao et al.[17].As a result,six active ingredients were selected for Kushen(Sophorae Flavescentis Radix),and no active ingredient was found for Baituling (Rhizoma Heterosmilacis Japonicae),resulting in a total of six active ingredients in the CKI-infused blood(Table1).
Targets of active ingredients
The aforementioned CKI active ingredients were input into TCMSP,Swiss Target Prediction,and STITCH databases.A total of 12,60,and 10 targets were obtained from the TCMSP,Swiss Target Prediction,and STITCH databases.After deduplication,a total of 44 targets of the active ingredients were obtained(Table2).
Active ingredient-target network
The ingredient-target network included 50 nodes (six ingredient nodes and 44 target nodes)and 82 edges,as shown in Figure1.In this network,each ingredient interacted with an average of 13.67 targets,and the top two,matrine and sophoridine,interacted with 30 and 12 targets,respectively.In addition,CHRNA7 and CHRNB2 interacted with five ingredients.Therefore,CKI displays the phenomenon in which the same active ingredient acts on multiple targets,and the same target acts on multiple active ingredients,reflecting the characteristic multi-ingredient and -target interactions of traditional Chinese medicine.
PPI network topology
The target gene name of CKI were input into the STRING program to get the PPI relationships of the target,and the Cytoscape 3.7.2 software was used for visualization (Figure2).Topological parameters,such as degree,betweenness centrality,and closeness centrality,were obtained using the topology analysis function of the Network Analyzer tool in the Cytoscape 3.7.2 software.Moreover,23 key targets were obtained using the average of degree as the condition for screening core targets,mainly involving interleukin-6 (IL-6),caspase 3 (CASP3),tumor necrosis factor (TNF),acetylcholinesterase,and androgen receptor.
GO and KEGG enrichment analyses
Seventy-seven biological processes (P<0.05) were obtained from the GO enrichment analysis and sorted according toPvalues in an ascending order.Moreover,15 processes related to immunity and inflammation are shown in Table3.The biological processes related to immunity mainly involved B cell activation and response to glucocorticoids.The biological processes related to the inflammatory responses mainly involved cellular responses to interleukin-1 (IL-1),the IL-6-mediated signaling pathway,positive regulation of chemokine production,and the cytokine-mediated signaling pathway.CKI’s therapeutic effect is therefore suggestively mediated by regulating biological processes,such as B-cell activation,IL-6,and other cytokine-mediated signaling pathways,to regulate immunity and suppress inflammation.
The KEGG pathway enrichment and screening resulted in 19 signal pathways (P<0.05) (Figure3).The COVID-19-associated pathways were mainly enriched in the TNF signaling pathway,steroid hormone biosynthesis,and natural killer cell-mediated cytotoxicity,where the TNF signaling pathway is the inflammatory factor in the TNF-mediated pathway related to inflammation,steroid hormone biosynthesis,and natural killer cell-mediated cytotoxicity,which are closely related to the body's immune responses[18-19].These findings suggest that CKI exerts its therapeutic effect by regulating body immunity and inhibiting inflammatory responses.
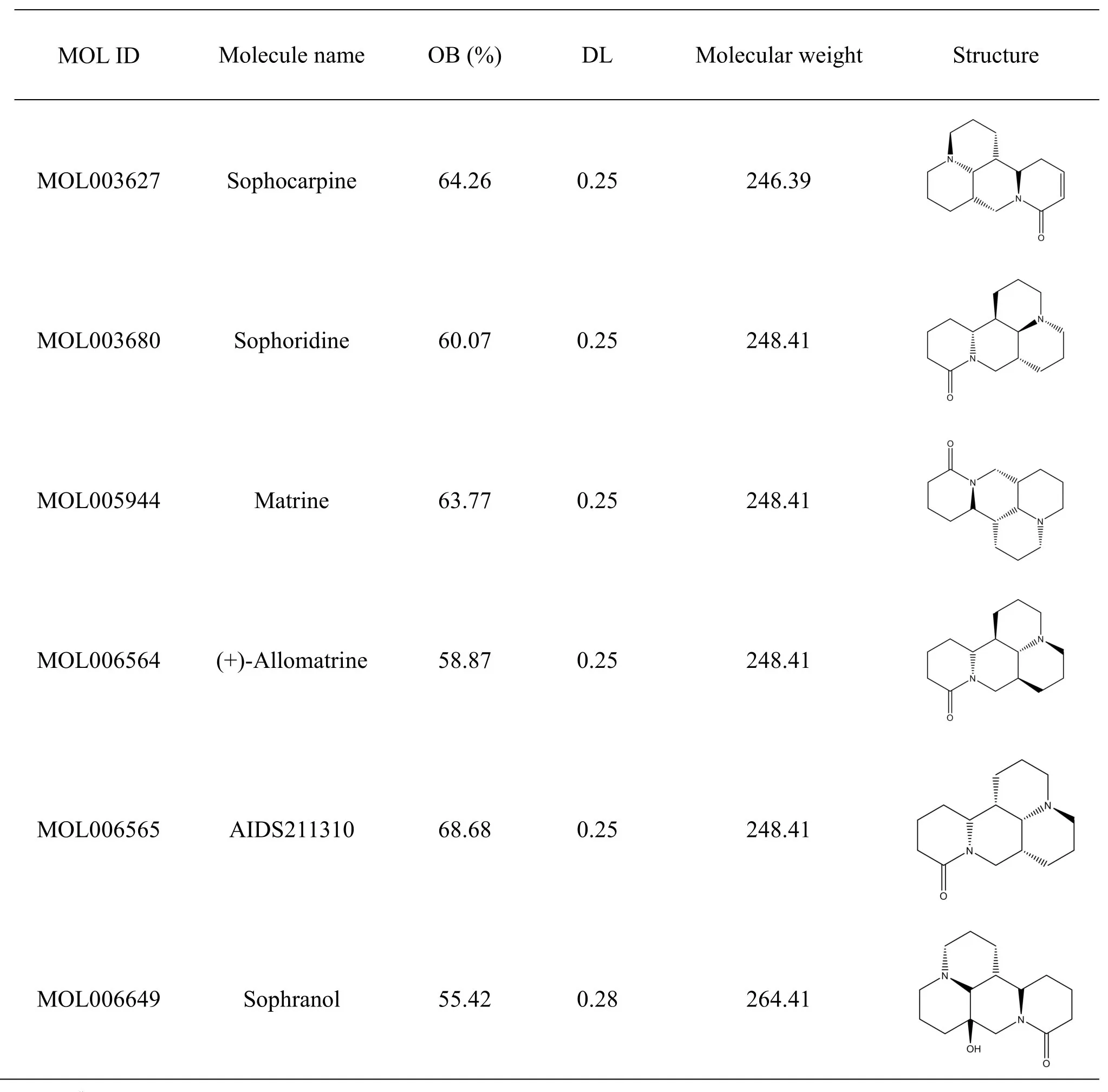
Table1 The active ingredients of CKI
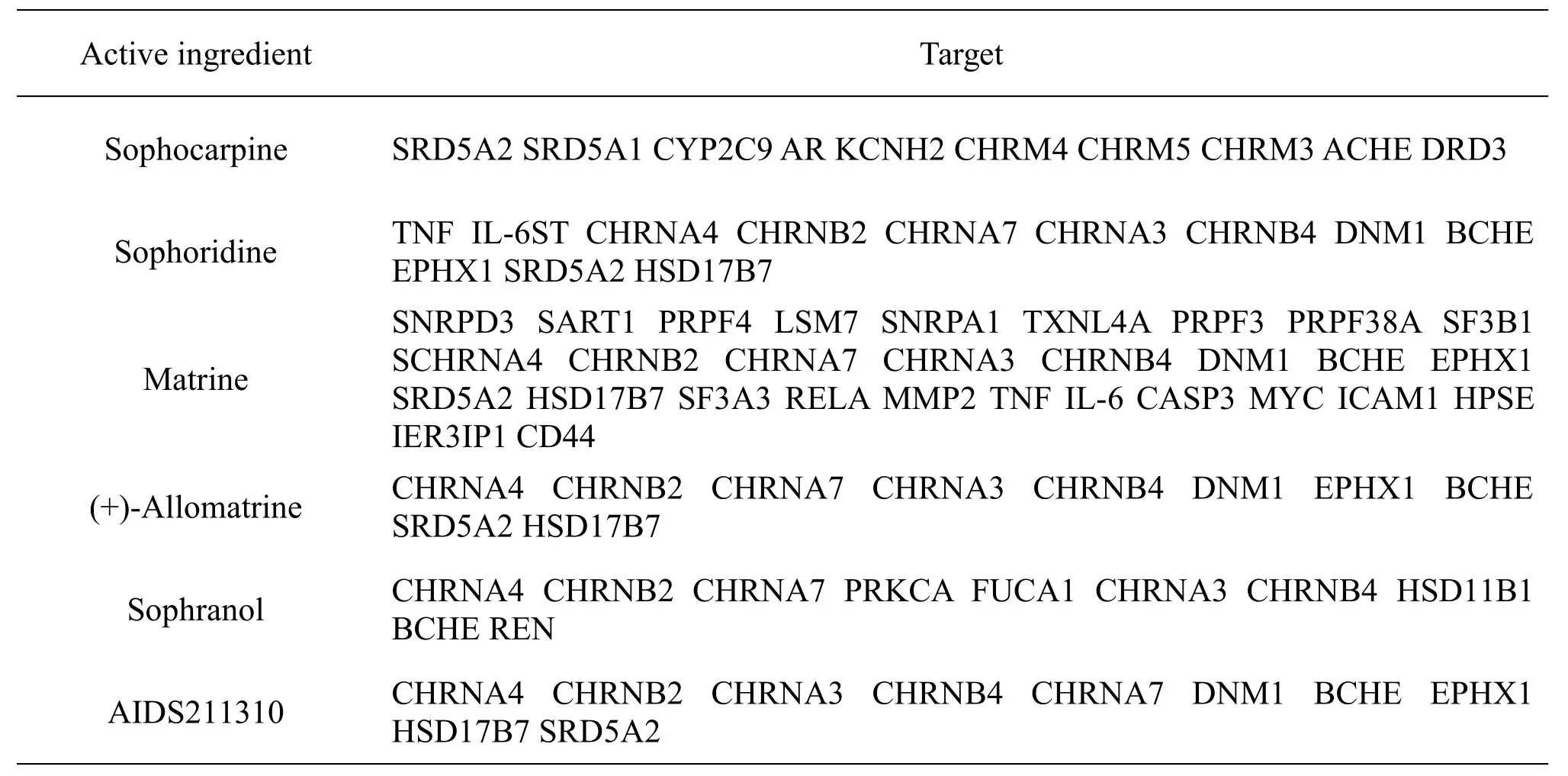
Table2 Target genes of active ingredients of CKI
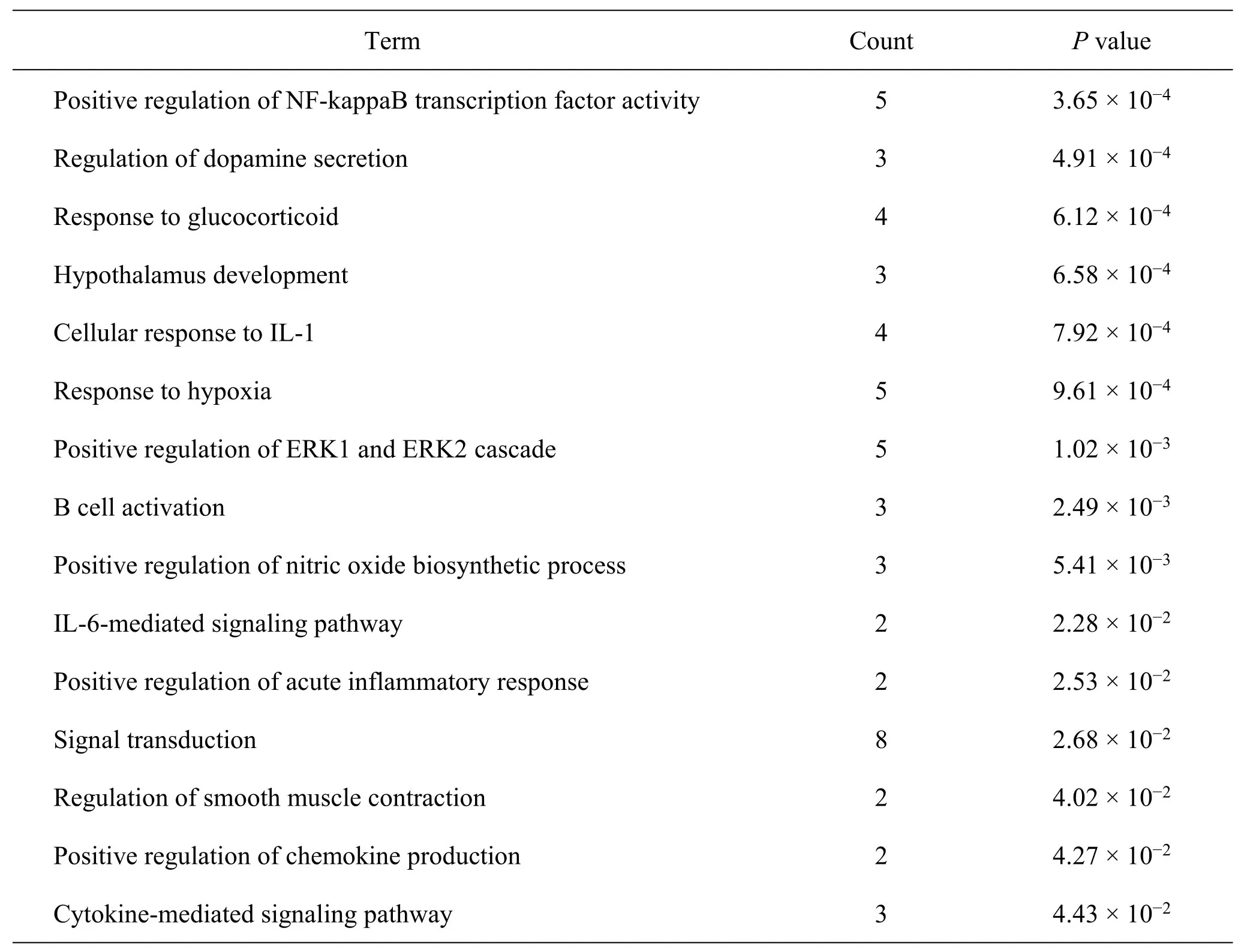
Table3 Enrichment analysis of GO biological processes of CKI
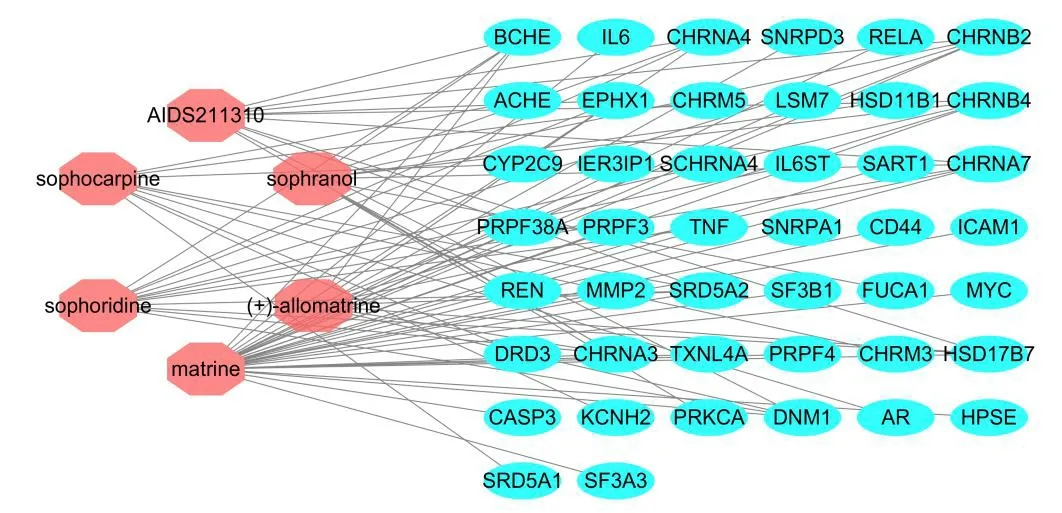
Figure1 Active ingredient-target network of CKI.Rose red bubbles,active ingredients; light blue bubbles,targets;lines,interactions among the ingredients and targets.
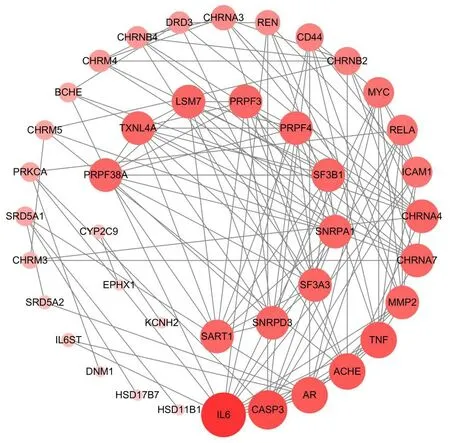
Figure2 PPI networks.The size and darkness of the node correspond to the size of the degree value(degree).
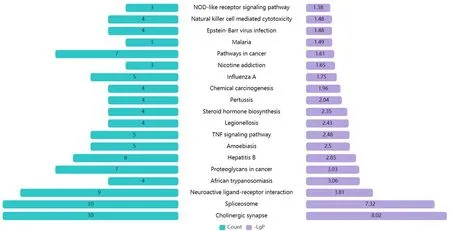
Figure3 KEGG enrichment pathway analysis for CKI.The number in each cyan bar indicates the number of genes,which also corresponds to the length of the bar.The number in each purple bar indicates the negative logarithm (-l g) of the P value with base 10,whereas the length of the bar corresponds to the -l g P value and the level of significance of the enrichment.
Molecular docking
Molecular docking of the active ingredients of CKI with SARS-CoV-2 3CL hydrolase and ACE2 was performed.Chloroquine,remdesivir,ribavirin,ritonavir,and other COVID-19 medications were used as positive controls.The molecular docking fraction was negatively correlated with the affinity of the docking between receptor and ligand.The results indicated that all of the docking fractions were less than -5.0 kJ/mol (Table4),whereas the affinity of(+)-allomatrine and AIDS211310 with SARS-CoV-2 and ACE2 was stronger than that of Western medications,such as ribavirin and favipiravir.When the docking mode with the lowest binding energy to SARS-CoV-2 3CL hydrolase and ACE2 (Figure4 and Figure5) was selected,AIDS211310 formed a hydrogen bond with the amino acid residue AGR-393,whereas (+)-allomatrine did not interact with SARS-CoV-2 3CL hydrolase to form hydrogen bonds.The results of the molecular docking showed that the active ingredients of CKI had high affinity for ACE2 and SARS-CoV-2 proteins.Therefore,CKI’s therapeutic effect may involve blocking viral translation and replication,as well as its binding to the ACE2 receptor through alkaloids,such as sophoridine and matrine.
Discussion
In this study,six active ingredients,including sophoridine,sophocarpine,matrine,(+)-allomatrine,AIDS211310,and sophranol,in CKI-infused blood were screened based on OB,DL,and related literature.Among these,sophocarpine [20,21],matrine [22,23],and sophoridine[24] have demonstrated efficacy in regulating immunity and inhibiting inflammation.AIDS211310,(+)-allomatrine,and other active ingredients showed good docking activity with SARS-CoV-2 and ACE2 proteins.These results indicate that these ingredients may directly act on SARS-CoV-2 3CL hydrolase to inhibit viral replication and proliferation.They may also act on ACE2 receptors of human cells to block viral invasion.
During viral infection,an inflammatory response is induced to engulf and isolate the virus.However,the excessive immune responses may release high levels of cytokines and inflammatory chemokines,such as TNF-α,IL-1,IL-6,and IL-8,causing uncontrollable inflammatory reactions and triggering a cytokine storm,which result in serious tissue and organ damage[25-27].Therefore,cytokine storms play an important role in disease progression in COVID-19 patients [28].Glucocorticoids have been currently used to treat refractory cytokine storms in which IL-6 receptor antagonists are ineffective [29].However,corticosteroids not only suppress inflammation in the lungs,but also suppress immune responses and viral clearance by the immune system,resulting in their controversial clinical applications [30].The results from the KEGG pathway analysis suggest that CKI may inhibit excessive immune responses through steroid hormone biosynthesis regulation,thereby inhibiting cytokine storm occurrences.
Suppressing the outbreak of inflammatory factors is of great significance COVID-19 prevention and treatment,as well as alleviating multiple organ damage COVID-19 patients [28].Based on the results of the PPI network topology,the key targets of CKI in COVID-19 treatment were IL-6,TNF,and CASP3.IL-6 plays an important role in the inflammatory process and B cell maturation,and is mainly produced at inflammation site.Wu et al.[31]reported that serum levels of inflammatory factors,such as IL-6,were negatively correlated with lung function indicators,such as forced lung capacity in the first-second vital capacity,peak expiratory flow,and respiratory velocity of lung capacity,in patients with mycoplasma pneumonia.Research by Jiang et al.has shown that the serum IL-6 levels in patients with asthma-COPD overlap syndrome,COPD,or asthma are higher than those of the control group,indicating that IL-6 is involved in airway inflammation and lung injury [32].The serum IL-6 and TNF-α levels in patients with cervical cancer receiving conventional radiotherapy and chemotherapy alone with external radiotherapy and chemotherapy than those in patients who were simultaneously supplemented with CKI,suggesting that CKI may therapeutically inhibit inflammatory responses[33].TNF is considered as the core of cytokine storms [34].TNF overproduction in the body can cause respiratory failure,septic shock,and even death in severe cases.The mortality rate is positively correlated with TNF level.Zhou et al.[35]stimulated angiotensin II in rats,observed contractions of the aortic ring,and concluded that TNF-α could affect blood pressure stability in early-stage septic shock by activating the inositol 1,4,5-trisphophate receptor pathway.CASP3 is the main apoptotic protein[36],which promotes the apoptosis of lymphocytes to downregulate or terminate inflammatory responses.The results from this study suggest that CKI’s anti-inflammatory effect may be mediated by downregulating IL-6 and TNF while simultaneously upregulating CASP3 expression.
During the clinical application of CKI,symptoms,such as nausea,vomiting,fever,chills,abdominal distension,and stomach discomfort,are occasionally observed.Occasional allergic reactions manifested by flushing,sweating,itching,and rashes on the skin of the head and neck may be related to the patient's specific constitution [37-38].Furthermore,local use is mildly irritating but well-absorbed.
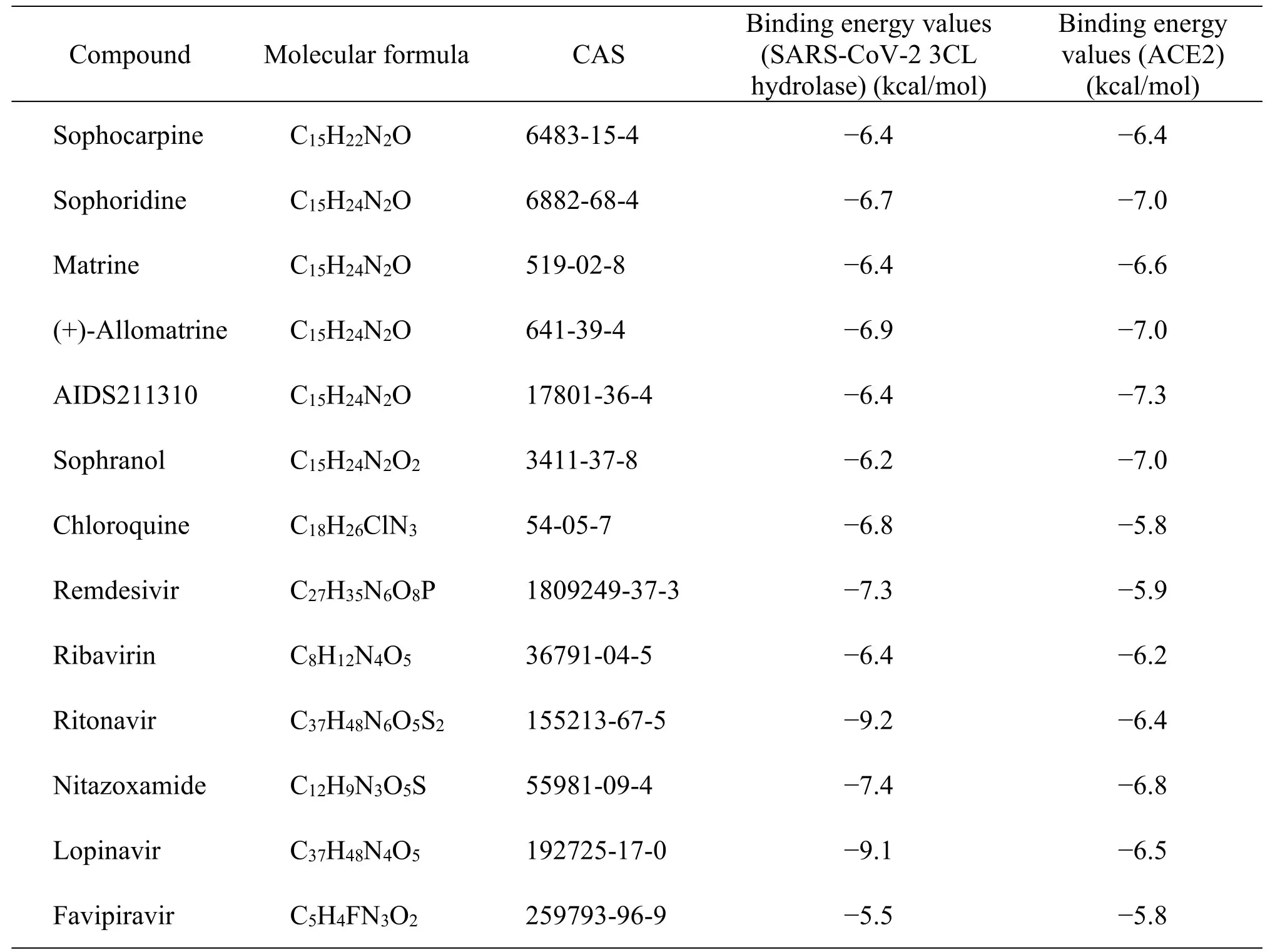
Table4 The binding energy values of the active ingredients and ACE2.
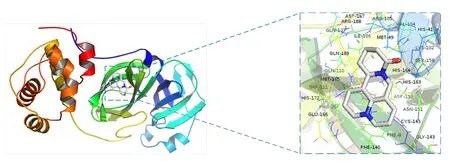
Figure4 Molecular docking pattern of (+)-allomatrine and SARS-CoV-2 3CL hydrolase.The dotted frame in the figure is an enlarged view of the location of the active pocket where the target protein and its receptor bind.SARS-CoV-2,severe acute respiratory syndrome coronavirus 2.
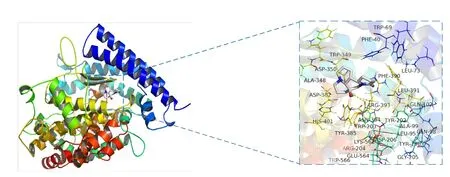
Figure5 Molecular docking pattern of AIDS211310 with ACE2.The dotted frame in the figure is an enlarged view of the location of the active pocket where the target protein and its receptor bind.The yellow dotted line is the hydrogen bond,and around the receptor molecule is the amino acid residue within 4A of the binding site.ACE2,angiotensin-converting enzyme 2.
Conclusion
In this study,the mechanism of CKI in COVID-19 treatment has been preliminarily explored in terms of its active ingredients,targets,and pathways using network pharmacology and molecular docking technology.Alkaloids in CKI,such as sophocarpine,sophoridine,and matrine,have been reported to block viral replication and its binding to SARS-CoV-2 3CL hydrolase and the ACE2 receptor,thereby regulating the IL-6-mediated signaling pathway,TNF signaling pathway,and steroid hormone biosynthesis,which inhibit IL-6- and TNF-mediated inflammatory responses to therapeutically protect the body against COVID-19.However,this study is based only on network pharmacology method for prediction,which has certain limitations and requires further experimental verification.
 Traditional Medicine Research2020年5期
Traditional Medicine Research2020年5期
- Traditional Medicine Research的其它文章
- Inner peace makes you live longer
- East to west:research progress in traditional Chinese medicine for antiaging strategies
- The effects of nutritional ketosis induced by Bigu-herbs regimen and ketogenic diet on diseases and aging
- From religious manual to herbal pharmacopoeia:a textual study of the formation and transformation of Shennong’s Classic of Materia Medica
- South Asian medicinal plants and chronic kidney disease
- Ethnoveterinary medicines used against various livestock disorders in the flora of Shamozai Valley,Swat,KP Pakistan
